Nucleolar GTP-Binding Protein 1-2 (NOG1-2) Interacts with Jasmonate-ZIMDomain Protein 9 (JAZ9) to Regulate Stomatal Aperture during Plant Immunity
Abstract
1. Introduction
2. Results
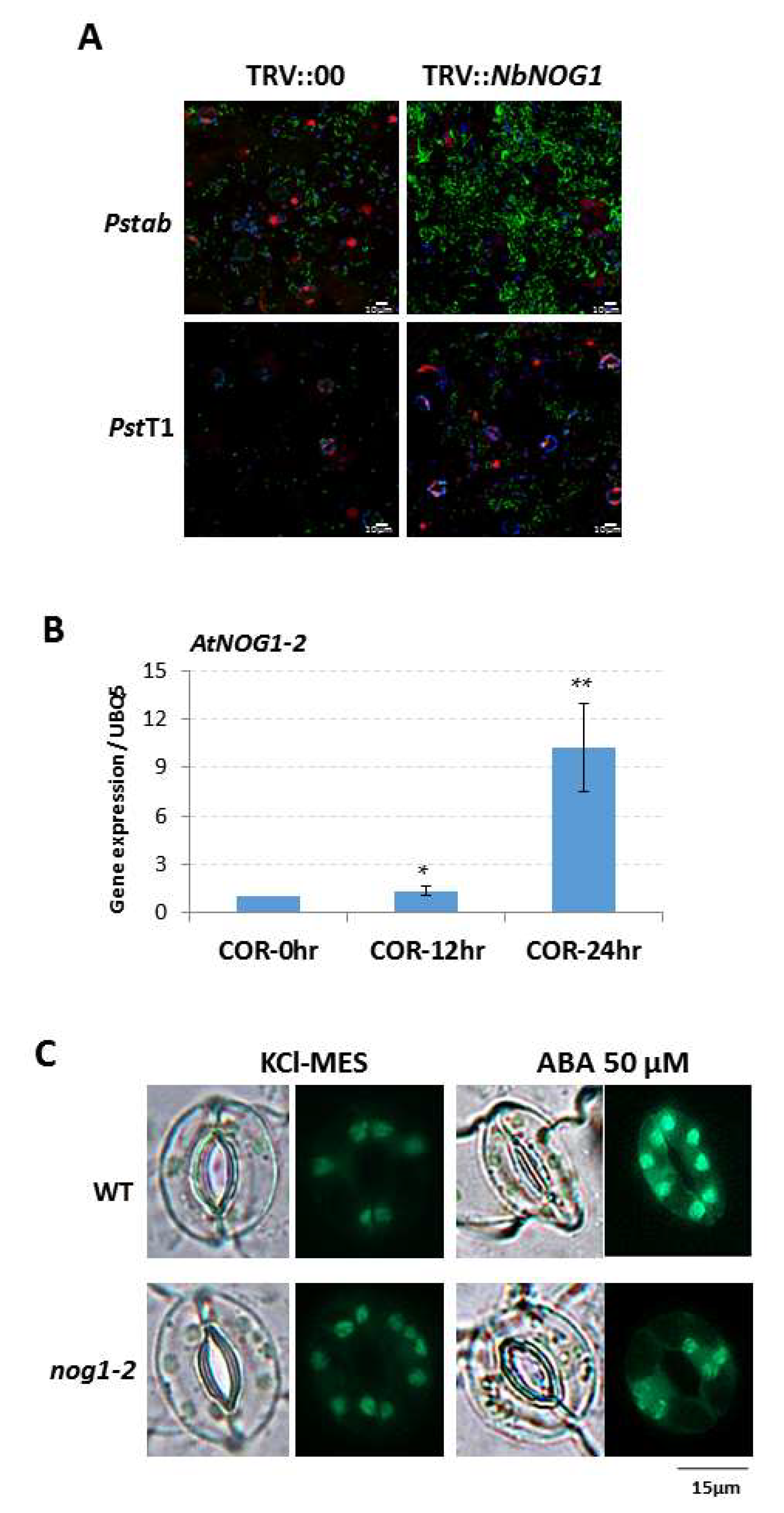
2.1. NOG1-2 Functions in Guard Cell Signaling
2.2. NOG1-2 Interacts with JAZ9, A Key Protein for Stomatal Closure
2.3. JAZ9 Alters GTPase Activity of NOG1-2
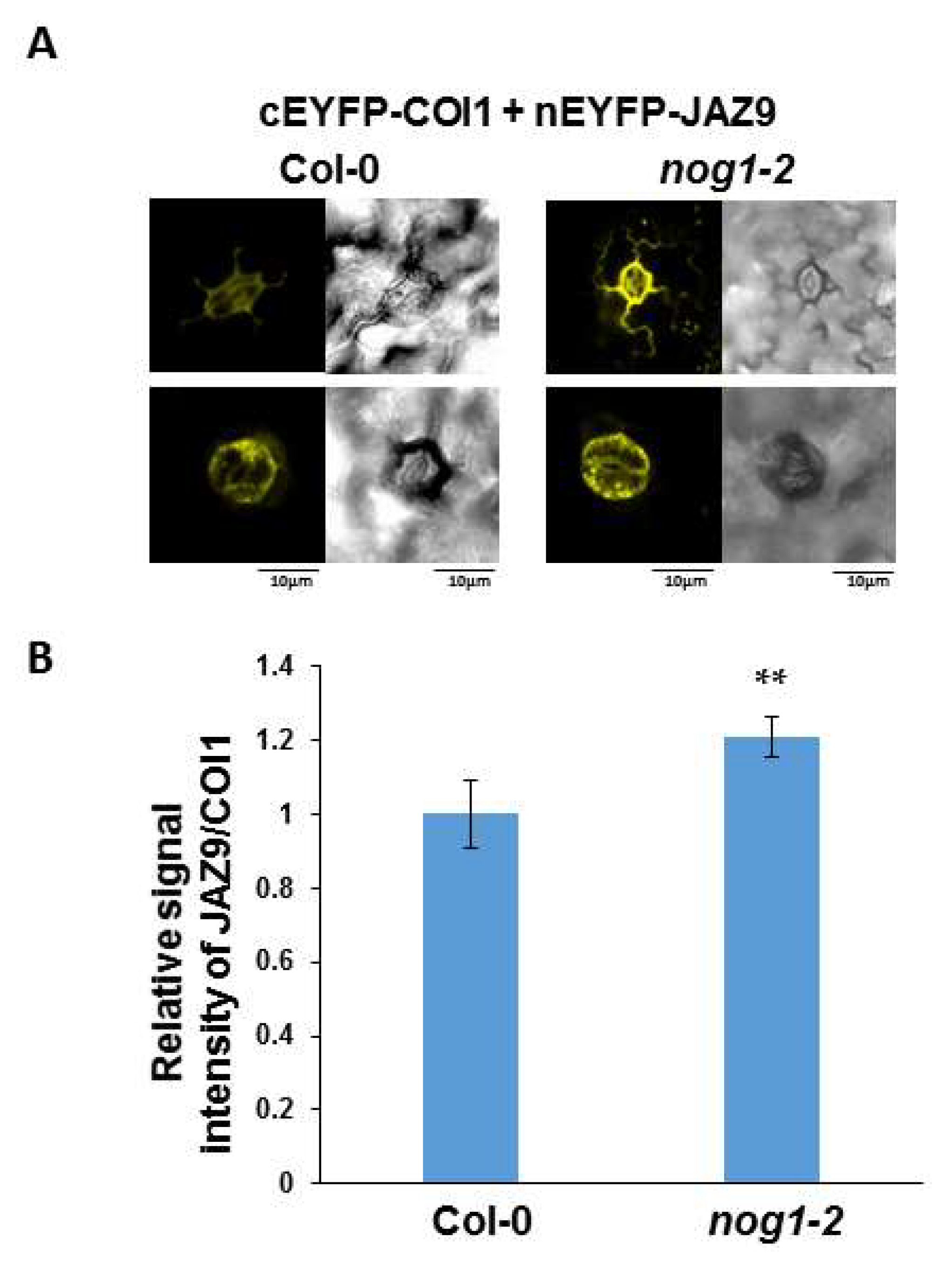
2.4. NOG1-2 May Interfere with Interaction between JAZ9 and COI1
2.5. NOG1-2 Positively Regulates Signaling Pathways Related to Stomatal Function
3. Discussion
4. Materials and Methods
4.1. Bacterial Entry Assay in Detached Leaf of NbNOG1-2 Silenced N. Benthamiana
4.2. Construction of the cDNA Library
4.3. Yeast Two-Hybrid Analysis
4.4. Bimolecular Fluorescence Complementation (Bifc) Analysis
4.5. In Vitro GTPase Activity Assay and Phosphate Release Assay
4.6. Semi-In Vivo Co-Immunoprecipitation
4.7. Transcriptome Analysis of nog1-2 and jaz9 Using Arabidopsis Microarray
4.8. Quantitative Real-Time RT-PCR
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Melotto, M.; Underwood, W.; Koczan, J.; Nomura, K.; He, S.Y. Plant stomata function in innate immunity against bacterial invasion. Cell 2006, 126, 969–980. [Google Scholar] [CrossRef] [PubMed]
- Kwak, J.M.; Mäser, P.; Schroeder, J.I. The Clickable Guard Cell, Version II: Interactive Model of Guard Cell Signal Transduction Mechanisms and Pathways. Arabidopsis Book 2008, 6, e0114. [Google Scholar] [CrossRef] [PubMed]
- Melotto, M.; Underwood, W.; He, S.Y. Role of stomata in plant innate immunity and foliar bacterial diseases. Annu. Rev. Phytopathol. 2008, 46, 101–122. [Google Scholar] [CrossRef] [PubMed]
- Underwood, W.; Melotto, M.; He, S.Y. Role of plant stomata in bacterial invasion. Cell Microbiol. 2007, 9, 1621–1629. [Google Scholar] [CrossRef] [PubMed]
- Boter, M.; Ruíz-Rivero, O.; Abdeen, A.; Prat, S. Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev. 2004, 18, 1577–1591. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Qi, T.; Huang, H.; Ren, Q.; Wu, D.; Chang, C.; Peng, W.; Liu, Y.; Peng, J.; Xie, D. The Jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect Jasmonate-regulated stamen development in Arabidopsis. Plant Cell 2011, 23, 1000–1013. [Google Scholar] [CrossRef] [PubMed]
