Overexpression of a SBP-Box Gene (VpSBP16) from Chinese Wild Vitis Species in Arabidopsis Improves Salinity and Drought Stress Tolerance
Abstract
:1. Introduction
2. Results
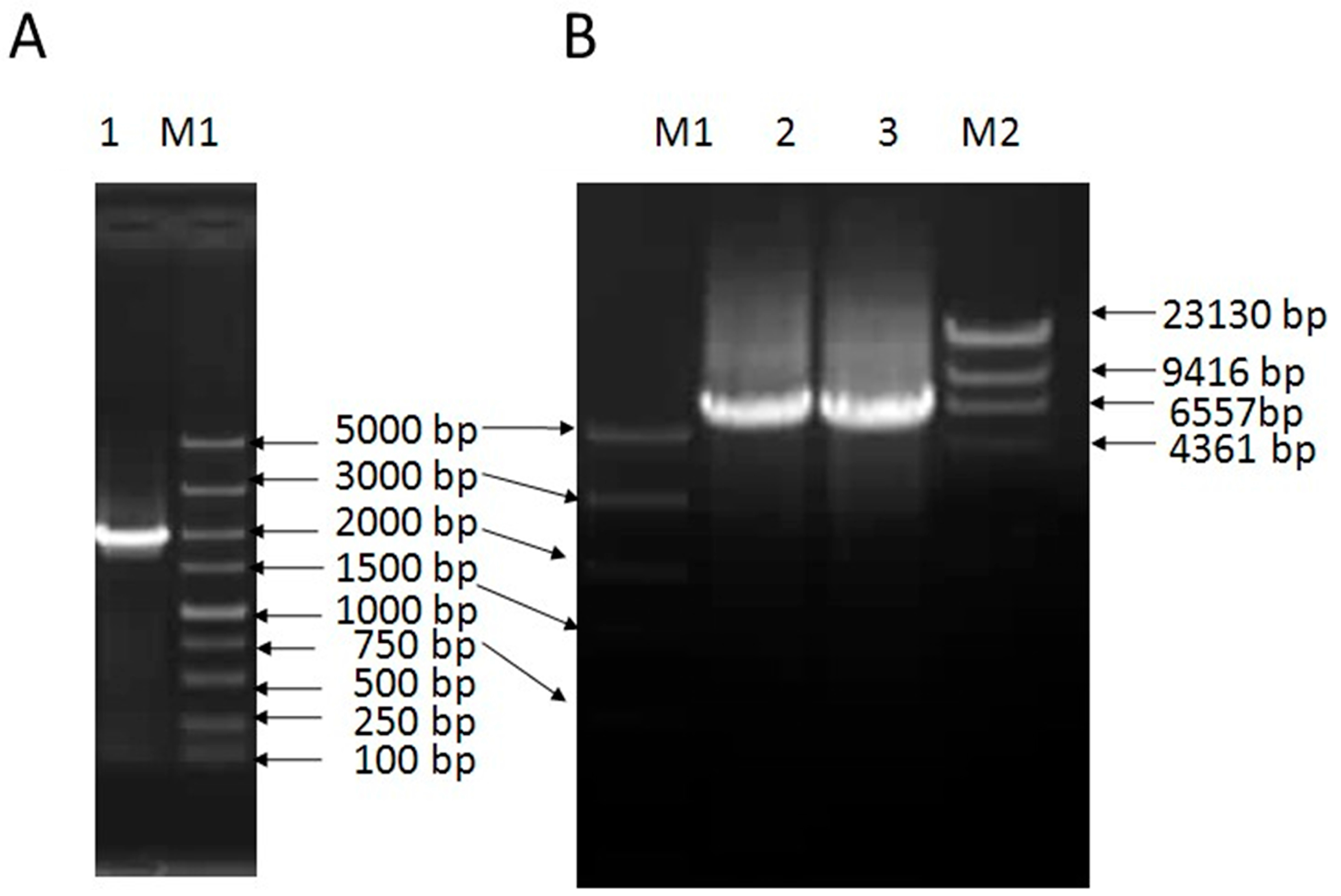
2.1. Cloning and Sequence Analysis of VpSBP16
2.2. Subcellular Localization and Function of VpSBP16 in Transcriptional Activation
2.3. Effect of Osmotic Stress on Seed Germination in Transgenic VpSBP16 Arabidopsis Lines
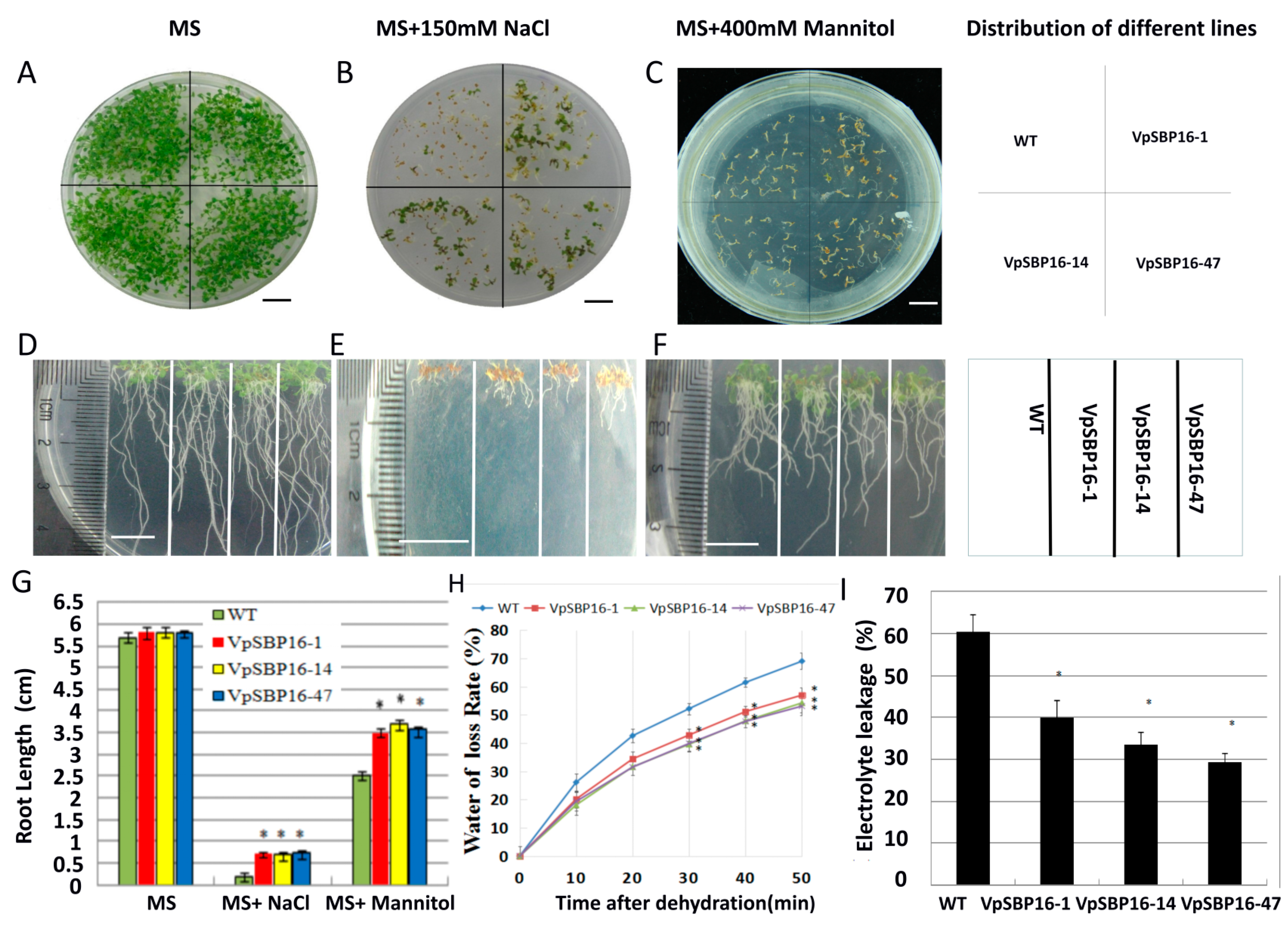
2.4. VpSBP16 Transgenic Arabidopsis Seedling Resistance to Osmotic Stress
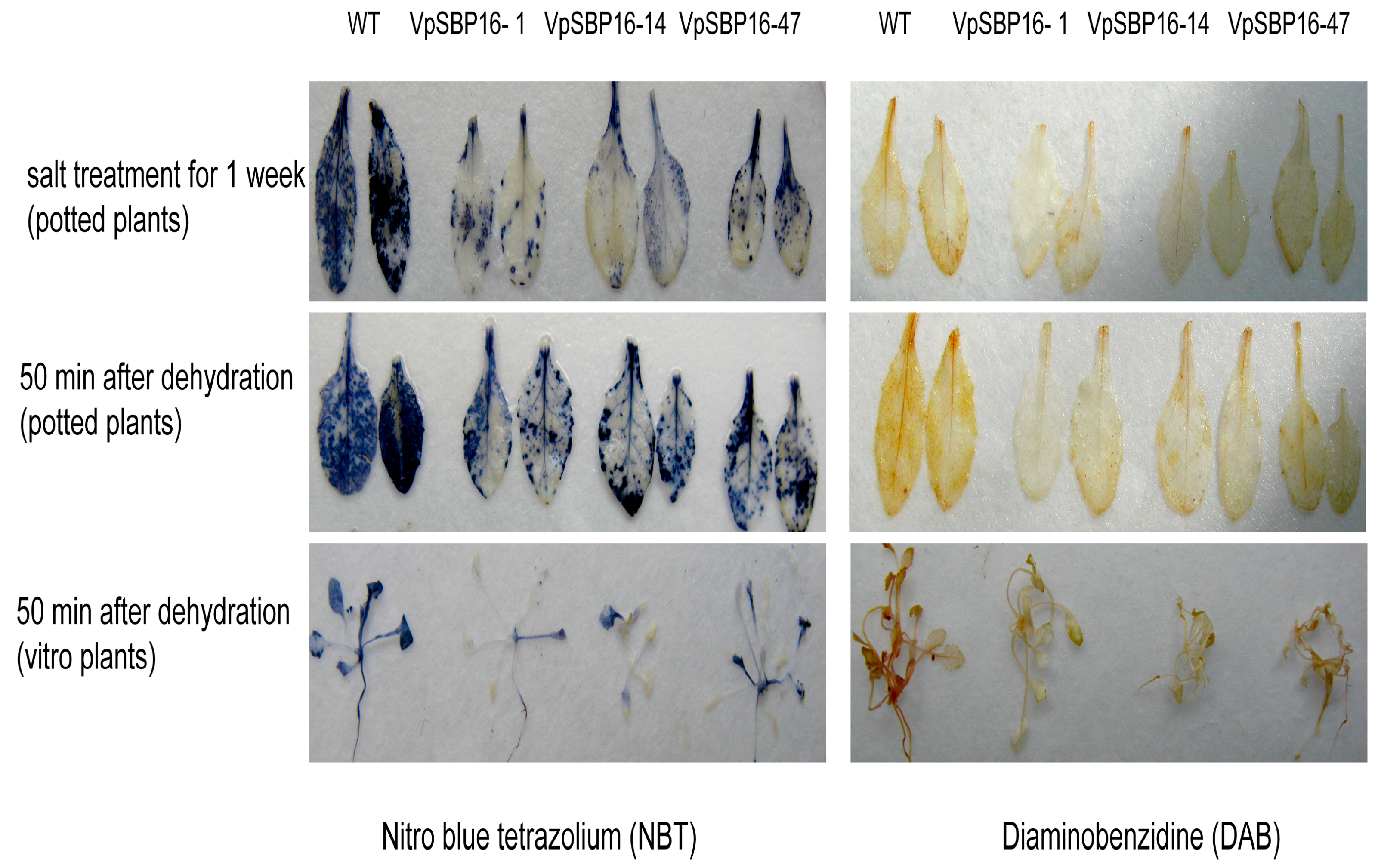
2.5. Response to Abiotic Stresses of VpSBP16 Transgenic Arabidopsis Plants
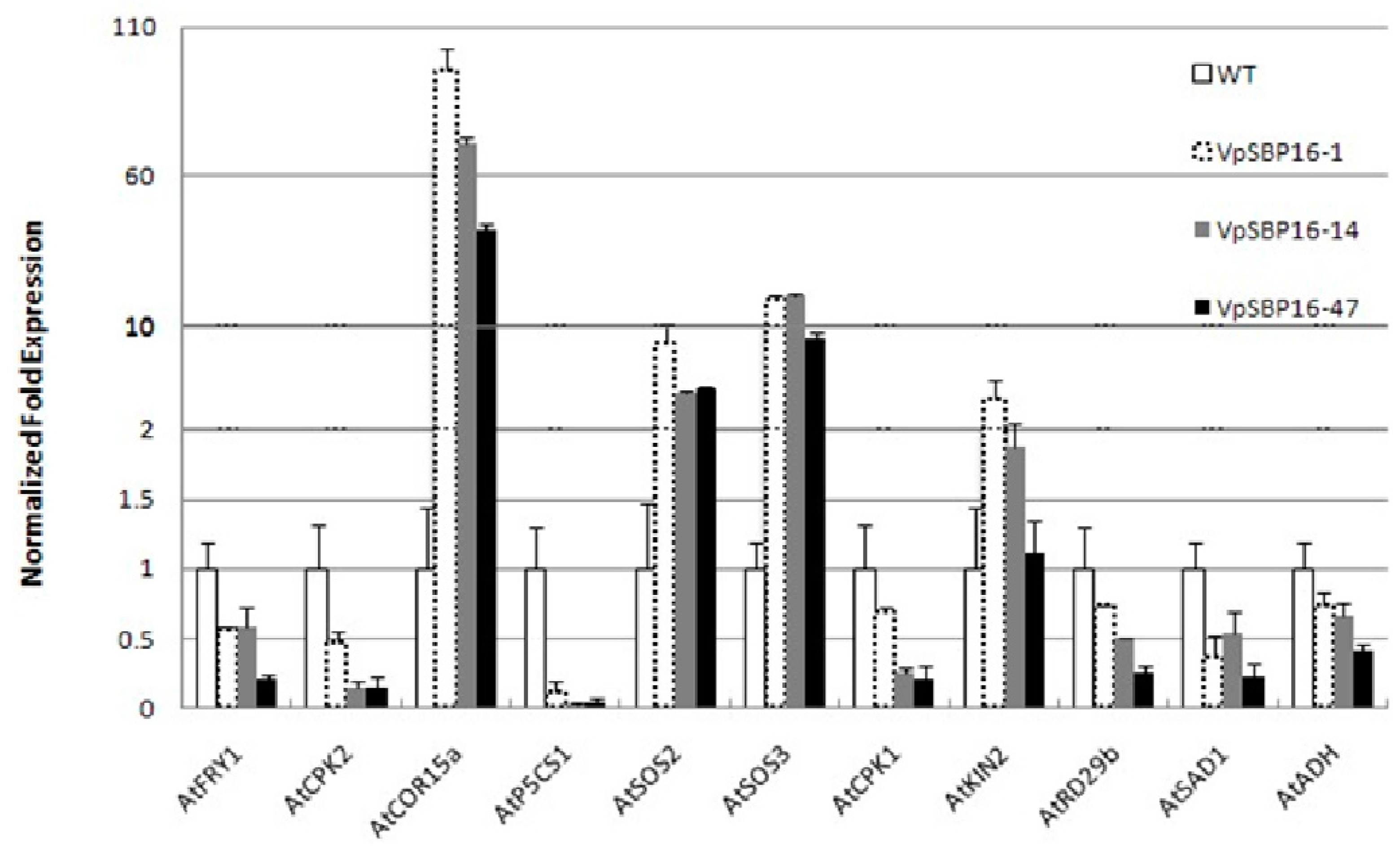
2.6. Altered Expression of Abiotic Stress Responsive Genes in Transgenic VpSBP16 Plants
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Isolation and Analysis of the VpSBP16 cDNA
4.3. Subcellular Localization and Trans-Activation Assay
4.4. Generation of Transgenic A. thaliana Plants Over-Expressing VpSBP16
4.5. Semi-Quantitative RT-PCR Analysis
4.6. Germination Assays
4.7. Salt and Drought Treatments for Wild Type (WT) and T3 Transgenic Lines
4.8. Determination of the Water Loss Rate and Electrolyte Leakage
4.9. Detection of H2O2 and O2−
4.10. Quantitative Real-Time RT-PCR Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| CaMV | Cauliflower mosaic virus |
| ORF | Open reading frame |
| RT-PCR | Reverse transcription-polymerase chain reaction |
| MS | Murashige and Skoog |
| NBT | Nitro blue tetrazolium |
| DAB | Diaminobenzidine |
| ROS | Reactive oxygen species |
| H2O2 | Hydrogen peroxide |
| O2− | Superoxide anion |
| SOS | Salt overly sensitive |
| SBP | Squamosa promoter binding protein |
| NLS | Nuclear localization signal |
| WT | Wild-type |
References
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) Function as Transcriptional Activators in Abscisic Acid Signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Li, H.; X, A.; X, L.; H, Q.; D, W. Transgenic expression of MYB15 confers enhanced sensitivity to abscisic acid and improved drought tolerance in Arabidopsis thaliana. J. Genet. Genom. 2009, 36, 17–29. [Google Scholar] [CrossRef]
