Wheat Gene TaATG8j Contributes to Stripe Rust Resistance
Abstract
:1. Introduction
2. Results
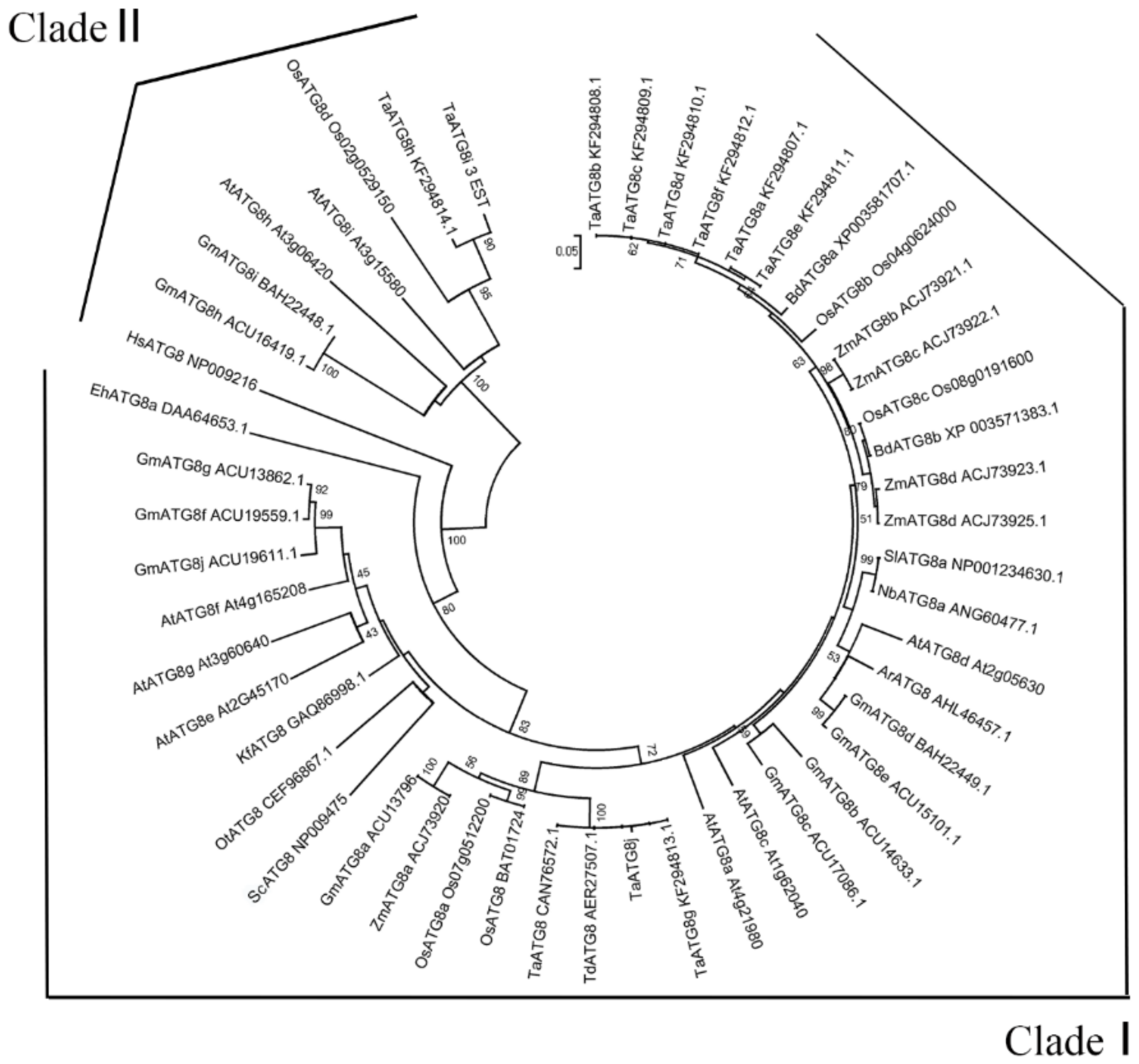
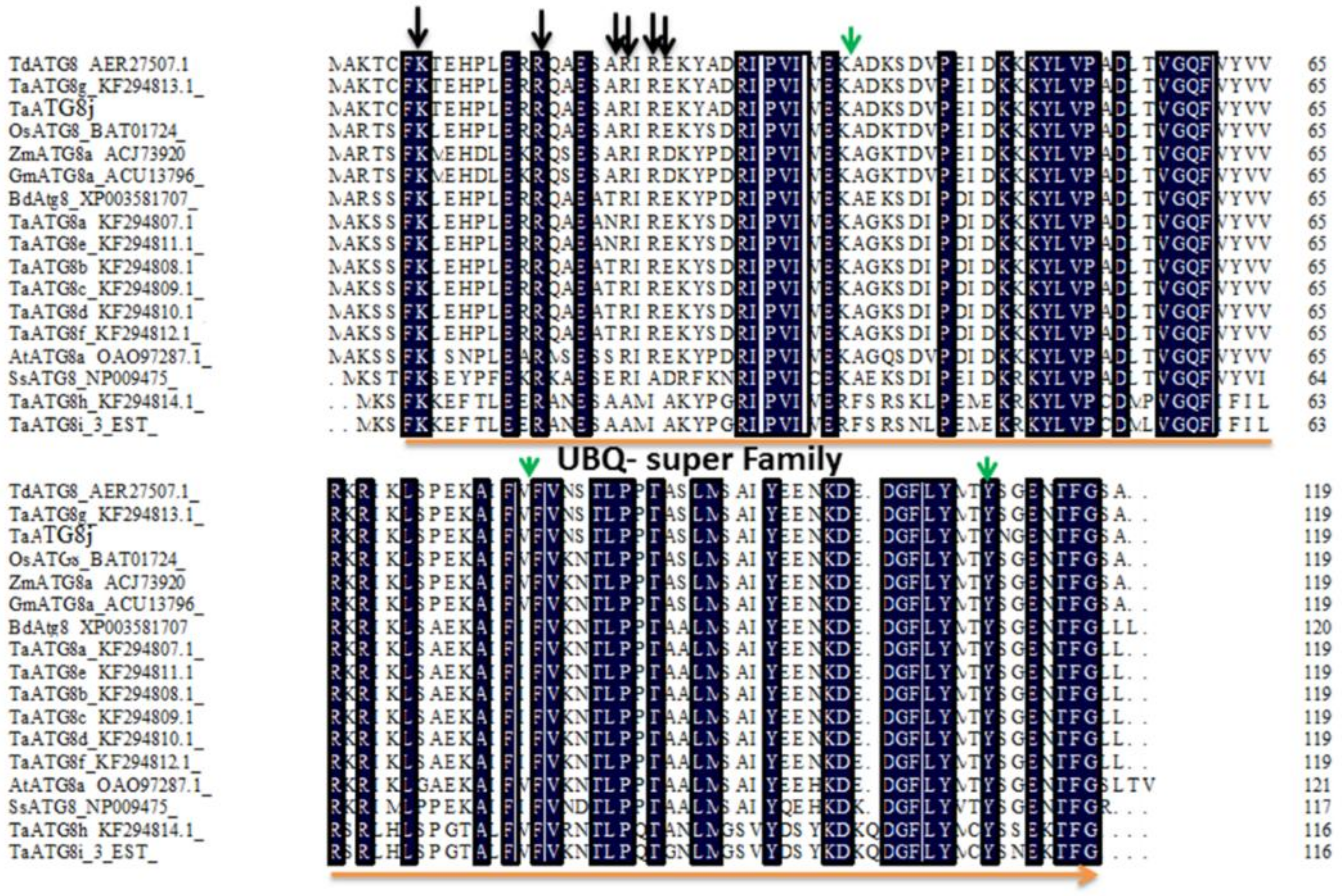
2.1. Identification and Isolation of the TaATG8j Gene
2.2. TaATG8j Is Induced upon Incompatible Pst Attack
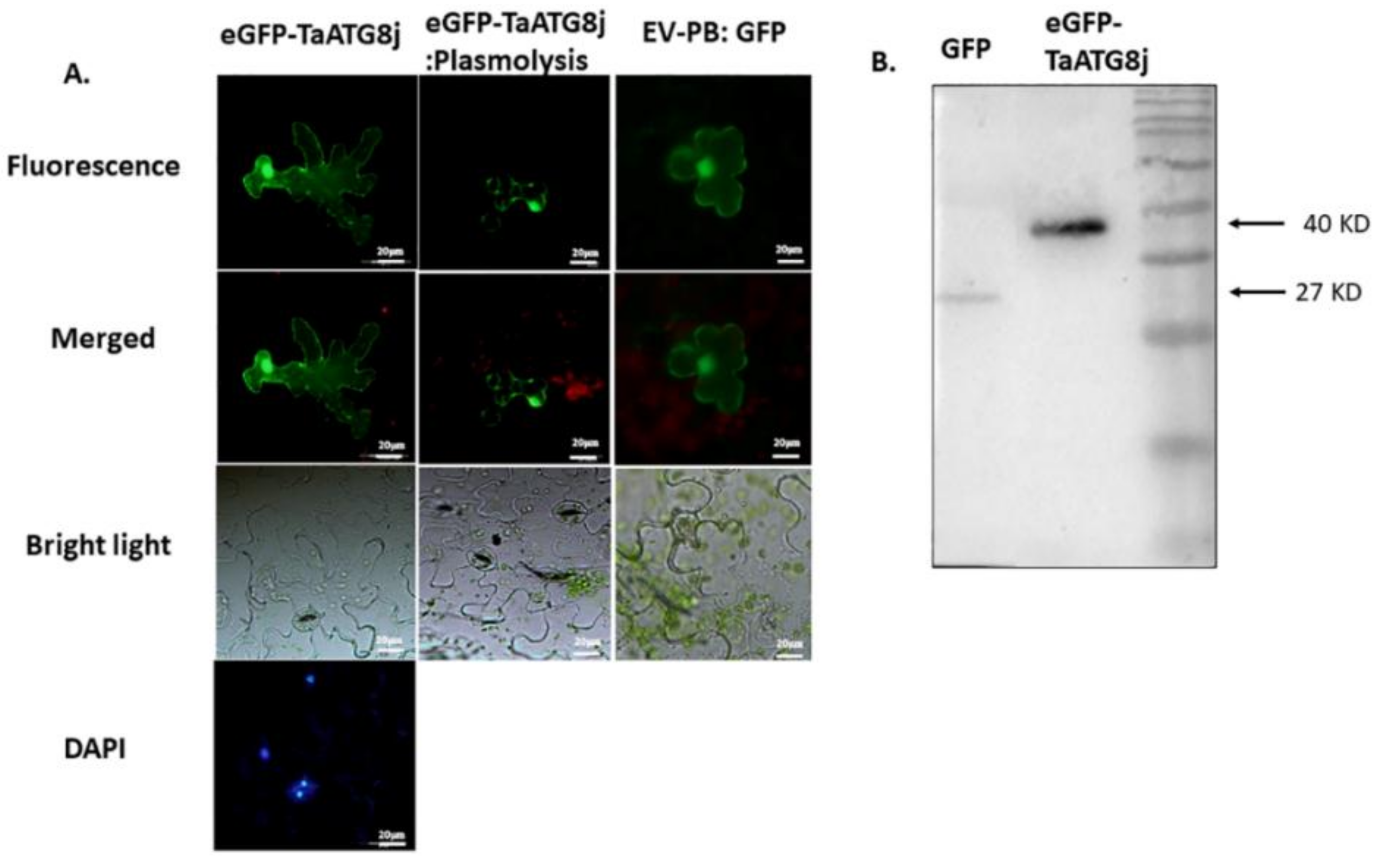
2.3. Protein TaATG8j Is Distributed throughout the Cytoplasm but Mainly in Nuclei and Plasma Membranes
2.4. TaATG8j Delays Cell Death Triggered by BAX in N. benthamiana
2.5. Overexpression of TaATG8j Induces Cell Death in Yeast
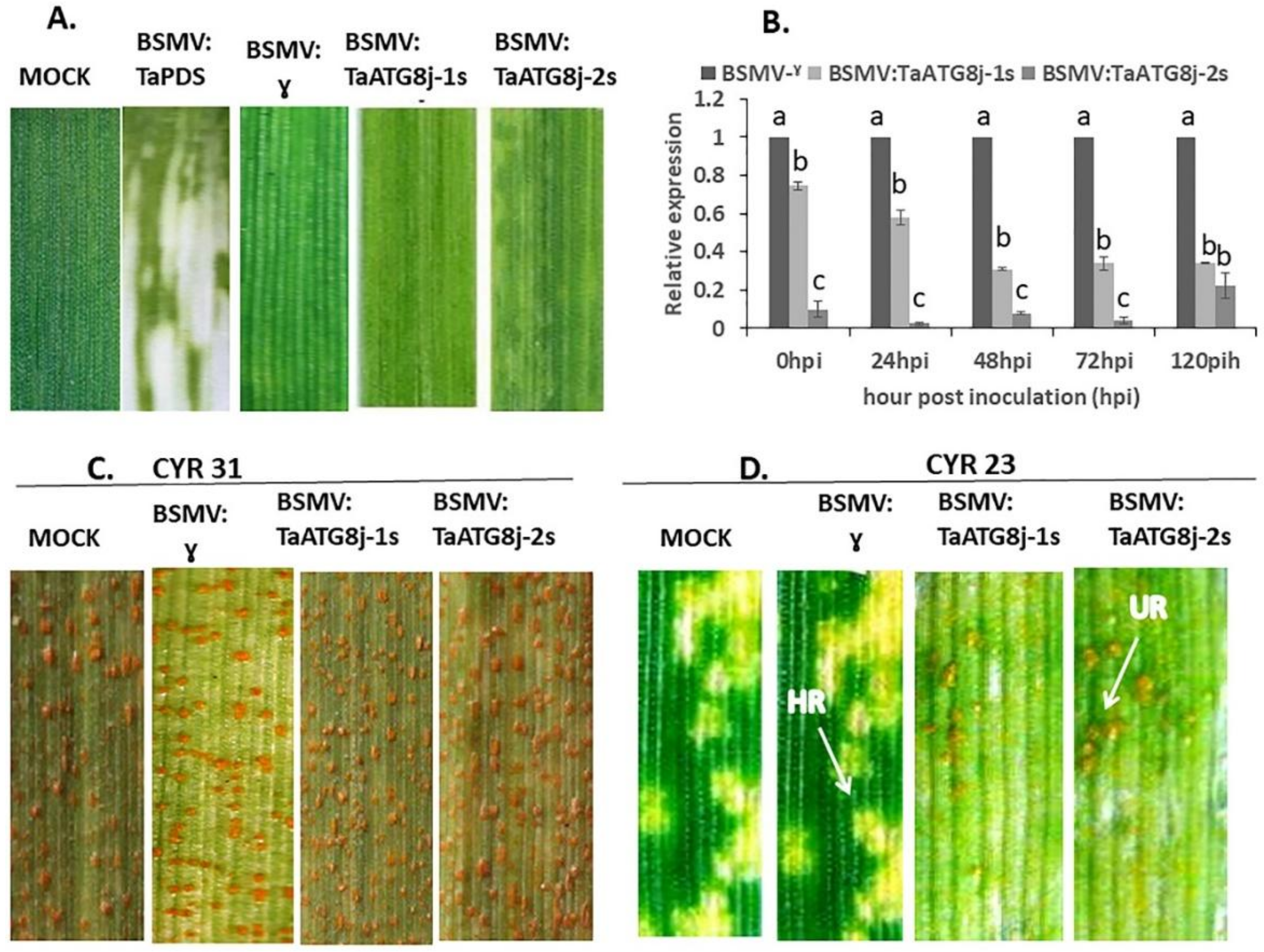
2.6. Knockdown of TaATG8j Enhances Wheat Susceptibility to Pst
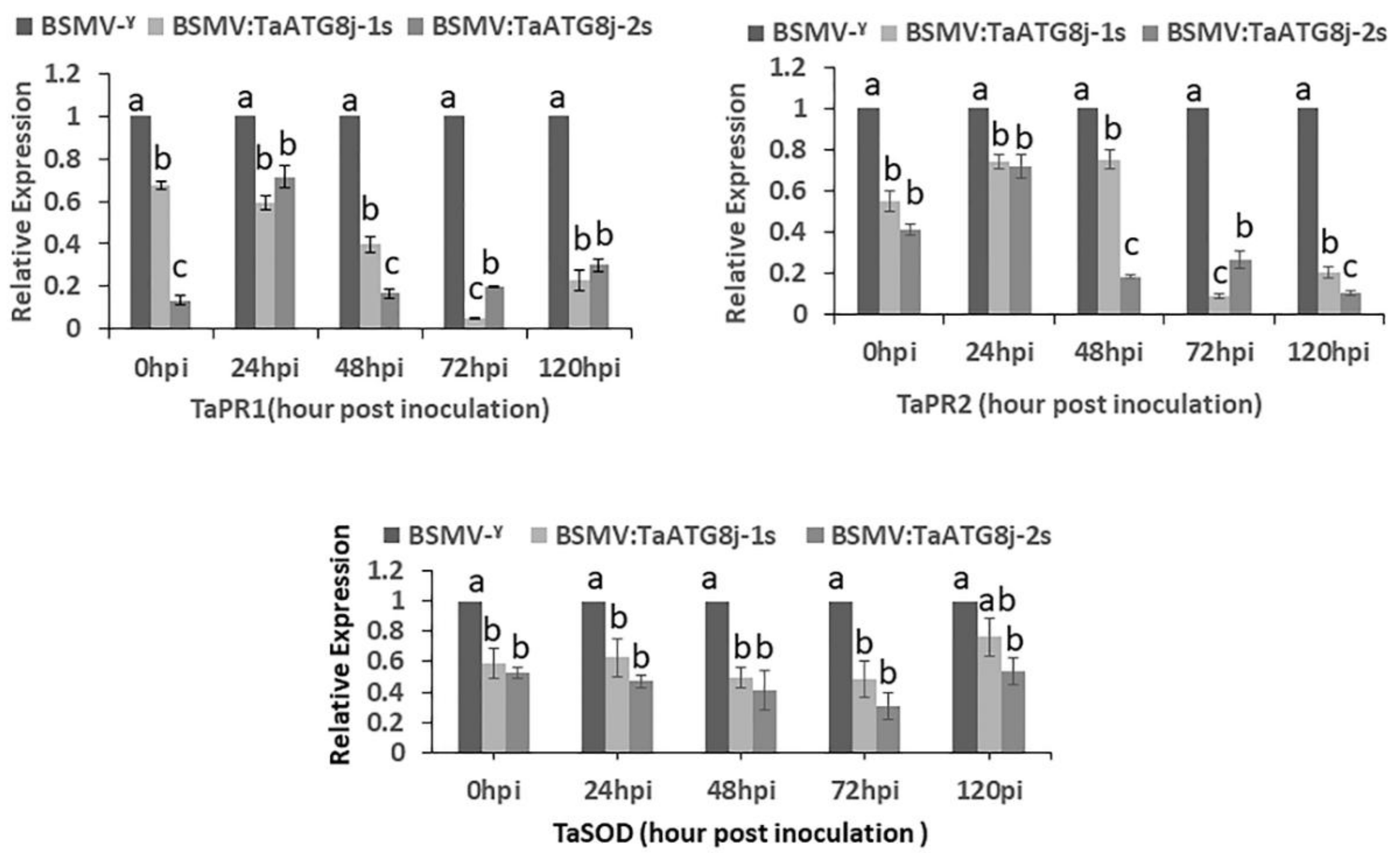
2.7. Suppression of Defense-Related Genes in TaATG8j-Knockdown Plants
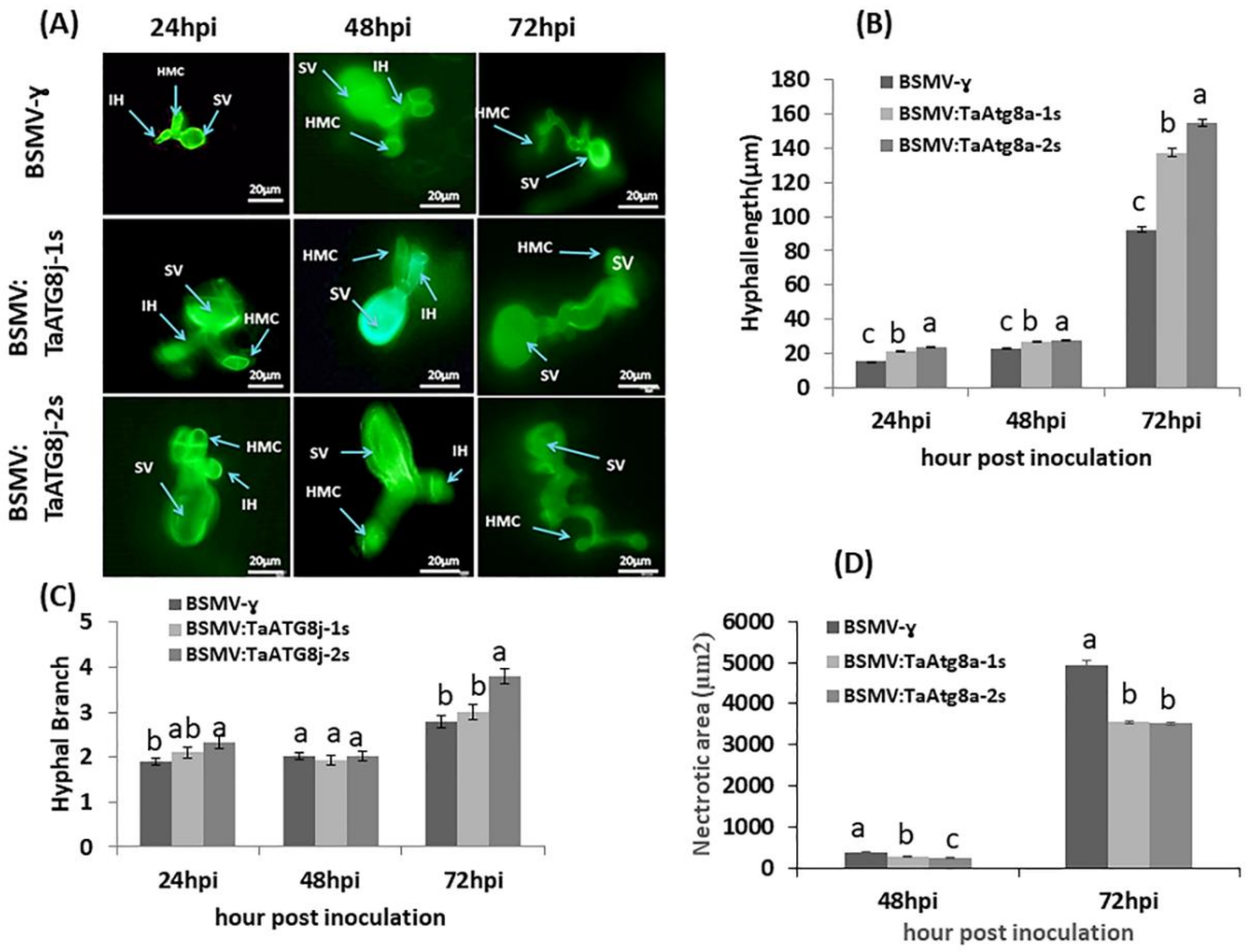
2.8. Histological Observation of Pst Growth and Host Necrotic Cell Death
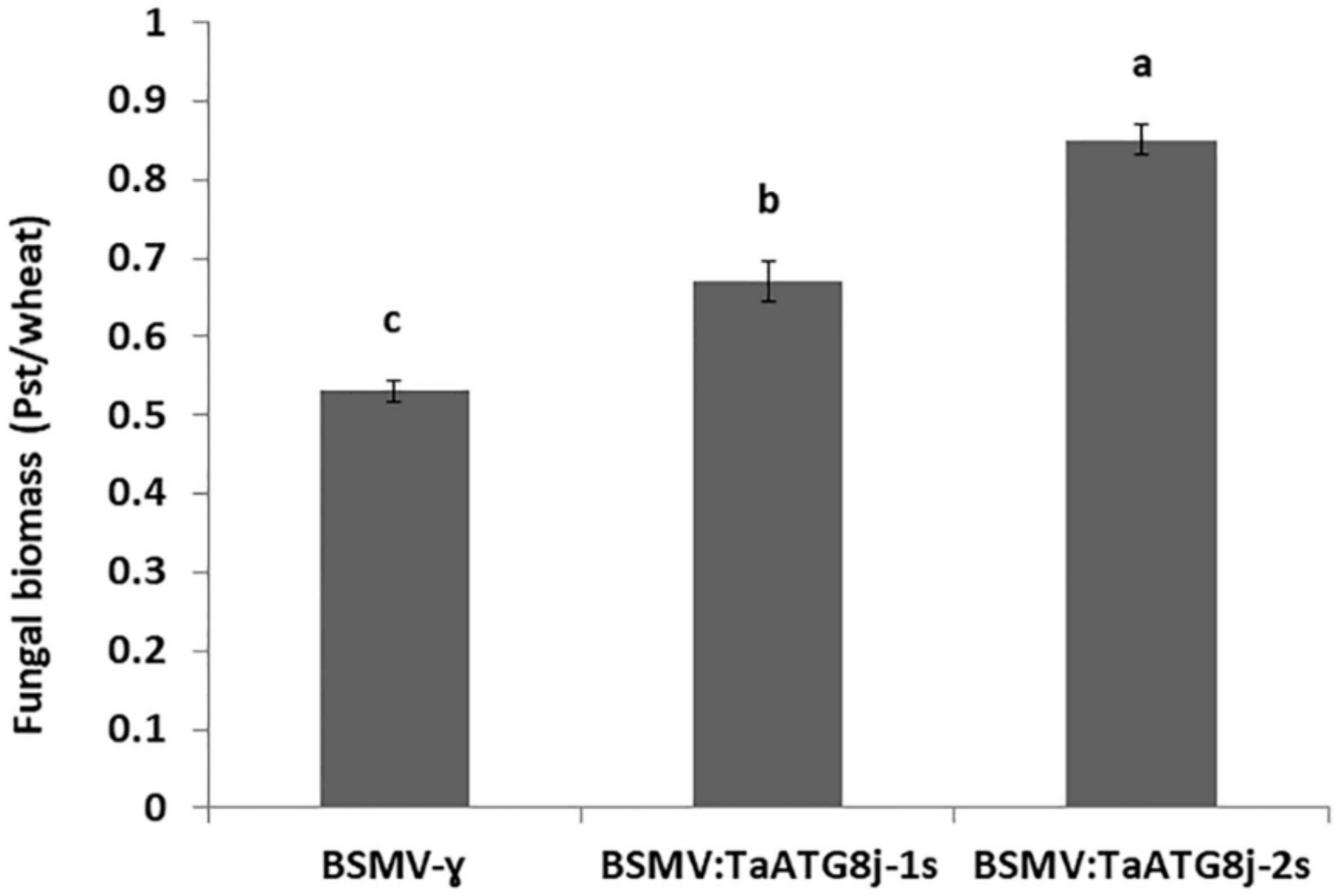
2.9. Increased Fungal Biomass in TaATG8j-Knockdown Plants
3. Discussion
4. Materials and Methods
4.1. Cloning and Sequence Analyses of TaATG8j
4.2. Plant and Fungal Materials
4.3. Extraction of RNA, cDNA Synthesis and qRT-PCR Analysis
4.4. Subcellular Localization and Immunoblotting of GFP-TaATG8j
4.5. Agrobacterium-Mediated Transient Expression of TaATG8j in N. benthamiana
4.6. Overexpression of TaATG8j in Yeast
4.7. BSMV-Mediated Silencing of TaATG8j in Wheat–Pst Interactions
4.8. Expression of TaATG8j and Pathogenesis-Related Protein (PR) Genes in TaATG8j-Knockdown Plants
4.9. Histological Study of Fungal Growth in TaATG8j-Knockdown Plants
4.10. Fungal Biomass Assay in TaATG8j-Knockdown Plants
4.11. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| ATG | autophagy-related gene |
