Some Metabolites Act as Second Messengers in Yeast Chronological Aging
Abstract
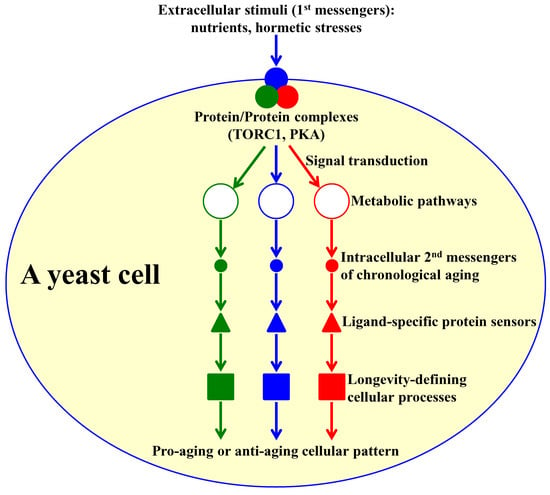
:1. Introduction
2. Concentrations of Some Metabolites Define the Rate of Chronological Aging in Yeast
2.1. NADPH
2.2. Glycerol
2.3. Trehalose
2.4. Hydrogen Peroxide (H2O2)
2.5. Amino Acids
2.6. Sphingolipids
2.7. Spermidine
2.8. Hydrogen Sulfide (H2S)
2.9. Acetic Acid
2.10. Ethanol
2.11. FFA and Diacylglycerol (DAG)
3. The Spatiotemporal Dynamics of Changes in Concentrations of Some Metabolites Define Yeast CLS
4. Conclusions
Acknowledgments
Conflicts of Interest
Abbreviations
| Asp | aspartate |
| Asn | asparagine |
| ATG | autophagy-related genes |
| ER | endoplasmic reticulum |
| ETC | electron transport chain |
| FFA | free (non-esterified) fatty acids |
| CLS | chronological lifespan |
| CR | caloric restriction |
| DAG | diacylglycerol |
| DDR | DNA damage response |
| Glu | glutamate |
| Gln | glutamine |
| GTR | glutathione reductase |
| LD | lipid droplets |
| PKA | protein kinase A |
| Pkh1 | Pkb-activating kinase homolog protein 1 |
| Pkh2 | Pkb-activating kinase homolog protein 2 |
| RCD | regulated cell death |
| ROS | reactive oxygen species |
| TCA | tricarboxylic acid |
| TORC1 | target of rapamycin complex 1 |
| TRR | thioredoxin reductase |
| TSP | transsulfuration pathway |
References
- Fontana, L.; Partridge, L.; Longo, V.D. Extending healthy life span—From yeast to humans. Science 2010, 328, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Longo, V.D.; Shadel, G.S.; Kaeberlein, M.; Kennedy, B. Replicative and chronological aging in Saccharomyces cerevisiae. Cell Metab. 2012, 16, 18–31. [Google Scholar] [CrossRef] [PubMed]
- Arlia-Ciommo, A.; Leonov, A.; Piano, A.; Svistkova, V.; Titorenko, V.I. Cell-autonomous mechanisms of chronological aging in the yeast Saccharomyces cerevisiae. Microb. Cell 2014, 1, 163–178. [Google Scholar] [CrossRef] [PubMed]
- Kaeberlein, M. Lessons on longevity from budding yeast. Nature 2010, 464, 513–519. [Google Scholar] [CrossRef] [PubMed]
- Váchová, L.; Cáp, M.; Palková, Z. Yeast colonies: A model for studies of aging, environmental adaptation, and longevity. Oxid. Med. Cell. Longev. 2012, 2012. [Google Scholar] [CrossRef] [PubMed]
- Denoth Lippuner, A.; Julou, T.; Barral, Y. Budding yeast as a model organism to study the effects of age. FEMS Microbiol. Rev. 2014, 38, 300–325. [Google Scholar] [CrossRef] [PubMed]
- Steinkraus, K.A.; Kaeberlein, M.; Kennedy, B.K. Replicative aging in yeast: The means to the end. Annu. Rev. Cell Dev. Biol. 2008, 24, 29–54. [Google Scholar] [CrossRef] [PubMed]
- Steffen, K.K.; Kennedy, B.K.; Kaeberlein, M. Measuring replicative life span in the budding yeast. J. Vis. Exp. 2009, 28, 1209. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.D.; Tsuchiya, M.; Fox, L.A.; Dang, N.; Hu, D.; Kerr, E.O.; Johnston, E.D.; Tchao, B.N.; Pak, D.N.; Welton, K.L.; et al. Quantitative evidence for conserved longevity pathways between divergent eukaryotic species. Genome Res. 2008, 18, 564–570. [Google Scholar] [CrossRef] [PubMed]