- Yang, O.S.; Popova, O.V.; Süthoff, U.; Lüking, I.; Dietz, K.J.; Golldack, D. The Arabidopsis basic leucine zipper transcription factor AtbZIP24 regulates complex transcriptional networks involved in abiotic stress resistance. Gene 2009, 436, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Lai, Z.; Shi, J.; Yong, X.; Chen, Z.; Xu, X. Roles of arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biol. 2010, 10, 281–296. [Google Scholar]
- Jiang, Y.; Deyholos, M.K. Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol. Biol. 2009, 69, 91–105. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Chen, Z.; Liu, Y.; Zhang, H.; Zhang, M.; Liu, Q.; Hong, X.; Zhu, J.K.; Gong, Z. ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 2010, 63, 417–429. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, R.; Schippers, J.H.; Mieulet, D.; Obata, T.; Fernie, A.R.; Guiderdoni, E.; Mueller-Roeber, B. MULTIPASS, a rice R2R3-type MYB transcription factor, regulates adaptive growth by integrating multiple hormonal pathways. Plant J. 2013, 76, 258–273. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, T.H.; Li, C.W.; Su, R.C.; Cheng, C.P.; Tsai, Y.C.; Chan, M.T. A tomato bZIP transcription factor, SlAREB, is involved in water deficit and salt stress response. Planta 2010, 231, 1459–1473. [Google Scholar] [CrossRef] [PubMed]
- Klein, J.; Saedler, H.; Huijser, P. A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol. Genet. Genom. 1996, 250, 7–16. [Google Scholar]
- Xie, K.; Wu, C.; Xiong, L. Genomic Organization, Differential Expression, and Interaction of SQUAMOSA Promoter-Binding-Like Transcription Factors and microRNA156 in Rice. Plant Physiol. 2006, 142, 280–293. [Google Scholar] [CrossRef] [PubMed]
- Preston, J.C.; Hileman, L.C. Functional Evolution in the Plant SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) Gene Family. Front. Plant Sci. 2013, 4, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Birkenbihl, R.P.; Jach, G.; Saedler, H.; Huijser, P. Functional Dissection of the Plant-specific SBP-Domain: Overlap of the DNA-binding and Nuclear Localization Domains. J. Mol. Biol. 2005, 352, 585–596. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, K.; Kigawa, T.; Inoue, M.; Yamasaki, T.; Yabuki, T.; Aoki, M.; Seki, E.; Matsuda, T.; Tomo, Y.; Terada, T. An Arabidopsis SBP-domain fragment with a disrupted C-terminal zinc-binding site retains its tertiary structure. FEBS Lett. 2006, 580, 2109–2116. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hu, Z.; Yang, Y.; Chen, X.; Chen, G. Function annotation of an SBP-box gene in Arabidopsis based on analysis of co-expression networks and promoters. Int. J. Mol. Sci. 2009, 10, 116–132. [Google Scholar] [CrossRef] [PubMed]
- Stone, J.M.; Liang, X.; Nekl, E.R.; Stiers, J.J. Arabidopsis AtSPL14, a plant-specific SBP-domain transcription factor, participates in plant development and sensitivity to fumonisin B1. Plant J. 2005, 41, 744–754. [Google Scholar] [CrossRef] [PubMed]
- Ning, K.; Chen, S.; Huang, H.; Jiang, J.; Yuan, H.; Li, H. Molecular characterization and expression analysis of the SPL gene family with BpSPL9 transgenic lines found to confer tolerance to abiotic stress in Betula platyphylla Suk. Plant Cell Tissue Organ Cult. 2017, 130, 1–13. [Google Scholar] [CrossRef]
- Mao, H.D.; Yu, L.J.; Li, Z.J.; Yan, Y.; Han, R.; Liu, H.; Ma, M. Genome-wide analysis of the SPL family transcription factors and their responses to abiotic stresses in maize. Plant Gene 2016, 6, 1–12. [Google Scholar] [CrossRef]
- Stief, A.; Altmann, S.; Hoffmann, K.; Pant, B.D.; Scheible, W.; Bäurle, I. Arabidopsis miR156 Regulates Tolerance to Recurring Environmental Stress through SPL Transcription Factors. Plant Cell 2014, 26, 1792–1807. [Google Scholar] [CrossRef] [PubMed]
- Cui, N.; Sun, X.; Sun, M.; Jia, B.; Duanmu, H.; Lv, D.; Duan, X.; Zhu, Y. Overexpression of OsmiR156k leads to reduced tolerance to cold stress in rice (Oryza Sativa). Mol. Breed. 2015, 35, 214–225. [Google Scholar] [CrossRef]
- Cui, L.G.; Shan, J.X.; Shi, M.; Gao, J.P.; Lin, H.X. The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J. 2014, 80, 1108–1117. [Google Scholar] [CrossRef] [PubMed]
