Evolutionary History of the Glycoside Hydrolase 3 (GH3) Family Based on the Sequenced Genomes of 48 Plants and Identification of Jasmonic Acid-Related GH3 Proteins in Solanum tuberosum
Abstract
:1. Introduction
2. Results and Discussion
2.1. The GH3 Family Identified in 48 Plant Species
2.2. Analysis of the Phylogenetic Tree and Duplication Patterns
2.3. Structural Analysis of GH3 Proteins
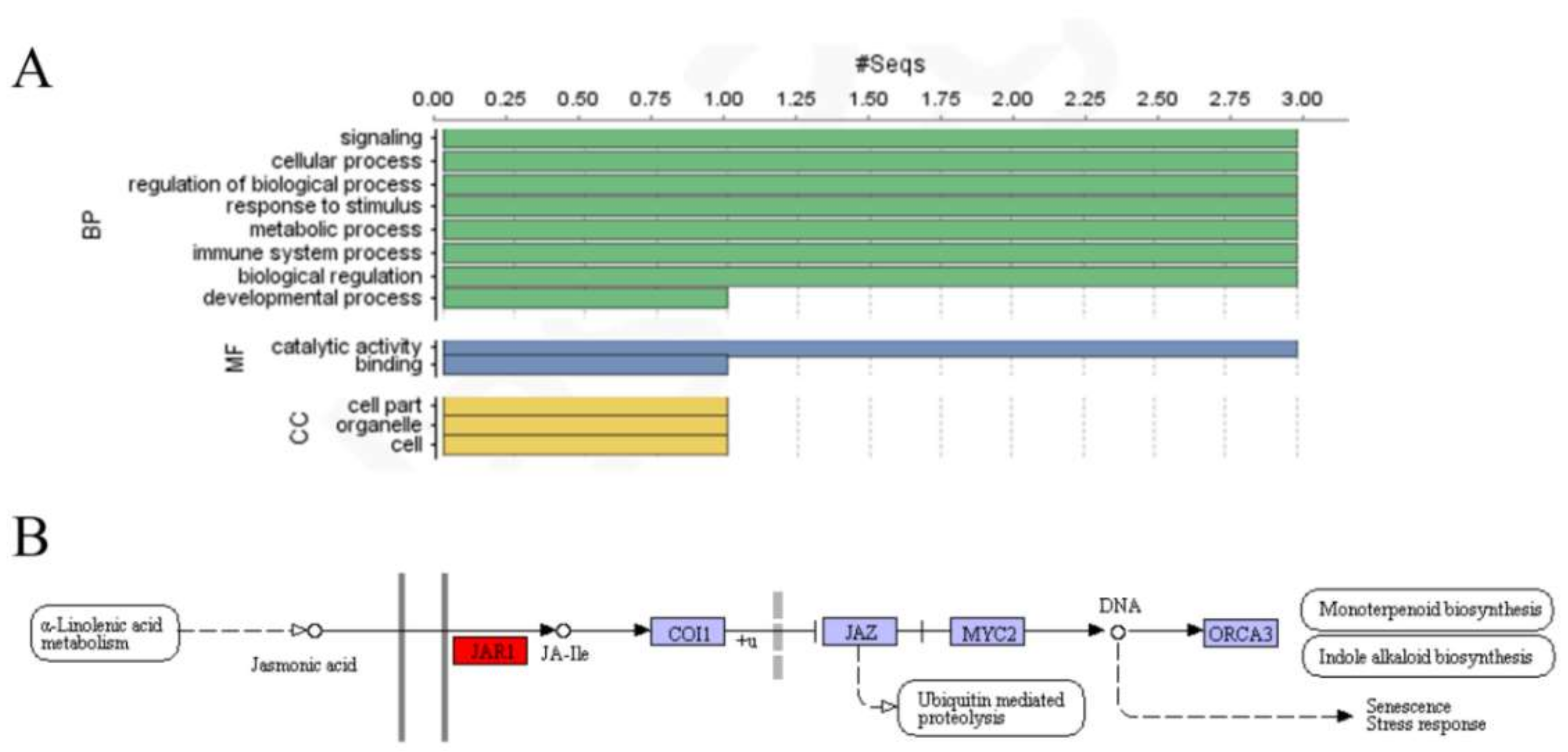
2.4. Gene Ontology Annotations and RNA-Sequence Data Analysis of StGH3 Proteins
2.5. Expression Analysis of StGH3 Group I Genes
3. Materials and Methods
3.1. Mining GH3 Genes from Various Species
3.2. Structure Analysis of GH3 Proteins
3.3. Construction of the Phylogenetic Tree
3.4. Analysis of Gene Duplications
3.5. GO Annotation and RNA-Seq Data Analysis
3.6. Plant Growth Conditions and Treatments
3.7. Expression Analysis of the StGH3 Genes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Hagen, G.; Guilfoyle, T.J. Rapid induction of selective transcription by auxins. Mol. Cell. Biol. 1985, 5, 1197–1203. [Google Scholar] [CrossRef] [PubMed]
- Roux, C.; Perrot-Rechenmann, C. Isolation by differential display and characterization of a tobacco auxin-responsive cDNA Nt-gh3, related to GH3. FEBS Lett. 1997, 419, 131–136. [Google Scholar] [CrossRef]
- Tanaka, S.; Mochizuki, N.; Nagatani, A. Expression of the AtGH3a gene, an Arabidopsis homologue of the soybean GH3 gene, is regulated by phytochrome B. Plant Cell Physiol. 2002, 43, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Kaur, N.; Tyagi, A.K.; Khurana, J.P. The auxin-responsive GH3 gene family in rice (Oryza sativa). Funct. Integr. Genom. 2006, 6, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Zhao, K.; Lei, H.; Shen, X.; Liu, Y.; Liao, X.; Li, T. Genome-wide analysis of the GH3 family in apple (Malus × domestica). BMC Genom. 2013, 14. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yue, R.; Sun, T.; Zhang, L.; Chen, W.; Zeng, H.; Wang, H.; Shen, C. Genome-wide identification, expression analysis of GH3 family genes in Medicago truncatula under stress-related hormones and Sinorhizobium meliloti infection. Appl. Microbial. Biotechnol. 2015, 99, 841–854. [Google Scholar] [CrossRef] [PubMed]
- Liao, D.; Chen, X.; Chen, A.; Wang, H.; Liu, J.; Liu, J.; Gu, M.; Sun, S.; Xu, G. The characterization of six auxin-induced tomato GH3 genes uncovers a member, SlGH3.4, strongly responsive to arbuscular mycorrhizal symbiosis. Plant Cell Physiol. 2015, 56, 674–687. [Google Scholar] [CrossRef] [PubMed]
- Westfall, C.S.; Zubieta, C.; Herrmann, J.; Kapp, U.; Nanao, M.H.; Jez, J.M. Structural basis for prereceptor modulation of plant hormones by GH3 proteins. Science 2012, 336, 1708–1711. [Google Scholar] [CrossRef] [PubMed]