- Ghavidel, A.; Baxi, K.; Ignatchenko, V.; Prusinkiewicz, M.; Arnason, T.G.; Kislinger, T.; Carvalho, C.E.; Harkness, T.A. A genome scale screen for mutants with delayed exit from mitosis: Ire1-independent induction of autophagy integrates ER homeostasis into mitotic lifespan. PLoS Genet. 2015, 11, e1005429. [Google Scholar] [CrossRef] [PubMed]
- McCormick, M.A.; Delaney, J.R.; Tsuchiya, M.; Tsuchiyama, S.; Shemorry, A.; Sim, S.; Chou, A.C.; Ahmed, U.; Carr, D.; Murakami, C.J.; et al. A comprehensive analysis of replicative lifespan in 4698 single-gene deletion strains uncovers conserved mechanisms of aging. Cell Metab. 2015, 22, 895–906. [Google Scholar] [CrossRef] [PubMed]
- Janssens, G.E.; Veenhoff, L.M. Evidence for the hallmarks of human aging in replicatively aging yeast. Microb. Cell 2016, 3, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Fabrizio, P.; Longo, V.D. The chronological life span of Saccharomyces cerevisiae. Methods Mol. Biol. 2007, 371, 89–95. [Google Scholar] [PubMed]
- Longo, V.D.; Fabrizio, P. Chronological aging in Saccharomyces cerevisiae. Subcell. Biochem. 2012, 57, 101–121. [Google Scholar] [PubMed]
- Burtner, C.R.; Murakami, C.J.; Kennedy, B.K.; Kaeberlein, M. A molecular mechanism of chronological aging in yeast. Cell Cycle 2009, 8, 1256–1270. [Google Scholar] [CrossRef] [PubMed]
- Longo, V.D.; Kennedy, B.K. Sirtuins in aging and age-related disease. Cell 2006, 126, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Murakami, C.; Delaney, J.R.; Chou, A.; Carr, D.; Schleit, J.; Sutphin, G.L.; An, E.H.; Castanza, A.S.; Fletcher, M.; Goswami, S.; et al. pH neutralization protects against reduction in replicative lifespan following chronological aging in yeast. Cell Cycle 2012, 11, 3087–3096. [Google Scholar] [CrossRef] [PubMed]
- Delaney, J.R.; Murakami, C.; Chou, A.; Carr, D.; Schleit, J.; Sutphin, G.L.; An, E.H.; Castanza, A.S.; Fletcher, M.; Goswami, S.; et al. Dietary restriction and mitochondrial function link replicative and chronological aging in Saccharomyces cerevisiae. Exp. Gerontol. 2013, 48, 1006–1013. [Google Scholar] [CrossRef] [PubMed]
- Molon, M.; Zadrag-Tecza, R.; Bilinski, T. The longevity in the yeast Saccharomyces cerevisiae: A comparison of two approaches for assessment the lifespan. Biochem. Biophys. Res. Commun. 2015, 460, 651–656. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Tu, B.P. Driving the cell cycle through metabolism. Annu. Rev. Cell Dev. Biol. 2012, 28, 59–87. [Google Scholar] [CrossRef] [PubMed]
- Titorenko, V.I.; Terlecky, S.R. Peroxisome metabolism and cellular aging. Traffic 2011, 12, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Beach, A.; Burstein, M.T.; Richard, V.R.; Leonov, A.; Levy, S.; Titorenko, V.I. Integration of peroxisomes into an endomembrane system that governs cellular aging. Front. Physiol. 2012, 3, 283. [Google Scholar] [CrossRef] [PubMed]
- Leonov, A.; Titorenko, V.I. A network of interorganellar communications underlies cellular aging. IUBMB Life 2013, 65, 665–674. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Withers, B.R.; Dickson, R.C. Sphingolipids and lifespan regulation. Biochim. Biophys. Acta 2014, 1841, 657–664. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, S.; Zimmermann, A.; Carmona-Gutierrez, D.; Eisenberg, T.; Ruckenstuhl, C.; Andryushkova, A.; Pendl, T.; Harger, A.; Madeo, F. Metabolites in aging and autophagy. Microb. Cell 2014, 1, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Pietrocola, F.; Galluzzi, L.; Bravo-San Pedro, J.M.; Madeo, F.; Kroemer, G. Acetyl coenzyme A: A central metabolite and second messenger. Cell Metab. 2015, 21, 805–821. [Google Scholar] [CrossRef] [PubMed]