- Pauwels, L.; Barbero, G.F.; Geerinck, J.; Tilleman, S.; Grunewald, W.; Perez, A.C.; Chico, J.M.; Bossche, R.V.; Sewell, J.; Gil, E.; et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 2010, 464, 788–791. [Google Scholar] [CrossRef] [PubMed]
- Sheard, L.B.; Tan, X.; Mao, H.; Withers, J.; Ben-Nissan, G.; Hinds, T.R.; Kobayashi, Y.; Hsu, F.F.; Sharon, M.; Browse, J.; et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 2010, 468, 400–405. [Google Scholar] [CrossRef] [PubMed]
- Assmann, S.M. Heterotrimeric and unconventional GTP binding proteins in plant cell signaling. Plant Cell 2002, 14 (Suppl. 1), S355–S373. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z. Small GTPases: Versatile signaling switches in plants. Plant Cell 2002, 14 (Suppl. 1), S375–S388. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Senthil-Kumar, M.; Kang, M.; Rojas, C.M.; Tang, Y.; Oh, S.; Choudhury, S.R.; Lee, H.-K.; Ishiga, Y.; Allen, R.D.; et al. The small GTPase, nucleolar GTP-binding protein 1 (NOG1), has a novel role in plant innate immunity. Sci Rep. 2017, 7, 9260. [Google Scholar] [CrossRef] [PubMed]
- Elizabeth, S.V.; Bender, C.L. The phytotoxin coronatine from Pseudomonas syringae pv. tomato DC3000 functions as a virulence factor and influences defence pathways in edible brassicas. Mol. Plant Pathol. 2007, 8, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Toum, L.; Torres, P.S.; Gallego, S.M.; Benavides, M.P.; Vojnov, A.A.; Gudesblat, G.E. Coronatine Inhibits Stomatal Closure through Guard Cell-Specific Inhibition of NADPH Oxidase-Dependent ROS Production. Front. Plant Sci. 2016, 7, 1851. [Google Scholar] [CrossRef] [PubMed]
- Melotto, M.; Mecey, C.; Niu, Y.; Chung, H.S.; Katsir, L.; Yao, J.; Zeng, W.; Thines, B.; Staswick, P.; Browse, J.; et al. A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J. 2008, 55, 979–988. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Kang, L.; Anand, A.; Lazarovits, G.; Mysore, K.S. Monitoring in planta bacterial infection at both cellular and whole-plant levels using the green fluorescent protein variant GFPuv. New Phytol. 2007, 174, 212–223. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Parihar, P.; Singh, S.; Mishra, R.K.; Singh, V.P.; Prasad, S.M. Reactive oxygen species signaling and stomatal movement: Current updates and future perspectives. Redox Biol. 2017, 11, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Halliwell, B.; Whiteman, M. Measuring reactive species and oxidative damage in vivo and in cell culture: How should you do it and what do the results mean? Br. J. Pharmacol. 2004, 142, 231–255. [Google Scholar] [CrossRef] [PubMed]
- Stein, M.; Dittgen, J.; Sanchez-Rodriguez, C.; Hou, B.H.; Molina, A.; Schulze-Lefert, P.; Lipka, V.; Somerville, S. Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell 2006, 18, 731–746. [Google Scholar] [CrossRef] [PubMed]
- Underwood, W.; Somerville, S.C. Perception of conserved pathogen elicitors at the plasma membrane leads to relocalization of the Arabidopsis PEN3 transporter. Proc. Natl. Acad. Sci. USA 2013, 110, 12492–12497. [Google Scholar] [CrossRef] [PubMed]
- Eisenach, C.; Chen, Z.H.; Grefen, C.; Blatt, M.R. The trafficking protein SYP121 of Arabidopsis connects programmed stomatal closure and K+ channel activity with vegetative growth. Plant J. 2012, 69, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Gimenez-Ibanez, S.; Boter, M.; Ortigosa, A.; García-Casado, G.; Chini, A.; Lewsey, M.G.; Ecker, J.R.; Ntoukakis, V.; Solano, R. JAZ2 controls stomata dynamics during bacterial invasion. New Phytol. 2017, 213, 1378–1392. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.S.; Howe, G.A. A Critical Role for the TIFY Motif in Repression of Jasmonate Signaling by a Stabilized Splice Variant of the JASMONATE ZIM-Domain Protein JAZ10 in Arabidopsis. Plant Cell 2009, 21, 131–145. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.L.; Yao, J.; Mei, C.S.; Tong, X.H.; Zeng, L.J.; Li, Q.; Xiao, L.T.; Sun, T.P.; Li, J.; Deng, X.W.; et al. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. USA 2012, 109, E1192–E1200. [Google Scholar] [CrossRef] [PubMed]
- Swarup, R.C.; Sona, P. Phosphatidic acid binding inhibits RGS1 activity to affect specific signaling pathways in Arabidopsis. Plant J. 2017, 90, 466–477. [Google Scholar]
- Shutes, A.; Der, C.J. Real-time in vitro measurement of GTP hydrolysis. Methods 2005, 37, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Li, J.F.; Park, E.; von Arnim, A.G.; Nebenfuhr, A. The FAST technique: A simplified Agrobacterium-based transformation method for transient gene expression analysis in seedlings of Arabidopsis and other plant species. Plant Methods 2009, 5, 6. [Google Scholar] [CrossRef] [PubMed]
