Transcriptional Activation of Human GD3 Synthase (hST8Sia I) Gene in Curcumin-Induced Autophagy in A549 Human Lung Carcinoma Cells
Abstract
1. Introduction
2. Results
2.1. Effect of Curcumin on Cell Growth
2.2. Curcumin Induces Autophagy in A549 Cells
2.3. Effect of Curcumin on hST8Sia I Gene Expression in A549 Cells
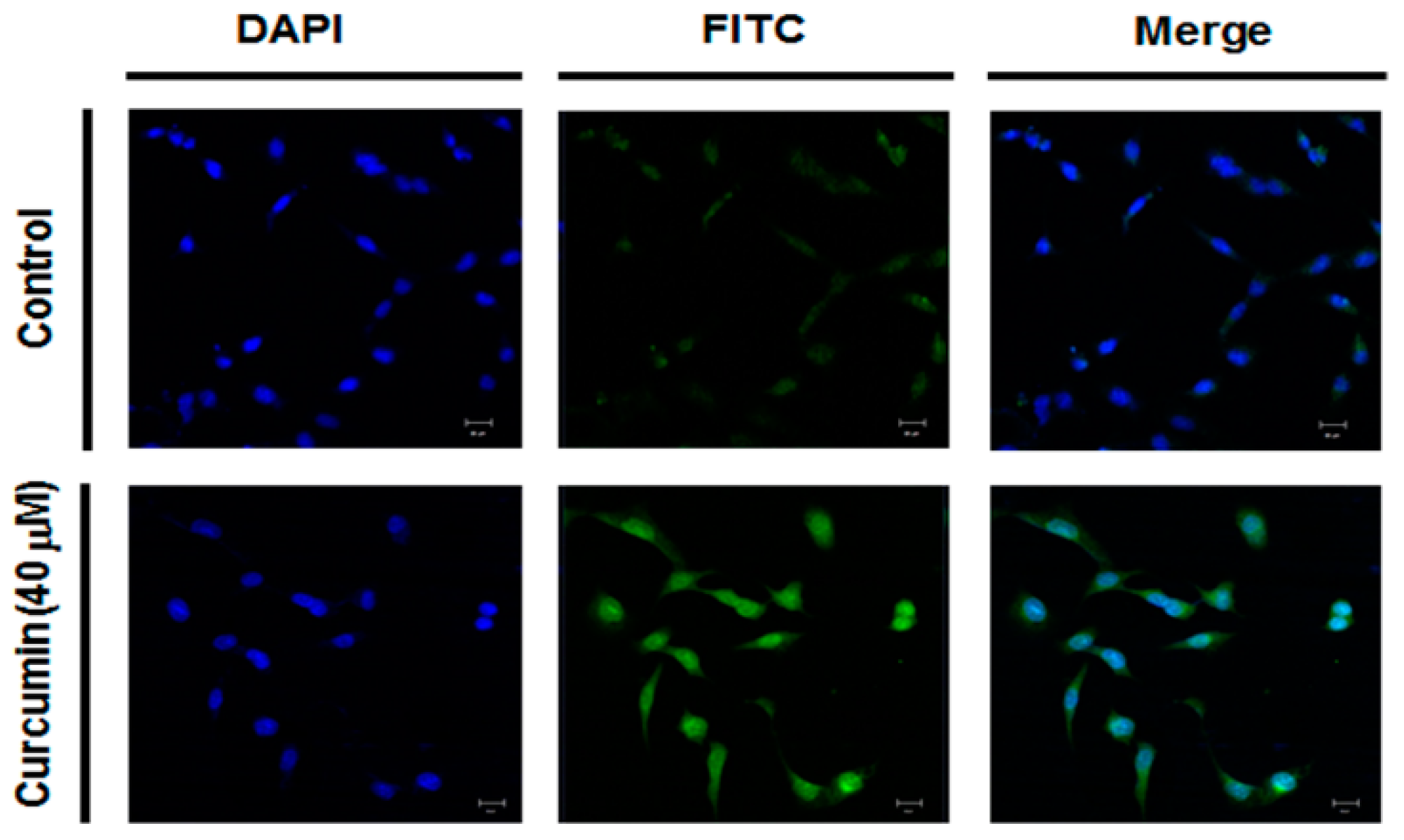
2.4. Effect of Curcumin on Ganglioside GD3 Expression in A549 Cells
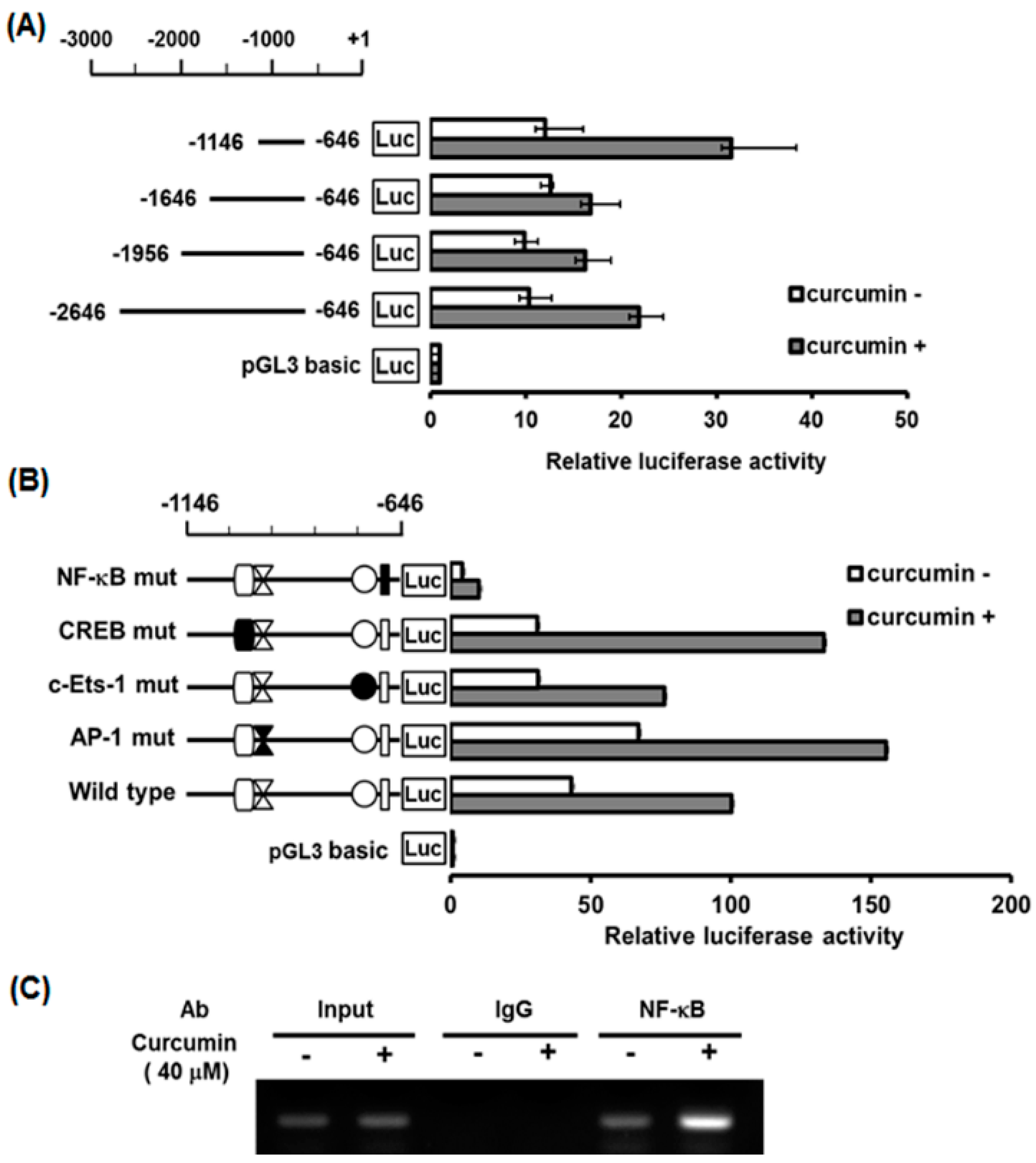
2.5. Characterization of Curcumin-Inducible hST8Sia I Promoter in A549 Cells
2.6. Identification of Curcumin-Responsive Element Controlling Inducible Expression of hST8Sia L in the Functional -1146/-646 Region of Its Promoter
2.7. Curcumin Mediates hST8Sia I Gene Transcription via AMPK Pathway in A549 Cells
3. Discussion
4. Experimental Section
4.1. Cell Cultures
4.2. MTT Cell Viability Assay
4.3. Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
4.4. Transfection and Luciferase Reporter Assays
4.5. Immunofluorescence
4.6. Chromatin Immunoprecipitation (ChIP) Assay
5. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| AMPK | AMP-activated protein kinase |
| AP-1 | Activator protein-1 |
| BSA | Bovine serum albumin |
| ChIP | Chromatin immunoprecipitation |
| CREB | cAMP response element-binding protein |
| DAPI | 4′,6-Diamidino-2-phenylindole |
| ERK | Extra cellular-signal-regulated kinase |
| FITC | Fluorescein isothiocyanate |
| IKK | IκB kinase |
| JNK | c-Jun N-terminal kinase |
| LAMP1 | Lysosomal-associated membrane protein 1 |
| LC3 | Microtubule-associated protein 1 light chain 3 |
| mAb | Monoclonal antibody |
| mTOR | Mechanistic target of rapamycin |
| NF-κB | Nuclear factor kappa B |
| PBS | Phosphate-buffered saline |
| PCR | Polymerase chain reaction |
| PI3K | Phosphatidylinositol 3-kinase |
| PKC | Protein kinase C |
| RPMI | Roswell Park Memorial Institute medium |
References
- Glick, D.; Barth, S.; Macleod, K.F. Autophagy: Cellular and molecular mechanisms. J. Pathol. 2010, 221, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Choi, A.M.; Ryter, S.W.; Levine, B. Autophagy in human health and disease. N. Engl. J. Med. 2013, 368, 1845–1846. [Google Scholar] [CrossRef] [PubMed]