- Dakik, P.; Titorenko, V.I. Communications between mitochondria, the nucleus, vacuoles, peroxisomes, the endoplasmic reticulum, the plasma membrane, lipid droplets, and the cytosol during yeast chronological aging. Front. Genet. 2016, 7, 177. [Google Scholar] [CrossRef] [PubMed]
- Eltschinger, S.; Loewith, R. TOR complexes and the maintenance of cellular homeostasis. Trends Cell Biol. 2016, 26, 148–159. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, V.; Costa, V. Unraveling the role of the target of rapamycin signaling in sphingolipid metabolism. Prog. Lipid Res. 2016, 61, 109–133. [Google Scholar] [CrossRef] [PubMed]
- Laxman, S. Conceptualizing eukaryotic metabolic sensing and signaling. J. Indian Inst. Sci. 2017, 97, 59–77. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Handee, W.; Kuo, M.H. The slim, the fat, and the obese: Guess who lives the longest? Curr. Genet. 2017, 63, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Mitrofanova, D.; Dakik, P.; McAuley, M.; Medkour, Y.; Mohammad, K.; Titorenko, V.I. Lipid metabolism and transport define longevity of the yeast Saccharomyces cerevisiae. Front. Biosci. 2018, 23, 1166–1194. [Google Scholar]
- Fraenkel, D.G. Yeast Intermediary Metabolism; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2011; ISBN 978-0-87969-797-6. [Google Scholar]
- Grant, C.M. Role of the glutathione/glutaredoxin and thioredoxin systems in yeast growth and response to stress conditions. Mol. Microbiol. 2001, 39, 533–541. [Google Scholar] [CrossRef] [PubMed]
- Brandes, N.; Tienson, H.; Lindemann, A.; Vitvitsky, V.; Reichmann, D.; Banerjee, R.; Jakob, U. Time line of redox events in aging postmitotic cells. Elife 2013, 2, e00306. [Google Scholar] [CrossRef] [PubMed]
- Wei, M.; Fabrizio, P.; Madia, F.; Hu, J.; Ge, H.; Li, L.M.; Longo, V.D. Tor1/Sch9-regulated carbon source substitution is as effective as calorie restriction in life span extension. PLoS Genet. 2009, 5, e1000467. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, A.A.; Bourque, S.D.; Kyryakov, P.; Gregg, C.; Boukh-Viner, T.; Beach, A.; Burstein, M.T.; Machkalyan, G.; Richard, V.; Rampersad, S.; et al. Effect of calorie restriction on the metabolic history of chronologically aging yeast. Exp. Gerontol. 2009, 44, 555–571. [Google Scholar] [CrossRef] [PubMed]
- François, J.; Parrou, J.L. Reserve carbohydrates metabolism in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2001, 25, 125–145. [Google Scholar] [CrossRef] [PubMed]
- Samokhvalov, V.; Ignatov, V.; Kondrashova, M. Reserve carbohydrates maintain the viability of Saccharomyces cerevisiae cells during chronological aging. Mech. Ageing Dev. 2004, 125, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Jiang, J.C.; Jazwinski, S.M. Gene regulatory changes in yeast during life extension by nutrient limitation. Exp. Gerontol. 2010, 45, 621–631. [Google Scholar] [CrossRef] [PubMed]