- Bierfreund, N.M.; Tintelnot, S.; Reski, R.; Decker, E.L. Loss of GH3 function does not affect phytochrome-mediated development in a moss, Physcomitrella patens. J. Plant Physiol. 2004, 161, 823–835. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.L.; Kohchi, T.; Yamato, K.T.; Jenkins, J.; Shu, S.; Ishizaki, K.; Yamaoka, S.; Nishihama, R.; Nakamura, Y.; Berger, F. Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 2017, 171, 287–304. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, T.; Fujita, T.; Shin, T.; Seki, M.; Nishide, H.; Uchiyama, I.; Kamiya, A.; Carninci, P.; Hayashizaki, Y.; Shinozaki, K.; et al. Comparative genomics of Physcomitrella patens gametophytic transcriptome and Arabidopsis thaliana: Implication for land plant evolution. Proc. Natl. Acad. Sci. USA 2003, 100, 8007–8012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mellor, N.; Band, L.R.; Pencik, A.; Novak, O.; Rashed, A.; Holman, T.; Wilson, M.H.; Voss, U.; Bishopp, A.; King, J.R.; et al. Dynamic regulation of auxin oxidase and conjugating enzymes AtDAO1 and GH3 modulates auxin homeostasis. Proc. Natl. Acad. Sci. USA 2016, 113, 11022–11027. [Google Scholar] [CrossRef] [PubMed]
- Staswick, P.E.; Tiryaki, I.; Rowe, M.L. Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 2002, 14, 1405–1415. [Google Scholar] [CrossRef] [PubMed]
- Okrent, R.A.; Wildermuth, M.C. Evolutionary history of the GH3 family of acyl adenylases in rosids. Plant Mol. Biol. 2011, 76, 489–505. [Google Scholar] [CrossRef] [PubMed]
- Jagadeeswaran, G.; Raina, S.; Acharya, B.R.; Maqbool, S.B.; Mosher, S.L.; Appel, H.M.; Schultz, J.C.; Klessig, D.F.; Raina, R. Arabidopsis GH3-LIKE DEFENSE GENE 1 is required for accumulation of salicylic acid, activation of defense responses and resistance to Pseudomonas syringae. Plant J. 2007, 51, 234–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Westfall, C.S.; Herrmann, J.; Chen, Q.; Wang, S.; Jez, J.M. Modulating plant hormones by enzyme action: The GH3 family of acyl acid amido synthetases. Plant Signal Behav. 2010, 5, 1607–1612. [Google Scholar] [CrossRef] [PubMed]
- Koo, A.J.; Gao, X.; Jones, A.D.; Howe, G.A. A rapid wound signal activates the systemic synthesis of bioactive jasmonates in Arabidopsis. Plant J. 2009, 59, 974–986. [Google Scholar] [CrossRef] [PubMed]
- Havko, N.E.; Major, I.T.; Jewell, J.B.; Attaran, E.; Browse, J.; Howe, G.A. Control of carbon assimilation and partitioning by jasmonate: An accounting of growth–defense tradeoffs. Plants 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Campos, M.L.; Yoshida, Y.; Major, I.T.; De, D.O.F.; Weraduwage, S.M.; Froehlich, J.E.; Johnson, B.F.; Kramer, D.M.; Jander, G.; Sharkey, T.D. Rewiring of jasmonate and phytochrome B signalling uncouples plant growth-defense tradeoffs. Nat. Commun. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Van Loon, L.C.; Bakker, P.A.; Pieterse, C.M. Systemic resistance induced by rhizosphere bacteria. Ann. Rev. Phytopathol. 1998, 36, 453–483. [Google Scholar] [CrossRef] [PubMed]