- Withers, J.; Yao, J.; Mecey, C.; Howe, G.A.; Melotto, M.; He, S.Y. Transcription factor-dependent nuclear localization of a transcriptional repressor in jasmonate hormone signaling. Proc. Natl. Acad. Sci. USA 2012, 109, 20148–20153. [Google Scholar] [CrossRef] [PubMed]
- Ueda, M.; Egoshi, S.; Dodo, K.; Ishimaru, Y.; Yamakoshi, H.; Nakano, T.; Takaoka, Y.; Tsukiji, S.; Sodeoka, M. Noncanonical Function of a Small-Molecular Virulence Factor Coronatine against Plant Immunity: An In Vivo Raman Imaging Approach. ACS Cent. Sci. 2017, 3, 462–472. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Wu, Y.; Yang, Y.; Du, M.; Zhang, X.; Guo, Y.; Li, C.; Zhou, J.-M. An Arabidopsis Plasma Membrane Proton ATPase Modulates JA Signaling and Is Exploited by the Pseudomonas syringae Effector Protein AvrB for Stomatal Invasion. Plant Cell 2015, 27, 2032–2041. [Google Scholar] [CrossRef] [PubMed]
- Chini, A.; Fonseca, S.; Chico, J.M.; Fernández-Calvo, P.; Solano, R. The ZIM domain mediates homo- and heteromeric interactions between Arabidopsis JAZ proteins. Plant J. 2009, 59, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.; Nomura, K.; He, S.Y.; Howe, G.A.; Browse, J. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 2007, 448, 661–665. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Wu, H.; Ma, S.; Xiang, D.; Liu, R.; Xiong, L. OsJAZ1 Attenuates Drought Resistance by Regulating JA and ABA Signaling in Rice. Front. Plant Sci. 2017, 8, 2108. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Meng, L.; Kong, W.; Yin, Z.; Wang, Y.; Schneider, J.D.; Chen, S. Quantitative proteomics reveals a role of JAZ7 in plant defense response to Pseudomonas syringae DC3000. J. Proteomics. 2018, 175, 114–126. [Google Scholar] [CrossRef] [PubMed]
- Wager, A.; Browse, J. Social Network: JAZ Protein Interactions Expand Our Knowledge of Jasmonate Signaling. Front. Plant Sci. 2012, 3, 41. [Google Scholar] [CrossRef] [PubMed]