| Cq | quantification cycle |
| DPI | days post-inoculation |
| hpi | hours post-inoculation |
| EST | expressed sequence tag |
| GFP | green fluorescent protein |
| HR | hypersensitive response |
| PCD | programmed cell death |
| PEG | polyethylene glycol |
| qRT-PCR | quantitative real-time PCR |
| Pst | Puccinia striiformis f. sp. tritici |
| VIGS | virus-induced gene silencing |
References
- Sharma, P.; Dubey, R.S. Drought induces oxidative stress and enhances the activities of antioxidant enzymes in growing rice seedlings. Plant Growth Regul. 2005, 46, 209–221. [Google Scholar] [CrossRef]
- Xie, Y.; Kang, R.; Sun, X.; Zhong, M.; Huang, J.; Klionsky, D.J.; Tang, D. Posttranslational modification of autophagy-related proteins in macroautophagy. Autophagy 2015, 11, 28–45. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Vierstra, R.D. Autophagy: A multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci. 2012, 17, 526–537. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Bassham, D.C. Autophagy: Pathways for Self-Eating in Plant Cells. Annu. Rev. Plant Biol. 2012, 63, 215–237. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J. Vacuolar import of proteins and organelles from the cytoplasm. Annu. Rev. Cell Dev. Biol. 1999, 15, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Hofius, D.; Schultz, L.T.; Joensen, J.; Tsitsigiannis, D.I.; Petersen, N.H.T.; Mattsson, O.; Jørgensen, L.B.; Jones, J.D.G.; Mundy, J.; Petersen, M. Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell 2009, 137, 773–783. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Schiff, M.; Czymmek, K.; Tallóczy, Z.; Levine, B.; Dinesh-Kumar, S.P. Autophagy regulates programmed cell death during the plant innate immune response. Cell 2005, 121, 567–577. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; Dinesh-Kumar, S.P. Arabidopsis ATG6 is required to limit the pathogen- associated cell death response. Autophagy 2008, 4, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Lenz, H.D.; Haller, E.; Melzer, E.; Kober, K.; Wurster, K.; Stahl, M.; Bassham, D.C.; Vierstra, R.D.; Parker, J.E.; Bautor, J.; et al. Autophagy differentially controls plant basal immunity to biotrophic and necrotrophic pathogens. Plant J. 2011, 66, 818–830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minina, E.A.; Bozhkov, P.V.; Hofius, D. Autophagy as initiator or executioner of cell death. Trends Plant Sci. 2014, 19, 692–697. [Google Scholar] [CrossRef] [PubMed]
- Yue, J.; Sun, H.; Zhang, W.; Pei, D.; He, Y.; Wang, H. Wheat heterologs of yeast ATG6 function in autophagy and are implicated in powdery mildew immunity. BMC Plant Biol. 2015, 15, 95. [Google Scholar] [CrossRef] [PubMed]
- Hafren, A.; Macia, J.L.; Love, A.J.; Milner, J.J.; Drucker, M.; Hofius, D. Selective autophagy limits cauliflower mosaic virus infection by NBR1-mediated targeting of viral capsid protein and particles. Proc. Natl. Acad. Sci. USA 2017, 114, 2026–2035. [Google Scholar] [CrossRef] [PubMed]
- Bassham, D.C. Function and regulation of macroautophagy in plants. Biochim. Biophys. Acta Mol. Cell Res. 2009, 1793, 1397–1403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitou, G.; Budak, H.; Gozuacik, D. Techniques to study autophagy in plants. Int. J. Plant Genom. 