- Levine, B.; Kroemer, G. Autophagy in the pathogenesis of disease. Cell 2008, 11, 27–42. [Google Scholar] [CrossRef] [PubMed]
- Goel, A.; Kunnumakkara, A.B.; Aggarwal, B.B. Curcumin as “curecumin”: From kitchen to clinic. Biochem. Pharmacol. 2008, 75, 787–809. [Google Scholar] [CrossRef] [PubMed]
- Fadus, M.C.; Lau, C.; Bikhchandani, J.; Lynch, H.T. Curcumin: An age-old anti-inflammatory and anti-neoplastic agent. J. Tradit. Complement. Med. 2016, 7, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Hussain, Z.; Thu, H.E.; Amjad, M.W.; Hussain, F.; Ahmed, T.A.; Khan, S. Exploring recent developments to improve antioxidant, anti-inflammatory and antimicrobial efficacy of curcumin: A review of new trends and future perspectives. Mater. Sci. Eng. C 2017, 77, 1316–1326. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, B.B.; Kumar, A.; Bharti, A.C. Anticancer potential of curcumin: Preclinical and clinical studies. Anticancer Res. 2003, 23, 363–398. [Google Scholar] [PubMed]
- Shishodia, S.; Chaturvedi, M.M.; Aggarwal, B.B. Role of curcumin in cancer therapy. Curr. Probl. Cancer 2007, 31, 243–305. [Google Scholar] [CrossRef] [PubMed]
- Aoki, H.; Takada, Y.; Kondo, S.; Sawaya, R.; Aggarwal, B.B.; Kondo, Y. Evidence that curcumin suppresses the growth of malignant gliomas in vitro and in vivo through induction of autophagy: Role of Akt and extracellular signal-regulated kinase signaling pathways. Mol. Pharmacol. 2007, 72, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Ma, X.; Wang, Z.; Zeng, X.; Hu, Z.; Ye, Z.; Shen, G. Curcumin induces apoptosis and protective autophagy in castration-resistant prostate cancer cells through iron chelation. Drug Des. Dev. Ther. 2017, 11, 431–439. [Google Scholar] [CrossRef] [PubMed]
- Guan, F.; Ding, Y.; Zhang, Y.; Zhou, Y.; Li, M.; Wang, C. Curcumin suppresses proliferation and migration of MDA-MB-231 breast cancer cells through autophagy-dependent Akt degradation. PLoS ONE 2016, 11, e0146553. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, P.; Hong, H.; Wang, L.; Zhou, Y.; Lang, Y. JNK pathway mediates curcumin-induced apoptosis and autophagy in osteosarcoma MG63 cells. Exp. Ther. Med. 2017, 14, 593–599. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Wang, C.; Yang, D.; Wei, Z.; Xu, J.; Hu, Z.; Zhang, Y.; Wang, W.; Yan, R.; Cai, Q. Curcumin regulates proliferation, autophagy, and apoptosis in gastric cancer cells by affecting PI3K and P53 signaling. J. Cell. Physiol. 2018, 233, 4634–4642. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Cho, T.J.; Woo, B.H.; Choi, K.U.; Lee, C.H.; Ryu, M.H.; Park, H.R. Curcumin-induced autophagy contributes to the decreased survival of oral cancer cells. Arch. Oral Biol. 2012, 57, 1018–1025. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Takeda, T.; Tsuiji, K.; Wong, T.F.; Tadakawa, M.; Kondo, A.; Nagase, S.; Yaegashi, N. Curcumin induces cross-regulation between autophagy and apoptosis in uterine leiomyosarcoma cells. Int. J. Gynecol. Cancer 2013, 23, 803–808. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Shan, Q.Q.; Gong, P.Y.; Wang, S.C. The autophagy induced by curcumin via MEK/ERK pathway plays an early anti-leukemia role in human Philadelphia chromosome-positive acute lymphoblastic leukemia SUP-B15 cells. J. Cancer Res. Ther. 2018, 14, S125–S131. [Google Scholar] [PubMed]
- Lee, Y.J.; Kim, N.Y.; Suh, Y.A.; Lee, C. Involvement of ROS in curcumin-induced autophagic cell death. Korean J. Physiol. Pharmacol. 2011, 15, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Mosieniak, G.; Adamowicz, M.; Alster, O.; Jaskowiak, H.; Szczepankiewicz, A.A.; Wilczynski, G.M.; Ciechomska, I.A.; Sikora, E. Curcumin induces permanent growth arrest of human colon cancer cells: Link between senescence and autophagy. Mech. Ageing Dev. 2012, 133, 444–455. [Google Scholar] [CrossRef] [PubMed]
- Xiao, K.; Jiang, J.; Guan, C.; Dong, C.; Wang, G.; Bai, L.; Sun, J.; Hu, C.; Bai, C. Curcumin induces autophagy via activating the AMPK signaling pathway in lung adenocarcinoma cells. J. Pharmacol. Sci. 2013, 123, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Gao, S.; Yang, Y.; Zhao, X.; Fan, Y.; Ma, W.; Yang, D.; Yang, A.; Yu, Y. Curcumin induced autophagy anticancer effects on human lung adenocarcinoma cell line A549. Oncol. Lett. 2017, 14, 2775–2782. [Google Scholar] [CrossRef] [PubMed]