- Kyryakov, P.; Beach, A.; Richard, V.R.; Burstein, M.T.; Leonov, A.; Levy, S.; Titorenko, V.I. Caloric restriction extends yeast chronological lifespan by altering a pattern of age-related changes in trehalose concentration. Front. Physiol. 2012, 3, 256. [Google Scholar] [CrossRef] [PubMed]
- Ocampo, A.; Liu, J.; Schroeder, E.A.; Shadel, G.S.; Barrientos, A. Mitochondrial respiratory thresholds regulate yeast chronological life span and its extension by caloric restriction. Cell Metab. 2012, 16, 55–67. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Tang, Y.; Quan, Z.; Zhang, Z.; Oliver, S.G.; Zhang, N. Chronological lifespan in yeast is dependent on the accumulation of storage carbohydrates mediated by Yak1, Mck1 and Rim15 kinases. PLoS Genet. 2016, 12, e1006458. [Google Scholar] [CrossRef] [PubMed]
- Svenkrtova, A.; Belicova, L.; Volejnikova, A.; Sigler, K.; Jazwinski, S.M.; Pichova, A. Stratification of yeast cells during chronological aging by size points to the role of trehalose in cell vitality. Biogerontology 2016, 17, 395–408. [Google Scholar] [CrossRef] [PubMed]
- Leonov, A.; Feldman, R.; Piano, A.; Arlia-Ciommo, A.; Lutchman, V.; Ahmadi, M.; Elsaser, S.; Fakim, H.; Heshmati-Moghaddam, M.; Hussain, A.; et al. Caloric restriction extends yeast chronological lifespan via a mechanism linking cellular aging to cell cycle regulation, maintenance of a quiescent state, entry into a non-quiescent state and survival in the non-quiescent state. Oncotarget 2017, 8, 69328–69350. [Google Scholar] [CrossRef] [PubMed]
- Singer, M.A.; Lindquist, S. Multiple effects of trehalose on protein folding in vitro and in vivo. Mol. Cell 1998, 1, 639–648. [Google Scholar] [CrossRef]
- Jain, N.K.; Roy, I. Effect of trehalose on protein structure. Protein Sci. 2009, 18, 24–36. [Google Scholar] [CrossRef] [PubMed]
- Giorgio, M.; Trinei, M.; Migliaccio, E.; Pelicci, P.G. Hydrogen peroxide: A metabolic by-product or a common mediator of ageing signals? Nat. Rev. Mol. Cell Biol. 2007, 8, 722–728. [Google Scholar] [CrossRef] [PubMed]
- Causton, H.C.; Ren, B.; Koh, S.S.; Harbison, C.T.; Kanin, E.; Jennings, E.G.; Lee, T.I.; True, H.L.; Lander, E.S.; Young, R.A. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 2001, 12, 323–337. [Google Scholar] [CrossRef] [PubMed]
- Fabrizio, P.; Pozza, F.; Pletcher, S.D.; Gendron, C.M.; Longo, V.D. Regulation of longevity and stress resistance by Sch9 in yeast. Science 2001, 292, 288–290. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, E.A.; Raimundo, N.; Shadel, G.S. Epigenetic silencing mediates mitochondria stress-induced longevity. Cell Metab. 2013, 17, 954–964. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, E.A.; Shadel, G.S. Crosstalk between mitochondrial stress signals regulates yeast chronological lifespan. Mech. Ageing Dev. 2014, 135, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Crespo, J.L.; Powers, T.; Fowler, B.; Hall, M.N. The TOR-controlled transcription activators GLN3, RTG1, and RTG3 are regulated in response to intracellular levels of glutamine. Proc. Natl. Acad. Sci. USA 2002, 99, 6784–6789. [Google Scholar] [CrossRef] [PubMed]