2009, 2009, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Reggiori, F.; Klionsky, D.J. Autophagic processes in yeast: Mechanism, machinery and regulation. Genetics 2013, 194, 341–361. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Klionsky, D.J. Autophagosome formation: Core machinery and adaptations. Nat. Cell Biol. 2007, 9, 1102–1109. [Google Scholar] [CrossRef] [PubMed]
- Nakatogawa, H.; Ichimura, Y.; Ohsumi, Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 2007, 130, 165–178. [Google Scholar] [CrossRef] [PubMed]
- Kirisako, T.; Ichimura, Y.; Okada, H.; Kabeya, Y.; Mizushima, N.; Yoshimori, T.; Ohsumi, M.; Takao, T.; Noda, T.; Ohsumi, Y. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J. Cell Biol. 2000, 151, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Nair, U.; Yen, W.L.; Mari, M.; Cao, Y.; Xie, Z.; Baba, M.; Reggiori, F.; Klionsky, D.J. A role for Atg8-PE deconjugation in autophagosome biogenesis. Autophagy 2012, 8, 780–793. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J.; Cregg, J.M.; Dunn, W.A.; Emr, S.D.; Sakai, Y.; Sandoval, N.V.; Sandoval, I.V. A unified nomenclature for yeast autophagy-related genes. Dev. Cell 2003, 5, 539–545. [Google Scholar] [CrossRef]
- Hayward, A.P.; Tsao, J.; Dinesh-Kumar, S.P. Autophagy and plant innate immunity: Defense through degradation. Semin. Cell Dev. Biol. 2009, 20, 1041–1047. [Google Scholar] [CrossRef] [PubMed]
- Seay, M.; Hayward, A.P.; Tsao, J.; Dinesh-Kumar, S.P. Something old, something new: Plant innate immunity and autophagy. Curr. Top. Microbiol. Immunol. 2009, 335, 287–306. [Google Scholar] [PubMed]
- Yoshimoto, K.; Hideki, H.; Shusei, S.; Tomohiko, K.; Tabata, S.; Noda, T.; Ohsumi, Y. Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 2004, 16, 2967–2983. [Google Scholar] [CrossRef] [PubMed]