- Arshad, M.; Feyissa, B.A.; Amyot, L.; Aung, B.; Hannoufa, A. MicroRNA156 improves drought stress tolerance in alfalfa (Medicago sativa) by silencing SPL13. Plant Sci. 2017, 258, 122–136. [Google Scholar] [CrossRef] [PubMed]
- Hou, H.; Li, J.; Gao, M.; Singer, S.D.; Wang, H.; Mao, L.; Fei, Z.; Wang, X. Genomic Organization, Phylogenetic Comparison and Differential Expression of the SBP-Box Family Genes in Grape. PLoS ONE 2013, 8, e59358. [Google Scholar] [CrossRef] [PubMed]
- Hou, H.; Yan, Q.; Wang, X.; Xu, H. A SBP-Box Gene VpSBP5 from Chinese Wild Vitis Species Responds to Erysiphe necator and Defense Signaling Molecules. Plant Mol. Biol. Rep. 2013, 31, 1261–1270. [Google Scholar] [CrossRef]
- Marè, C.; Mazzucotelli, E.; Crosatti, C.; Francia, E.; Stanca, A.M.; Cattivelli, L. Hv-WRKY38: A new transcription factor involved in cold- and drought-response in barley. Plant Mol. Biol. 2004, 55, 399–416. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; He, C. Regulation of plant reactive oxygen species (ROS) in stress responses: Learning from AtRBOHD. Plant Cell Rep. 2016, 35, 995–1007. [Google Scholar] [CrossRef] [PubMed]
- Cardon, G.; Höhmann, S.; Klein, J.; Nettesheim, K.; Saedler, H.; Huijser, P. Molecular characterisation of the Arabidopsis SBP-box genes. Gene 1999, 237, 91–104. [Google Scholar] [CrossRef]
- And, P.M.H.; Bressan, R.A.; Zhu, J.K.; Bohnert, H.J. Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Biol. 2000, 51, 463–499. [Google Scholar] [CrossRef]
- Mittova, V.; Tal, M.; Volokita, M.; Guy, M. Salt stress induces up-regulation of an efficient chloroplast antioxidant system in the salt-tolerant wild tomato species Lycopersicon pennellii but not in the cultivated species. Physiol. Plant. 2002, 115, 393–400. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.; Dai, X.; Zhang, W. A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J. Exp. Bot. 2012, 63, 2541–2556. [Google Scholar] [CrossRef] [PubMed]
- Fukao, T.; Bailey-Serres, J. The submergence tolerance regulator SUB1A mediates crosstalk between submergence and drought tolerance in rice. Plant Cell 2011, 23, 412–427. [Google Scholar] [CrossRef] [PubMed]