- Powers, R.W., 3rd; Kaeberlein, M.; Caldwell, S.D.; Kennedy, B.K.; Fields, S. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 2006, 20, 174–184. [Google Scholar] [CrossRef] [PubMed]
- Conrad, M.; Schothorst, J.; Kankipati, H.N.; Van Zeebroeck, G.; Rubio-Texeira, M.; Thevelein, J.M. Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2014, 38, 254–299. [Google Scholar] [CrossRef] [PubMed]
- Swinnen, E.; Ghillebert, R.; Wilms, T.; Winderickx, J. Molecular mechanisms linking the evolutionary conserved TORC1-Sch9 nutrient signalling branch to lifespan regulation in Saccharomyces cerevisiae. FEMS Yeast Res. 2014, 14, 17–32. [Google Scholar] [CrossRef] [PubMed]
- Urban, J.; Soulard, A.; Huber, A.; Lippman, S.; Mukhopadhyay, D.; Deloche, O.; Wanke, V.; Anrather, D.; Ammerer, G.; Riezman, H.; et al. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol. Cell 2007, 26, 663–674. [Google Scholar] [CrossRef] [PubMed]
- Huber, A.; Bodenmiller, B.; Uotila, A.; Stahl, M.; Wanka, S.; Gerrits, B.; Aebersold, R.; Loewith, R. Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev. 2009, 23, 1929–1943. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Moir, R.D.; Willis, I.M. Regulation of RNA polymerase III transcription involves SCH9-dependent and SCH9-independent branches of the target of rapamycin (TOR) pathway. J. Biol. Chem. 2009, 284, 12604–12608. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Zheng, X.F. Sch9 partially mediates TORC1 signaling to control ribosomal RNA synthesis. Cell Cycle 2009, 8, 4085–4090. [Google Scholar] [CrossRef] [PubMed]
- Bonawitz, N.D.; Chatenay-Lapointe, M.; Pan, Y.; Shadel, G.S. Reduced TOR signaling extends chronological life span via increased respiration and upregulation of mitochondrial gene expression. Cell Metab. 2007, 5, 265–277. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Shadel, G.S. Extension of chronological life span by reduced TOR signaling requires down-regulation of Sch9p and involves increased mitochondrial OXPHOS complex density. Aging 2009, 1, 131–145. [Google Scholar] [CrossRef] [PubMed]
- Roosen, J.; Engelen, K.; Marchal, K.; Mathys, J.; Griffioen, G.; Cameroni, E.; Thevelein, J.M.; de Virgilio, C.; de Moor, B.; Winderickx, J. PKA and Sch9 control a molecular switch important for the proper adaptation to nutrient availability. Mol. Microbiol. 2005, 55, 862–880. [Google Scholar] [CrossRef] [PubMed]
- Wanke, V.; Cameroni, E.; Uotila, A.; Piccolis, M.; Urban, J.; Loewith, R.; de Virgilio, C. Caffeine extends yeast lifespan by targeting TORC1. Mol. Microbiol. 2008, 69, 277–285. [Google Scholar] [CrossRef] [PubMed]
- Smets, B.; Ghillebert, R.; de Snijder, P.; Binda, M.; Swinnen, E.; de Virgilio, C.; Winderickx, J. Life in the midst of scarcity: Adaptations to nutrient availability in Saccharomyces cerevisiae. Curr. Genet. 2010, 56, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Medvedik, O.; Lamming, D.W.; Kim, K.D.; Sinclair, D.A. MSN2 and MSN4 link calorie restriction and TOR to sirtuin-mediated lifespan extension in Saccharomyces cerevisiae. PLoS Biol. 2007, 5, e261. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.; Cho, B.R.; Joo, H.S.; Hahn, J.S. Yeast Yak1 kinase, a bridge between PKA and stress-responsive transcription factors, Hsf1 and Msn2/Msn4. Mol. Microbiol. 2008, 70, 882–895. [Google Scholar] [CrossRef] [PubMed]