- Rao, M.V.; Hyungil, L.; Creelman, R.A.; Mullet, J.E.; Davis, K.R. Jasmonic acid signaling modulates ozone-induced hypersensitive cell death. Plant Cell 2000, 12, 1633–1646. [Google Scholar] [CrossRef] [PubMed]
- Heitz, T.; Widemann, E.; Lugan, R.; Miesch, L.; Ullmann, P.; Désaubry, L.; Holder, E.; Grausem, B.; Kandel, S.; Miesch, M. Cytochromes P450 CYP94C1 and CYP94B3 catalyze two successive oxidation steps of plant hormone Jasmonoyl-isoleucine for catabolic turnover. J. Biol. Chem. 2012, 287, 6296–6306. [Google Scholar] [CrossRef] [PubMed]
- Westfall, C.S.; Sherp, A.M.; Zubieta, C.; Alvarez, S.; Schraft, E.; Marcellin, R.; Ramirez, L.; Jez, J.M. Arabidopsis thaliana GH3.5 acyl acid amido synthetase mediates metabolic crosstalk in auxin and salicylic acid homeostasis. Proc. Natl. Acad. Sci. USA 2016, 113, 13917–13922. [Google Scholar] [CrossRef] [PubMed]
- Okrent, R.A.; Brooks, M.D.; Wildermuth, M.C. Arabidopsis GH3.12 (PBS3) conjugates amino acids to 4-substituted benzoates and is inhibited by salicylate. J. Biol. Chem. 2009, 284, 9742–9754. [Google Scholar] [CrossRef] [PubMed]
- Takase, T.; Nakazawa, M.; Ishikawa, A.; Kawashima, M.; Ichikawa, T.; Takahashi, N.; Shimada, H.; Manabe, K.; Matsui, M. ydk1-D, an auxin-responsive GH3 mutant that is involved in hypocotyl and root elongation. Plant J. 2004, 37, 471–483. [Google Scholar] [CrossRef] [PubMed]
- Nakazawa, M.; Yabe, N.; Ichikawa, T.; Yamamoto, Y.Y.; Yoshizumi, T.; Hasunuma, K.; Matsui, M. DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. Plant J. 2001, 25, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Stone, J.M. Arabidopsis thaliana GH3.9 influences primary root growth. Planta 2007, 226, 21–34. [Google Scholar] [CrossRef] [PubMed]
- Paponov, I.A.; Teale, W.; Lang, D.; Paponov, M.; Reski, R.; Rensing, S.A.; Palme, K. The evolution of nuclear auxin signalling. BMC Evol. Biol. 2009, 9. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Gray, W.M. SAUR proteins as effectors of hormonal and environmental signals in plant growth. Mol. Plant 2015, 8, 1153–1164. [Google Scholar] [CrossRef] [PubMed]
- Withers, J.; Yao, J.; Mecey, C.; Howe, G.A.; Melotto, M.; He, S.Y. Transcription factor-dependent nuclear localization of a transcriptional repressor in jasmonate hormone signaling. Proc. Natl. Acad. Sci. USA 2012, 109, 20148–20153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terol, J.; Domingo, C.; Talon, M. The GH3 family in plants: Genome wide analysis in rice and evolutionary history based on EST analysis. Gene 2006, 371, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Nystedt, B.; Street, N.R.; Wetterbom, A.; Zuccolo, A.; Lin, Y.C.; Scofield, D.G.; Vezzi, F.; Delhomme, N.; Giacomello, S.; Alexeyenko, A. The Norway spruce genome sequence and conifer genome evolution. Nature 2013, 497, 579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stumpe, M.; Göbel, C.; Faltin, B.; Beike, A.K.; Hause, B.; Himmelsbach, K.; Bode, J.; Kramell, R.; Wasternack, C.; Frank, W. The moss Physcomitrella patens contains cyclopentenones but no jasmonates: Mutations in allene oxide cyclase lead to reduced fertility and altered sporophyte morphology. New Phytol. 