- Hakomori, S. Structure, organization, and function of glycosphingolipids in membrane. Curr. Opin. Hematol. 2003, 10, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Sonnino, S.; Prinetti, A. Gangliosides as regulators of cell membrane organization and functions. Adv. Exp. Med. Biol. 2010, 688, 165–184. [Google Scholar] [PubMed]
- Bektas, M.; Spiegel, S. Glycosphingolipids and cell death. Glycoconj. J. 2004, 20, 39–47. [Google Scholar] [CrossRef] [PubMed]
- D’Angelo, G.; Capasso, S.; Sticco, L.; Russo, D. Glycosphingolipids: Synthesis and functions. FEBS J. 2013, 280, 6338–6353. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.; Lee, S.; Lee, J.T.; Kwon, T.K.; Kim, D.R.; Kim, H.; Park, H.C.; Suk, K. Gangliosides induce autophagic cell death in astrocytes. Br. J. Pharmacol. 2010, 159, 586–603. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.; Lee, H.J.; Lee, W.H.; Suk, K. NF-κB as a common signaling pathway in ganglioside-induced autophagic cell death and activation of astrocytes. J. Neuroimmunol. 2010, 226, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Matarrese, P.; Garofalo, T.; Manganelli, V.; Gambardella, L.; Marconi, M.; Grasso, M.; Tinari, A.; Misasi, R.; Malorni, W.; Sorice, M. Evidence for the involvement of GD3 ganglioside in autophagosome formation and maturation. Autophagy 2014, 10, 750–765. [Google Scholar] [CrossRef] [PubMed]
- Kang, N.Y.; Kang, S.K.; Lee, Y.C.; Choi, H.J.; Lee, Y.S.; Cho, S.Y.; Kim, Y.S.; Ko, J.H.; Kim, C.H. Transcriptional regulation of the human GD3 synthase gene expression in Fas-induced Jurkat T cells: A critical role of transcription factor NF-kB in regulated expression. Glycobiology 2006, 16, 375–389. [Google Scholar] [CrossRef] [PubMed]
- Kang, N.Y.; Kim, C.H.; Kim, K.S.; Ko, J.H.; Lee, J.H.; Jeong, Y.K.; Lee, Y.C. Expression of the human CMP-NeuAc:GM3 alpha2,8-sialyltransferase (GD3 synthase) gene through the NF-κB activation in human melanoma SK-MEL-2 cells. Biochim. Biophys. Acta 2007, 1769, 622–630. [Google Scholar] [CrossRef] [PubMed]