- Dagdas, Y.F.; Pooja, P.; Nattapong, S.; Yasin, T.; Khaoula, B.; Cian, D.; Maria, E.S.; Sophien, K.; Tolga, O.B. Host autophagosomes are diverted to a plant-pathogen interface. bioRxiv 2017. [Google Scholar] [CrossRef]
- Duan, Y.; Guo, J.; Shi, X.; Guan, X.; Liu, F.; Bai, P.; Huang, L.; Kang, Z. Wheat hypersensitive induced reaction genes TaHIR1 and TaHIR3 are involved in response to stripe rust fungus infection and abiotic stresses. Plant Cell Rep. 2013, 32, 273–283. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, X.; Duan, Y.; Yin, S.; Zhang, H.; Huang, L.; Kang, Z. TaAbc1, a member of Abc1-like family involved in hypersensitive response against the stripe rust fungal pathogen in wheat. PLoS ONE 2013, 8, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Chandra, C.R.; Kishor, S.N.; Mithilesh, K.; Rajeev, K. Wheat Genotypes (Triticum aestivum L.) vary widely in their responses of Fertility traits to high Temperature at Anthesis. Int. Res. J. Biol. Sci. 2014, 3, 54–60. [Google Scholar]
- Langridge, P. Genomics: Decoding our daily bread. Nature 2012, 491, 678–680. [Google Scholar] [CrossRef] [PubMed]
- Pei, D.; Zhang, W.; Sun, H.; Wei, X.; Yue, J.; Wang, H. Identification of autophagy-related genes ATG4 and ATG8 from wheat (Triticum aestivum L.) and profiling of their expression patterns responding to biotic and abiotic stresses. Plant Cell Rep. 2014, 33, 1697–1710. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Wei, J.; Han, Q.; Liu, R.; Duan, X.; Fu, Y.; Huang, X.; Wang, X.; Kang, Z. PsANT, the adenine nucleotide translocase of Puccinia striiformis, promotes cell death and fungal growth. Sci. Rep. 2015, 5, 11241. [Google Scholar] [CrossRef] [PubMed]