- Broach, J.R. Nutritional control of growth and development in yeast. Genetics 2012, 192, 73–105. [Google Scholar] [CrossRef] [PubMed]
- Laplante, M.; Sabatini, D.M. mTOR signaling in growth control and disease. Cell 2012, 149, 274–293. [Google Scholar] [CrossRef] [PubMed]
- Alers, S.; Wesselborg, S.; Stork, B. ATG13: Just a companion, or an executor of the autophagic program? Autophagy 2014, 10, 944–956. [Google Scholar] [CrossRef] [PubMed]
- Yorimitsu, T.; Zaman, S.; Broach, J.R.; Klionsky, D.J. Protein kinase A and Sch9 cooperatively regulate induction of autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell 2007, 18, 4180–4189. [Google Scholar] [CrossRef] [PubMed]
- Stephan, J.S.; Yeh, Y.Y.; Ramachandran, V.; Deminoff, S.J.; Herman, P.K. The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc. Natl. Acad. Sci. USA 2009, 106, 17049–17054. [Google Scholar] [CrossRef] [PubMed]
- Stephan, J.S.; Yeh, Y.Y.; Ramachandran, V.; Deminoff, S.J.; Herman, P.K. The Tor and cAMP-dependent protein kinase signaling pathways coordinately control autophagy in Saccharomyces cerevisiae. Autophagy 2010, 6, 294–295. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.E.; Johnson, F.B. Methionine restriction activates the retrograde response and confers both stress tolerance and lifespan extension to yeast, mouse and human cells. PLoS ONE 2014, 9, e97729. [Google Scholar] [CrossRef] [PubMed]
- Ruckenstuhl, C.; Netzberger, C.; Entfellner, I.; Carmona-Gutierrez, D.; Kickenweiz, T.; Stekovic, S.; Gleixner, C.; Schmid, C.; Klug, L.; Sorgo, A.G.; et al. Lifespan extension by methionine restriction requires autophagy-dependent vacuolar acidification. PLoS Genet. 2014, 10, e1004347. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Song, L.; Liu, S.Q.; Huang, D. Independent and additive effects of glutamic acid and methionine on yeast longevity. PLoS ONE 2013, 8, e79319. [Google Scholar] [CrossRef] [PubMed]
- Ruckenstuhl, C.; Netzberger, C.; Entfellner, I.; Carmona-Gutierrez, D.; Kickenweiz, T.; Stekovic, S.; Gleixner, C.; Schmid, C.; Klug, L.; Hajnal, I.; et al. Autophagy extends lifespan via vacuolar acidification. Microb. Cell 2014, 1, 160–162. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Zhang, X.; Lester, R.L.; Dickson, R.C. The sphingoid long chain base phytosphingosine activates AGC-type protein kinases in Saccharomyces cerevisiae including Ypk1, Ypk2, and Sch9. J. Biol. Chem. 2005, 280, 22679–22687. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Zhang, X.; Sumanasekera, C.; Lester, R.L.; Dickson, R.C. Signalling functions for sphingolipid long-chain bases in Saccharomyces cerevisiae. Biochem. Soc. Trans. 2005, 33, 1170–1173. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Liu, J.; Dickson, R.C. Down-regulating sphingolipid synthesis increases yeast lifespan. PLoS Genet. 2012, 8, e1002493. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Huang, X.; Withers, B.R.; Blalock, E.; Liu, K.; Dickson, R.C. Reducing sphingolipid synthesis orchestrates global changes to extend yeast lifespan. Aging Cell 2013, 12, 833–841. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, T.; Knauer, H.; Schauer, A.; Büttner, S.; Ruckenstuhl, C.; Carmona-Gutierrez, D.; Ring, J.; Schroeder, S.; Magnes, C.; Antonacci, L.; et al. Induction of autophagy by spermidine promotes longevity. Nat. Cell Biol. 2009, 11, 1305–1314. [Google Scholar] [CrossRef] [PubMed]