2010, 188, 740–749. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Ohshika, J.; Takahashi, T.; Ishizaki, K.; Kohchi, T.; Matusuura, H.; Takahashi, K. Functional analysis of allene oxide cyclase, MpAOC, in the liverwort Marchantia polymorpha. Phytochemistry 2015, 116, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Böttcher, C.; Boss, P.K.; Davies, C. Acyl substrate preferences of an IAA-amido synthetase account for variations in grape (Vitis vinifera L.) berry ripening caused by different auxinic compounds indicating the importance of auxin conjugation in plant development. J. Exp. Bot. 2011, 62, 4267–4280. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Yue, R.; Tao, S.; Yang, Y.; Zhang, L.; Xu, M.; Wang, H.; Shen, C. Genome-wide identification, expression analysis of auxin-responsive GH3 family genes in maize (Zea mays L.) under abiotic stresses. J. Integr. Plant Biol. 2015, 57, 783–795. [Google Scholar] [CrossRef] [PubMed]
- Große-Veldmann, B.; Korotkova, T.; Reinken, J.A.; Lobin, H.; Barthlott, W. Amborella trichopoda—Cultivation of the most ancestral angiosperm in Botanic Gardens. Veldmann 2011, 19, 143–155. [Google Scholar]
- Kuang, J.F.; Zhang, Y.; Chen, J.Y.; Chen, Q.J.; Jiang, Y.M.; Lin, H.T.; Xu, S.J.; Lu, W.J. Two GH3 genes from longan are differentially regulated during fruit growth and development. Gene 2011, 485, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.K.; Jain, M.; Garg, R. Genome-wide analysis and expression profiling suggest diverse roles of GH3 genes during development and abiotic stress responses in legumes. Front. Plant Sci. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wang, D.; Yang, C.; Kong, N.; Shi, Z.; Zhao, P.; Nan, Y.; Nie, T.; Wang, R.; Ma, H.; et al. Genome-wide identification of the potato WRKY transcription factor family. PLoS ONE 2017, 12. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Zhang, C.; Gu, M.; Bai, Z.; Zhang, W.; Qi, T.; Cheng, Z.; Peng, W.; Luo, H.; Nan, F. The Arabidopsis CORONATINE INSENSITIVE1 Protein Is a Jasmonate Receptor. Plant Cell 2009, 21, 2220–2236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, L.; Liu, F.; Lechner, E.; Genschik, P.; Crosby, W.L.; Ma, H.; Peng, W.; Huang, D.; Xie, D. The SCF( COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 2002, 14, 1919–1935. [Google Scholar] [CrossRef] [PubMed]
- Goossens, J.; Swinnen, G.; Vanden Bossche, R.; Pauwels, L.; Goossens, A. Change of a conserved amino acid in the MYC2 and MYC3 transcription factors leads to release of JAZ repression and increased activity. New Phytol. 2015, 206, 1229–1237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van der Fits, L.; Memelink, J. ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science 2000, 289, 295–297. [Google Scholar]