- Delaney, T.P. Genetic dissection of acquired resistance to disease. Plant Physiol. 1997, 113, 5–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Loon, L.C.; Van Kammen, A. Polyacrylamide disc electrophoresis of the soluble leaf proteins from Nicotiana tabacum var ‘Samsun’ and Samsun NN. II. Changes in protein constitution after infection with tobacco mosaic virus. Virology 1970, 40, 199–211. [Google Scholar]
- Baehrecke, E.H. Autophagy: Dual roles in life and death? Nat. Rev. Mol. Cell Biol. 2005, 6, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Kroemer, G.; Levine, B. Autophagic cell death: The story of a misnomer. Nat. Rev. Mol. Cell Biol. 2008, 9, 1004–1010. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Vicencio, J.M.; Kepp, O.; Tasdemir, E.; Maiuri, M.C.; Kroemer, G. To die or not to die: That is the autophagic question. Curr. Mol. Med. 2008, 8, 78–91. [Google Scholar] [PubMed]
- Kourtis, N.; Tavernarakis, N. Autophagy and cell death in model organisms. Cell Death Differ. 2009, 16, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Üstün, S.; Hafrén, A.; Hofius, D. Autophagy as a mediator of life and death in plants. Curr. Opin. Plant Biol. 2017, 40, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Dangl, J.L.; Horvath, D.M.; Staskawicz, B.J. Pivoting the plant immune system from dissection to deployment. Science 2013, 341, 746–751. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Ohsumi, Y. Current knowledge of the pre-autophagosomal structure (PAS). FEBS Lett. 2010, 584, 1280–1286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, J.; Huang, X.; Wang, X.; Chen, X.; Qu, Z.; Huang, L.; Kang, Z. Identification of expressed genes during compatible interaction between stripe rust (Puccinia striiformis) and wheat using a cDNA library. BMC Genom. 2009, 10, 586. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Myo, T.; Ma, W.; Lan, D.; Qi, T.; Guo, J.; Song, P.; Guo, J.; Kang, Z. TaTypA, a ribosome-binding GTPase protein, positively regulates wheat resistance to the stripe rust fungus. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Kang, Z.; Li, Z. Discovery of a normal T. type new pathogenic strain to Lovrin10. Acta Cllegii Septentr. Occident. Agric. 1984, 4, 18–28. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Kucsera, J.; Yarita, K.; Takeo, K. Simple detection method for distinguishing dead and living yeast colonies. J. Micro Biol. Methods 2000, 41, 19–21. [Google Scholar] [CrossRef]
- Scofield, S.R.; Huang, L.; Brandt, A.S.; Gill, B.S. Development of a virus-induced gene-silencing system for hexaploid wheat and its use in functional analysis of the Lr21-mediated leaf rust resistance pathway 1. Plant Physiol. 2005, 138, 2165–2173. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Duan, X.; Tang, C.; Li, X.; Voegele, R.T.; Wang, X.; Wei, G.; Kang, Z. TaADF7, an actin depolymerizing factor, contributes to wheat resistance against Puccinia striiformis f. sp. tritici. Plant J. 2014, 78, 16–30. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.F.; Huang, L.; Buchenauer, H.; Han, Q.M.; Zhang, H.C.; Kang, Z.S. Histochemical studies on the accumulation of reactive oxygen species (O−2 and H2O2) in the incompatible and compatible interaction of wheat: Puccinia striiformis f. sp. tritici. Physiol. Mol. Plant Pathol. 2007, 71, 230–239. [Google Scholar] [CrossRef]