- Pan, I.; Li, C.W.; Su, R.C.; Cheng, C.P.; Lin, C.S.; Chan, M.T. Ectopic expression of an EAR motif deletion mutant of SlERF3 enhances tolerance to salt stress and Ralstonia solanacearum in tomato. Planta 2010, 232, 1075–1086. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Zhang, H.; Pan, X.; Chen, X.; Zhang, Z.; Lu, X.; Huang, R. Overexpression of ethylene response factor TERF2 confers cold tolerance in rice seedlings. Transgenic Res. 2011, 20, 857–866. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Zhang, Z.; Zhang, H.; Wang, X.C.; Huang, R. Transcriptional modulation of ethylene response factor protein JERF3 in the oxidative stress response enhances tolerance of tobacco seedlings to salt, drought, and freezing. Plant Physiol. 2008, 148, 1953–1963. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Pardo, J.M.; Batelli, G.; Van Oosten, M.J.; Bressan, R.A.; Li, X. The Salt Overly Sensitive (SOS) Pathway: Established and Emerging Roles. Mol. Plant 2013, 6, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.G.; Ma, Q.J.; Sun, C.H.; Sun, M.H.; You, C.X.; Hao, Y.J. Overexpression of MdSOS2L1, a CIPK protein kinase, increases the antioxidant metabolites to enhance salt tolerance in apple and tomato. Physiol. Plant. 2015, 156, 201–214. [Google Scholar] [CrossRef] [PubMed]
- Huertas, R.; Olías, R.; Eljakaoui, Z.; Gálvez, F.J.; Li, J.; De Morales, P.A.; Belver, A.; Rodríguezrosales, M.P. Overexpression of SlSOS2 (SlCIPK24) confers salt tolerance to transgenic tomato. Plant Cell Environ. 2012, 35, 1467–1482. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Wang, J.; Bi, Y.; Wang, L.; Tang, L.; Xiang, Y.; Ohtani, M.; Demura, T.; Qiang, Z. Overexpression of PtSOS2 enhances salt tolerance in transgenic poplars. Plant Mol. Biol. Rep. 2014, 32, 185–197. [Google Scholar]
- Kesari, R.; Lasky, J.R.; Villamor, J.G.; Des Marais, D.L.; Chen, Y.J.; Liu, T.W. Intron-mediated alternative splicing of arabidopsis p5cs1 and its association with natural variation in proline and climate adaptation. Proc. Natl. Acad. Sci. USA 2012, 109, 9197–9202. [Google Scholar] [CrossRef] [PubMed]