- Massa, A.N.; Childs, K.L.; Buell, C.R. Abiotic and biotic stress responses in Solanum tuberosum group Phureja DM1-3 516 R44 as measured through whole transcriptome sequencing. Plant Genome 2013. [Google Scholar] [CrossRef]
- Takahashi, K.; Fujino, K.; Kikuta, Y.; Koda, Y. Expansion of potato cells in response to jasmonic acid. Plant Sci. 1994, 100, 3–8. [Google Scholar] [CrossRef]
- Koda, Y.; Kikuta, Y.; Tazaki, H.; Tsujino, Y.; Sakamura, S.; Yoshihara, T. Potato tuber-inducing activities of jasmonic acid and related compounds. Phytochemistry 1991, 30, 1435–1438. [Google Scholar] [CrossRef]
- Hannapel, D.J. A model system of development regulated by the long-distance transport of mRNA. J. Integr. Plant Biol. 2010, 52, 40–52. [Google Scholar] [CrossRef] [PubMed]
- Ulloa, R.M.; Raíces, M.; Macintosh, G.C.; Maldonado, S.; Télleziñón, M.T. Jasmonic acid affects plant morphology and calcium-dependent protein kinase expression and activity in Solanum tuberosum. Physiol. Plant 2002, 115. [Google Scholar] [CrossRef]
- Sarkar, D.; Pandey, S.K.; Sharma, S. Cytokinins antagonize the jasmonates action on the regulation of potato (Solanum tuberosum) tuber formation in vitro. Plant Cell Tissue Organ. C. 2006, 87. [Google Scholar] [CrossRef]
- Kong, H.; Landherr, L.L.; Frohlich, M.W.; Leebens-Mack, J.; Ma, H.; DePamphilis, C.W. Patterns of gene duplication in the plant SKP1 gene family in angiosperms: Evidence for multiple mechanisms of rapid gene birth. Plant J. 2007, 50, 873–885. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrermata, A.; Sánchezdelbarrio, J.C.; Guiraorico, S.; Librado, P.; Ramosonsins, S.E.; Sánchezgracia, A. DnaSP 6: DNA sequence polymorphism analysis of large datasets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Pan, S.; Cheng, S.; Zhang, B.; Mu, D.; Ni, P.; Zhang, G.; Yang, S.; Li, R.; Wang, J.; et al. Genome sequence and analysis of the tuber crop potato. Nature 2011, 475, 189–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Wang, X.; Xu, Y.; Deng, X.; Xu, Q. Genome-wide analysis of the R2R3-MYB transcription factor gene family in sweet orange (Citrus sinensis). Mol. Biol. Rep. 2014, 41, 6769–6785. [Google Scholar] [CrossRef] [PubMed]
- Nicot, N.; Hausman, J.F.; Hoffmann, L.; Evers, D. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J. Exp. Bot. 2005, 56, 2907–2914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, B.; Zhang, G.; Murphy, A.; De, K.D.; Tai, H.; Bizimungu, B.; Si, H.; Li, X.Q. Differences between the bud end and stem end of potatoes in dry matter content, starch granule size, and carbohydrate metabolic gene expression at the growing and sprouting stages. J. Agric. Food Chem. 2016, 64, 1176–1185. [Google Scholar] [CrossRef] [PubMed]