- Zheng, W.; Huang, L.; Huang, J.; Wang, X.; Chen, X.; Zhao, J. High genome Heterozygosity and endemic genetic recombination in the wheat stripe rust fungus. Nat. Commun. 2013, 4, 1–10. [Google Scholar] [CrossRef] [PubMed]



| Gene | Chromosomal | cDNA | ORF | Protein | DNA | Identity (%) a |
|---|---|---|---|---|---|---|
| Location | Size (bp) | Size (bp) | Size (aa) | Size (bp) | Identity | |
| TaATG8j-2AS | 2AS | 731 | 360 | 119 | 2438 | 99.3 |
| TaATG8j-2BS | 2BS | 784 | 360 | 119 | 2746 | 98.4 |
| TaATG8j-2DS | 2DS | 813 | 360 | 119 | 2810 | 98.6 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mamun, M.A.-A.; Tang, C.; Sun, Y.; Islam, M.N.; Liu, P.; Wang, X.; Kang, Z. Wheat Gene TaATG8j Contributes to Stripe Rust Resistance. Int. J. Mol. Sci. 2018, 19, 1666. https://doi.org/10.3390/ijms19061666
Mamun MA-A, Tang C, Sun Y, Islam MN, Liu P, Wang X, Kang Z. Wheat Gene TaATG8j Contributes to Stripe Rust Resistance. International Journal of Molecular Sciences. 2018; 19(6):1666. https://doi.org/10.3390/ijms19061666
Chicago/Turabian StyleMamun, Md. Abdullah-Al, Chunlei Tang, Yingchao Sun, Md. Nazrul Islam, Peng Liu, Xiaojie Wang, and Zhensheng Kang. 2018. "Wheat Gene TaATG8j Contributes to Stripe Rust Resistance" International Journal of Molecular Sciences 19, no. 6: 1666. https://doi.org/10.3390/ijms19061666
APA StyleMamun, M. A.-A., Tang, C., Sun, Y., Islam, M. N., Liu, P., Wang, X., & Kang, Z. (2018). Wheat Gene TaATG8j Contributes to Stripe Rust Resistance. International Journal of Molecular Sciences, 19(6), 1666. https://doi.org/10.3390/ijms19061666