- De Torres Zabala, M.; Zhai, B.; Jayaraman, S.; Eleftheriadou, G.; Winsbury, R.; Yang, R.; Truman, W.; Tang, S.; Smirnoff, N.; Grant, M. Novel JAZ co-operativity and unexpected JA dynamics underpin Arabidopsis defence responses to Pseudomonas syringae infection. New Phytol. 2016, 209, 1120–1134. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.; Figueroa, P.; Browse, J. Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J. Exp. Bot. 2011, 62, 2143–2154. [Google Scholar] [CrossRef] [PubMed]
- Chini, A.; Boter, M.; Solano, R. Plant oxylipins: COI1/JAZs/MYC2 as the core jasmonic acid-signalling module. FEBS J. 2009, 276, 4682–4692. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Q.; Yan, L.; Tan, D.; Chen, R.; Sun, J.; Gao, L.; Dong, M.Q.; Wang, Y.; Li, C. Phosphorylation-coupled proteolysis of the transcription factor MYC2 is important for jasmonate-signaled plant immunity. PLoS Genet. 2013, 9, e1003422. [Google Scholar] [CrossRef] [PubMed]
- Sethi, V.; Raghuram, B.; Sinha, A.K.; Chattopadhyay, S. A mitogen-activated protein kinase cascade module, MKK3-MPK6 and MYC2, is involved in blue light-mediated seedling development in Arabidopsis. Plant Cell 2014, 26, 3343–3357. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Zhang, W.; Stanley, B.A.; Assmann, S.M. Functional Proteomics of Arabidopsis thaliana Guard Cells Uncovers New Stomatal Signaling Pathways. Plant Cell 2008, 20, 3210–3226. [Google Scholar] [CrossRef] [PubMed]
- Jammes, F.; Song, C.; Shin, D.; Munemasa, S.; Takeda, K.; Gu, D.; Cho, D.; Lee, S.; Giordo, R.; Sritubtim, S.; et al. MAP kinases MPK9 and MPK12 are preferentially expressed in guard cells and positively regulate ROS-mediated ABA signaling. Proc. Natl. Acad. Sci. USA 2009, 106, 20520–20525. [Google Scholar] [CrossRef] [PubMed]
- Khokon, M.A.; Salam, M.A.; Jammes, F.; Ye, W.; Hossain, M.A.; Uraji, M.; Nakamura, Y.; Mori, I.C.; Kwak, J.M.; Murata, Y. Two guard cell mitogen-activated protein kinases, MPK9 and MPK12, function in methyl jasmonate-induced stomatal closure in Arabidopsis thaliana. Plant Biol. 2015, 17, 946–952. [Google Scholar] [CrossRef] [PubMed]
- Katou, S.; Yoshioka, H.; Kawakita, K.; Rowland, O.; Jones, J.D.; Mori, H.; Doke, N. Involvement of PPS3 phosphorylated by elicitor-responsive mitogen-activated protein kinases in the regulation of plant cell death. Plant Physiol. 2005, 139, 1914–1926. [Google Scholar] [CrossRef] [PubMed]