| Lineage | Organism | Genome Total Length (Mb) 1 | Numbers of GH3 Protein | Numbers of Group I 2 | Numbers of Group II 3 | Numbers of Group III | Tandem Duplication (Pairs) 4 | Segmental Duplication (Pairs) 5 | References | |
|---|---|---|---|---|---|---|---|---|---|---|
| Algae | Chlamydomonas reinhardtii | 120.405 | 0 | 0 | 0 | 0 | 0 | 0 | This study | |
| Volvox carteri | 137.684 | 0 | 0 | 0 | 0 | 0 | 0 | This study | ||
| Mosses | ||||||||||
| Marchantia polymorpha L. | 205.718 | 2 | 1 | 1 | 0 | 0 | 0 | Bowman et al. [10] | ||
| Physcomitrella patens | 477.948 | 2 | 2 | 0 | 0 | 0 | 0 | Nicole et al. [9] | ||
| Ferns | ||||||||||
| Selaginella moellendorffii | 212.502 | 18 | 9 | 9 | 0 | 2 | 1 | This study | ||
| Gymnosperms | ||||||||||
| Picea abies | 28354 | 7 | 1 | 6 | 0 | 0 | 0 | This study | ||
| Angiosperms | ||||||||||
| Amborellaceae | Amborella trichopoda # | 706.495 | 6 | 2 | 4 | 0 | 0 | 0 | This study | |
| Eudicots | ||||||||||
| Chenopodiaceae | Beta vulgaris | 482.053 | 5 | 1 | 4 | 0 | 0 | 1 | This study | |
| Brassicaceae | Arabidopsis lyrata | 204.898 | 17 | 2 | 8 | 7 | 10 | 0 | This study | |
| Arabidopsis thaliana | 116.846 | 19 | 2 | 8 | 9 | 11 | 2 | Jain et al. [4] | ||
| Brassica oleracea | 501.692 | 35 | 4 | 11 | 20 | 10 | 13 | This study | ||
| Brassica rapa | 284.129 | 38 | 5 | 11 | 22 | 8 | 13 | This study | ||
| Capsella rubella | 133.064 | 20 | 2 | 8 | 10 | 12 | 3 | This study | ||
| Solanaceae | Capsicum annuum | 2935.88 | 11 | 4 | 7 | 0 | 0 | 1 | This study | |
| Solanum lycopersicum | 760.067 | 17 | 11 | 6 | 0 | 6 | 4 | This study | ||
| Solanum tuberosum | 705.934 | 13 | 4 | 9 | 0 | 3 | 4 | This study | ||
| Leguminosae sp. | Cajanus cajan | 620.626 | 12 | 4 | 8 | 0 | 0 | 0 | This study | |
| Cicer arietinum | 520.885 | 7 | 2 | 5 | 0 | 0 | 1 | This study | ||
| Glycine max | 953.339 | 24 | 8 | 16 | 0 | 0 | 36 | This study | ||
| Lotus japonicus | 394.455 | 7 | 1 | 6 | 0 | 0 | 0 | This study | ||
| Medicago truncatula | 412.924 | 17 * | 6 | 7 | 0 | 7 | 2 | Yang et al. [6] | ||
| Phaseolus vulgaris | 535.413 | 12 | 4 | 8 | 0 | 0 | 7 | This study | ||
| Rosids | Fragaria vesca | 214.373 | 8 | 2 | 6 | 0 | 0 | 0 | This study | |
| Malus domestica | 1288.87 | 15 | 5 | 10 | 0 | 1 | 4 | Yuan et al. [5] | ||
| Prunus mume | 234.03 | 7 | 2 | 5 | 0 | 0 | 1 | This study | ||
| Prunus persica | 212.767 | 7 | 2 | 5 | 0 | 0 | 1 | This study | ||
| Pyrus bretschneideri | 508.551 | 15 | 2 | 13 | 0 | 0 | 4 | This study | ||
| Cucurbitaceae | Citrullus lanatus | 321.047 | 8 | 2 | 6 | 0 | 1 | 0 | This study | |
| Cucumis sativus | 323.986 | 10 | 3 | 7 | 0 | 1 | 0 | This study | ||
| Actinidiaceae | Actinidia chinensis | 604.217 | 15 | 4 | 11 | 0 | 0 | 3 | This study | |
| Caricaceae | Carica papaya | 370.419 | 6 | 2 | 4 | 0 | 0 | 0 | This study | |
| Euphorbiaceae | Ricinus communis | 350.622 | 7 | 2 | 5 | 0 | 0 | 0 | This study | |