- Des Marais, D.L.; Mckay, J.K.; Richards, J.H.; Sen, S.; Wayne, T.; Juenger, T.E. Physiological genomics of response to soil drying in diverse Arabidopsis accessions. Plant Cell 2012, 24, 893–914. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.Y.; Wu, W.H. Atcpk23 functions in Arabidopsis responses to drought and salt stresses. Plant Mol. Biol. 2007, 65, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, Y.; Maruyama, K.; Osakabe, Y.; Qin, F.; Seki, M.; Shinozaki, K. Functional analysis of an arabidopsis transcription factor, dreb2a, involved in drought-responsive gene expression. Plant Cell 2006, 18, 1292–1309. [Google Scholar] [CrossRef] [PubMed]
- Guiltinan, M.J.; Marcotte, W.R., Jr.; Quatrano, R.S. A plant leucine zipper protein that recognizes an abscisic acid response element. Science 1990, 250, 267–271. [Google Scholar] [CrossRef] [PubMed]
- Msanne, J.; Lin, J.; Stone, J.M.; Awada, T. Characterization of abiotic stress-responsive Arabidopsis thaliana rd29a and rd29b genes and evaluation of transgenes. Planta 2011, 234, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.M.; Gong, Z.Z.; Christopher, D.R.; Senthil, S.; Guo, Y.; Xu, W.Y.; Galbraith, D.; Zhu, J.K. Modulation of abscisic acid signal transduction and biosynthesis by an Sm-like protein in Arabidopsis. Dev. Cell 2001, 1, 771–781. [Google Scholar] [CrossRef]
- Lu, G.; Paul, A.L.; Mccarty, D.R.; Ferl, R.J. Transcription factor veracity: Is GBF3 responsible for aba-regulated expression of Arabidopsis Adh? Plant Cell 1996, 8, 847–857. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.M.; Lee, B.; Ishitani, M.; Lee, H.; Zhang, C.; Zhu, J.K. Fiery1 encoding an inositol polyphosphate 1-phosphatase is a negative regulator of abscisic acid and stress signaling in Arabidopsis. Gene Dev. 2001, 15, 1971–1984. [Google Scholar] [CrossRef] [PubMed]