- Morselli, E.; Galluzzi, L.; Kepp, O.; Criollo, A.; Maiuri, M.C.; Tavernarakis, N.; Madeo, F.; Kroemer, G. Autophagy mediates pharmacological lifespan extension by spermidine and resveratrol. Aging 2009, 1, 961–970. [Google Scholar] [CrossRef] [PubMed]
- Minois, N.; Carmona-Gutierrez, D.; Madeo, F. Polyamines in aging and disease. Aging 2011, 3, 716–732. [Google Scholar] [CrossRef] [PubMed]
- Hine, C.; Harputlugil, E.; Zhang, Y.; Ruckenstuhl, C.; Lee, B.C.; Brace, L.; Longchamp, A.; Treviño-Villarreal, J.H.; Mejia, P.; Ozaki, C.K.; et al. Endogenous hydrogen sulfide production is essential for dietary restriction benefits. Cell 2015, 160, 132–144. [Google Scholar] [CrossRef] [PubMed]
- De Cabo, R.; Carmona-Gutierrez, D.; Bernier, M.; Hall, M.N.; Madeo, F. The search for antiaging interventions: From elixirs to fasting regimens. Cell 2014, 157, 1515–1526. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Longo, V. Dietary restriction with and without caloric restriction for healthy aging. F1000Res. 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Hine, C.; Mitchell, J.R. Calorie restriction and methionine restriction in control of endogenous hydrogen sulfide production by the transsulfuration pathway. Exp. Gerontol. 2015, 68, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Shim, H.S.; Longo, V.D. A protein restriction-dependent sulfur code for longevity. Cell 2015, 160, 15–17. [Google Scholar] [CrossRef] [PubMed]
- Burhans, W.C.; Weinberger, M. Acetic acid effects on aging in budding yeast: Are they relevant to aging in higher eukaryotes? Cell Cycle 2009, 8, 2300–2302. [Google Scholar] [CrossRef] [PubMed]
- Burtner, C.R.; Murakami, C.J.; Olsen, B.; Kennedy, B.K.; Kaeberlein, M. A genomic analysis of chronological longevity factors in budding yeast. Cell Cycle 2011, 10, 1385–1396. [Google Scholar] [CrossRef] [PubMed]
- Mirisola, M.G.; Longo, V.D. Acetic acid and acidification accelerate chronological and replicative aging in yeast. Cell Cycle 2012, 11, 3532–3533. [Google Scholar] [CrossRef] [PubMed]
- Giannattasio, S.; Guaragnella, N.; Zdralević, M.; Marra, E. Molecular mechanisms of Saccharomyces cerevisiae stress adaptation and programmed cell death in response to acetic acid. Front. Microbiol. 2013, 4, 33. [Google Scholar] [CrossRef] [PubMed]
- Falcone, C.; Mazzoni, C. External and internal triggers of cell death in yeast. Cell. Mol. Life Sci. 2016, 73, 2237–2250. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, T.; Schroeder, S.; Andryushkova, A.; Pendl, T.; Küttner, V.; Bhukel, A.; Mariño, G.; Pietrocola, F.; Harger, A.; Zimmermann, A.; et al. Nucleocytosolic depletion of the energy metabolite acetyl-coenzyme A stimulates autophagy and prolongs lifespan. Cell Metab. 2014, 19, 431–444. [Google Scholar] [CrossRef] [PubMed]