| Lentibulariaceae | Utricularia gibba | 100.689 | 6 | 2 | 4 | 0 | 0 | 1 | This study | |
| Malvaceae | Theobroma cacao | 335.437 | 19 | 2 | 17 | 0 | 8 | 3 | This study | |
| Gossypium raimondii | 767.667 | 32 | 6 | 10 | 16 | 12 | 11 | This study | ||
| Eucalyptus grandis | 691.43 | 9 | 3 | 6 | 0 | 0 | 2 | This study | ||
| Nelumbonaceae | Nelumbo nucifera | 797.494 | 11 | 4 | 7 | 0 | 0 | 4 | This study | |
| Rutaceae | Citrus sinensis | 323.528 | 9 | 2 | 7 | 0 | 1 | 0 | This study | |
| Salicaceae | Populus trichocarpa | 417.287 | 13 | 4 | 9 | 0 | 0 | 8 | This study | |
| Vitaceae | Vitis vinifera | 486.197 | 9 | 3 | 6 | 0 | 0 | 1 | Bottcher et al. [35] | |
| Monocots | ||||||||||
| Poaceae | Brachypodium distachyon | 218.345 | 9 | 2 | 6 | 1 | 0 | 1 | This study | |
| Hordeum vulgare | 1825.17 | 5 | 1 | 4 | 0 | 0 | 0 | This study | ||
| Oryza sativa | 374.423 | 13 | 4 | 9 | 0 | 0 | 1 | Jain et al. [4] | ||
| Triticum urartu | 3747.05 | 9 | 3 | 6 | 0 | 0 | 0 | This study | ||
| Zea mays | 2145.45 | 13 | 5 | 8 | 0 | 1 | 3 | Feng et al. [36] | ||
| Arecaceae | Elaeis guineensis | 1017.1 | 16 | 5 | 11 | 0 | 0 | 2 | This study | |
| Musaceae | Musa acuminata | 472.231 | 18 | 8 | 10 | 0 | 1 | 18 | This study | |
| Orchidaceae | Phalaenopsis equestris | 1064.2 | 6 | 3 | 3 | 0 | 0 | 0 | This study | |
| Total | 579 | 159 | 339 | 85 | ||||||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, C.; Zhang, L.; Wang, D.; Ma, H.; Liu, B.; Shi, Z.; Ma, X.; Chen, Y.; Chen, Q. Evolutionary History of the Glycoside Hydrolase 3 (GH3) Family Based on the Sequenced Genomes of 48 Plants and Identification of Jasmonic Acid-Related GH3 Proteins in Solanum tuberosum. Int. J. Mol. Sci. 2018, 19, 1850. https://doi.org/10.3390/ijms19071850
Zhang C, Zhang L, Wang D, Ma H, Liu B, Shi Z, Ma X, Chen Y, Chen Q. Evolutionary History of the Glycoside Hydrolase 3 (GH3) Family Based on the Sequenced Genomes of 48 Plants and Identification of Jasmonic Acid-Related GH3 Proteins in Solanum tuberosum. International Journal of Molecular Sciences. 2018; 19(7):1850. https://doi.org/10.3390/ijms19071850
Chicago/Turabian StyleZhang, Chao, Leilei Zhang, Dongdong Wang, Haoli Ma, Bailin Liu, Zheng Shi, Xiaohui Ma, Yue Chen, and Qin Chen. 2018. "Evolutionary History of the Glycoside Hydrolase 3 (GH3) Family Based on the Sequenced Genomes of 48 Plants and Identification of Jasmonic Acid-Related GH3 Proteins in Solanum tuberosum" International Journal of Molecular Sciences 19, no. 7: 1850. https://doi.org/10.3390/ijms19071850
APA StyleZhang, C., Zhang, L., Wang, D., Ma, H., Liu, B., Shi, Z., Ma, X., Chen, Y., & Chen, Q. (2018). Evolutionary History of the Glycoside Hydrolase 3 (GH3) Family Based on the Sequenced Genomes of 48 Plants and Identification of Jasmonic Acid-Related GH3 Proteins in Solanum tuberosum. International Journal of Molecular Sciences, 19(7), 1850. https://doi.org/10.3390/ijms19071850