- Fujita, M.; Fujita, Y.; Maruyama, K.; Seki, M.; Hiratsu, K.; Ohme-Takagi, M.; Tran, L.S.P.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 2004, 39, 863–876. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.; Bent, A. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Saleki, R.; Young, P.G.; Lefebvre, D.D. Mutants of Arabidopsis thaliana Capable of Germination under Saline Conditions. Plant Physiol. 1993, 101, 839–845. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhao, G.; Xia, C.; Jia, J.; Liu, X.; Kong, X. A wheat R2R3-MYB gene, TaMYB30-B, improves drought stress tolerance in transgenic Arabidopsis. J. Exp. Bot. 2012, 63, 5873–5885. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.H.; Liu, J.; He, X.J.; Mu, R.L.; Zhou, H.L.; Chen, S.Y. Modulation of ethylene responses affects plant salt-stress responses. Plant Physiol. 2007, 143, 707–719. [Google Scholar] [CrossRef] [PubMed]
- Kotchoni, S.O.; Kuhns, C.; Ditzer, A.; Kirch, H.H.; Bartels, D. Over-expression of different aldehyde dehydrogenase genes in Arabidopsis thaliana confers tolerance to abiotic stress and protects plants against lipid peroxidation and oxidative stress. Plant Cell Environ. 2006, 29, 1033–1048. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Woo, D.H.; Kim, J.M.; Lee, S.Y.; Chung, W.S.; Moon, Y.H. Arabidopsis MKK4 mediates osmotic-stress response via its regulation of MPK3 activity. Biochem. Biophys. Res. Commun. 2011, 412, 150–154. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Niu, J.; Zhao, S.; Jiao, C.; Xu, W.; Fei, Z.; Wang, X. Characterization of Erysiphe necator-Responsive Genes in Chinese Wild Vitis quinquangularis. Int. J. Mol. Sci. 2012, 13, 11497–11519. [Google Scholar] [CrossRef] [PubMed]




| Primer Pairs | Forward and Reverse Primers (5′-3′) | Restriction Enzyme Cutting Site |
|---|---|---|
| VpSBP16-F | F: GCCCAATTCGTCTGCAAGAAGT | none |
| VpSBP16-R | R: CACCACCCTTGCCACATGAAACA | none |
| VpSBP16-F1 | F: ATGGGGTCTTGGAGCTACG | none |
| VpSBP16-R1 | R: TTACGTTATCCGGAGGC | none |
| AtSOS2-F | F: ATTGAGGCTGTAGCGAAC | none |
| AtSOS2-R | R: GGTATTCCTTCTGTTGCC | none |
| AtSOS3-F | F: GGAGGAATCTCTTCGCTG | none |
| AtSOS3-R | R: CACGAAAGCCTTATCCACC | none |
| AtFRY1-F | F: CGCAGTAGCACTAGGATTG | none |
| AtFRY1-R | R: TTGACACCGAGTTTATTGG | none |
| AtADH-F | F: CTCTTGGTGCTGTTGGTTTAGG | none |
| AtADH-R | R: AATTGGCTTGTCATGGTCTTTC | none |