- Mariño, G.; Pietrocola, F.; Eisenberg, T.; Kong, Y.; Malik, S.A.; Andryushkova, A.; Schroeder, S.; Pendl, T.; Harger, A.; Niso-Santano, M.; et al. Regulation of autophagy by cytosolic acetyl-coenzyme A. Mol. Cell 2014, 53, 710–725. [Google Scholar] [CrossRef] [PubMed]
- Fabrizio, P.; Gattazzo, C.; Battistella, L.; Wei, M.; Cheng, C.; McGrew, K.; Longo, V.D. Sir2 blocks extreme life-span extension. Cell 2005, 123, 655–667. [Google Scholar] [CrossRef] [PubMed]
- Richard, V.R.; Beach, A.; Piano, A.; Leonov, A.; Feldman, R.; Burstein, M.T.; Kyryakov, P.; Gomez-Perez, A.; Arlia-Ciommo, A.; Baptista, S.; et al. Mechanism of liponecrosis, a distinct mode of programmed cell death. Cell Cycle 2014, 13, 3707–3726. [Google Scholar] [CrossRef] [PubMed]
- Sheibani, S.; Richard, V.R.; Beach, A.; Leonov, A.; Feldman, R.; Mattie, S.; Khelghatybana, L.; Piano, A.; Greenwood, M.; Vali, H.; et al. Macromitophagy, neutral lipids synthesis, and peroxisomal fatty acid oxidation protect yeast from “liponecrosis”, a previously unknown form of programmed cell death. Cell Cycle 2014, 13, 138–147. [Google Scholar] [CrossRef] [PubMed]
- Mohammad, K.; Dakik, P.; Medkour, Y.; McAuley, M.; Mitrofanova, D.; Titorenko, V.I. Yeast cells exposed to exogenous palmitoleic acid either adapt to stress and survive or commit to regulated liponecrosis and die. Oxid. Med. Cell. Longev. 2018, 2018. [Google Scholar] [CrossRef]
- Beach, A.; Titorenko, V.I. In search of housekeeping pathways that regulate longevity. Cell Cycle 2011, 10, 3042–3044. [Google Scholar] [CrossRef] [PubMed]
- Medkour, Y.; Svistkova, V.; Titorenko, V.I. Cell-nonautonomous mechanisms underlying cellular and organismal aging. Int. Rev. Cell Mol. Biol. 2016, 321, 259–297. [Google Scholar] [PubMed]
- Burstein, M.T.; Kyryakov, P.; Beach, A.; Richard, V.R.; Koupaki, O.; Gomez-Perez, A.; Leonov, A.; Levy, S.; Noohi, F.; Titorenko, V.I. Lithocholic acid extends longevity of chronologically aging yeast only if added at certain critical periods of their lifespan. Cell Cycle 2012, 11, 3443–3462. [Google Scholar] [CrossRef] [PubMed]
- Cantley, L.; Hunter, T.; Sever, R.; Thorner, J. Signal Transduction: Principles, Pathways, and Processes; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2014; ISBN 978-0-87969-901-7. [Google Scholar]
- Lim, W.; Mayer, B.; Pawson, T. Cell Signaling: Principles and Mechanisms; Garland Science: New York, NY, USA, 2015; ISBN 978-0-8153-4244-1. [Google Scholar]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohammad, K.; Dakik, P.; Medkour, Y.; McAuley, M.; Mitrofanova, D.; Titorenko, V.I. Some Metabolites Act as Second Messengers in Yeast Chronological Aging. Int. J. Mol. Sci. 2018, 19, 860. https://doi.org/10.3390/ijms19030860
Mohammad K, Dakik P, Medkour Y, McAuley M, Mitrofanova D, Titorenko VI. Some Metabolites Act as Second Messengers in Yeast Chronological Aging. International Journal of Molecular Sciences. 2018; 19(3):860. https://doi.org/10.3390/ijms19030860
Chicago/Turabian StyleMohammad, Karamat, Paméla Dakik, Younes Medkour, Mélissa McAuley, Darya Mitrofanova, and Vladimir I. Titorenko. 2018. "Some Metabolites Act as Second Messengers in Yeast Chronological Aging" International Journal of Molecular Sciences 19, no. 3: 860. https://doi.org/10.3390/ijms19030860