- Zulawski, M.; Braginets, R.; Schulze, W.X. PhosPhAt goes kinases—searchable protein kinase target information in the plant phosphorylation site database PhosPhAt. Nucleic Acids Res. 2013, 41, D1176–D1184. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, A.P.T.; Moore, D.J. Understanding the GTPase Activity of LRRK2: Regulation, Function, and Neurotoxicity. Adv. Neurobiol. 2017, 14, 71–88. [Google Scholar] [PubMed]
- Liu, Z.; Mobley, J.A.; DeLucas, L.J.; Kahn, R.A.; West, A.B. LRRK2 autophosphorylation enhances its GTPase activity. FASEB J. 2016, 30, 336–347. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, S.R.; Westfall, C.S.; Hackenberg, D.; Pandey, S. Measurement of GTP-binding and GTPase activity of heterotrimeric Galpha proteins. In G Protein-Coupled Receptor Signaling in Plants; Humana Press: Totowa, NJ, USA, 2013; Volume 1043, pp. 13–20. [Google Scholar]



© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.; Rojas, C.M.; Oh, S.; Kang, M.; Choudhury, S.R.; Lee, H.-K.; Allen, R.D.; Pandey, S.; Mysore, K.S. Nucleolar GTP-Binding Protein 1-2 (NOG1-2) Interacts with Jasmonate-ZIMDomain Protein 9 (JAZ9) to Regulate Stomatal Aperture during Plant Immunity. Int. J. Mol. Sci. 2018, 19, 1922. https://doi.org/10.3390/ijms19071922
Lee S, Rojas CM, Oh S, Kang M, Choudhury SR, Lee H-K, Allen RD, Pandey S, Mysore KS. Nucleolar GTP-Binding Protein 1-2 (NOG1-2) Interacts with Jasmonate-ZIMDomain Protein 9 (JAZ9) to Regulate Stomatal Aperture during Plant Immunity. International Journal of Molecular Sciences. 2018; 19(7):1922. https://doi.org/10.3390/ijms19071922
Chicago/Turabian StyleLee, Seonghee, Clemencia M. Rojas, Sunhee Oh, Miyoung Kang, Swarup Roy Choudhury, Hee-Kyung Lee, Randy D. Allen, Sona Pandey, and Kirankumar S. Mysore. 2018. "Nucleolar GTP-Binding Protein 1-2 (NOG1-2) Interacts with Jasmonate-ZIMDomain Protein 9 (JAZ9) to Regulate Stomatal Aperture during Plant Immunity" International Journal of Molecular Sciences 19, no. 7: 1922. https://doi.org/10.3390/ijms19071922
APA StyleLee, S., Rojas, C. M., Oh, S., Kang, M., Choudhury, S. R., Lee, H.-K., Allen, R. D., Pandey, S., & Mysore, K. S. (2018). Nucleolar GTP-Binding Protein 1-2 (NOG1-2) Interacts with Jasmonate-ZIMDomain Protein 9 (JAZ9) to Regulate Stomatal Aperture during Plant Immunity. International Journal of Molecular Sciences, 19(7), 1922. https://doi.org/10.3390/ijms19071922