| AtCOR15a-F | F: GCGAACAATCCTTCACAG | none |
| AtCOR15a-R | R: CTTCGGGAGACCCACCT | none |
| AtKIN2-F | F: GTCAGAGACCAACAAGAATGCC | none |
| AtKIN2-R | R: TGACTCGAATCGCTACTTGTTC | none |
| AtP5CS1-F | F: TTCTCAGATGGTTTCCAGGTTG | none |
| AtP5CS1-R | R: TGGGAATGTCCTGATGGGTG | none |
| AtRD29B-F | F: GTGAAGATGACTATCTCGGTGGTC | none |
| AtRD29B-R | R: TACCAAGAGACTCAGCAATCTCTG | none |
| AtCDPK1-F | F: CCGTTTTGGGCTGAGACTGAA | none |
| AtCDPK1-R | R: CCATGGGTGAGCTAACACTTGC | none |
| AtCDPK2-F | F: GTCCATTACCTTCCCGGCATAT | none |
| AtCDPK2-R | R: CAGCAATTACCCGTAATGCCATT | none |
| Atactin1-F | F: AGGCACCTCTTAACCCTAAAGC | none |
| Atactin1-R | R: GGACAACGGAATCTCTCAGC | none |
| VpSBP16-F2 | F: CGCTCTAGAATGGGGTCTTGGAGCTACG | XbaI site underlined |
| VpSBP16-R2 | R: ATAGGTACCCGTTATCCGGAGGCAGAAAG | KpnI site underlined |
| VpSBP16-F3 | F: TATCCCGGGATGGGGTCTTGGAGCTACG | XmaI site underlined |
| VpSBP16-R3 | R: GGCGGATCCTTACGTTATCCGGAGGCAGA | BamHI site underlined |
| Gal4-F | F: CCATGGTAATGAAGCTACTGTCTTCTAT | NcoI site underlined |
| Gal4-R | R: GGATCCTTACTCTTTTTTTGGGTTTG | BamHI site underlined |
| VpSBP16-F4 | F: CACGGATCCATGGGGTCTTGGAGCTA | BamHI site underlined |
| VpSBP16-R4 | R: GGCGGTACCTTACGTTATCCGGAGGC | KpnI site underlined |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hou, H.; Jia, H.; Yan, Q.; Wang, X. Overexpression of a SBP-Box Gene (VpSBP16) from Chinese Wild Vitis Species in Arabidopsis Improves Salinity and Drought Stress Tolerance. Int. J. Mol. Sci. 2018, 19, 940. https://doi.org/10.3390/ijms19040940
Hou H, Jia H, Yan Q, Wang X. Overexpression of a SBP-Box Gene (VpSBP16) from Chinese Wild Vitis Species in Arabidopsis Improves Salinity and Drought Stress Tolerance. International Journal of Molecular Sciences. 2018; 19(4):940. https://doi.org/10.3390/ijms19040940
Chicago/Turabian StyleHou, Hongmin, Hui Jia, Qin Yan, and Xiping Wang. 2018. "Overexpression of a SBP-Box Gene (VpSBP16) from Chinese Wild Vitis Species in Arabidopsis Improves Salinity and Drought Stress Tolerance" International Journal of Molecular Sciences 19, no. 4: 940. https://doi.org/10.3390/ijms19040940