- Kwon, H.Y.; Dae, H.M.; Song, N.R.; Kim, K.S.; Kim, C.H.; Lee, Y.C. Valproic acid induces transcriptional activation of human GD3 synthase (hST8Sia I) in SK-N-BE(2)-C human neuroblastoma cells. Mol. Cells 2009, 27, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Kwon, H.Y.; Kim, S.J.; Kim, C.H.; Son, S.W.; Kim, K.S.; Lee, J.H.; Do, S.I.; Lee, Y.C. Triptolide downregulates human GD3 synthase (hST8Sia I) gene expression in SK-MEL-2 human melanoma cells. Exp. Mol. Med. 2010, 42, 849–855. [Google Scholar] [CrossRef] [PubMed]
- Baik, J.S.; Kim, K.S.; Moon, H.I.; An, H.K.; Park, S.J.; Kim, C.H.; Lee, Y.C. Cordycepin-mediated transcriptional regulation of human GD3 synthase (hST8Sia I) in human neuroblastoma SK-N-BE(2)-C cells. Acta Biochim. Biophys. Sin. 2014, 46, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Baichwal, V.R.; Baeuerle, P.A. Activate NF-κB or die? Curr. Biol. 1997, 7, R94–R96. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, X.; Li, J.J. The role of NF-κB in the regulation of cell stress responses. Int. Immunopharmacol. 2002, 2, 1509–1520. [Google Scholar] [CrossRef]
- Ravindran, J.; Prasad, S.; Aggarwal, B.B. Curcumin and cancer cells: How many ways can curry kill tumor cells selectively? AAPS J. 2009, 11, 495–510. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Aggarwal, B.B. Activation of transcription factor NF-κB is suppressed by curcumin (diferuloylmethane). J. Biol. Chem. 1995, 270, 24995–25000. [Google Scholar] [CrossRef] [PubMed]
- Bharti, A.C.; Donato, N.; Singh, S.; Aggarwal, B.B. Curcumin (diferuloylmethane) down-regulates the constitutive activation of nuclear factor-κB and IκBa kinase in human multiple myeloma cells, leading to suppression of proliferation and induction of apoptosis. Blood 2003, 101, 1053–1062. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, S.; Takada, Y.; Singh, S.; Myers, J.N.; Aggarwal, B.B. Inhibition of growth and survival of human head and neck squamous cell carcinoma cells by curcumin via modulation of nuclear factor-κB signaling. Int. J. Cancer 2004, 111, 679–692. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Yao, Y.; Wu, J.; Liu, Q.; Zhang, J.; Pu, X.; Zhang, Q.; Xia, L. Curcumin inhibits gastric cancer-derived mesenchymal stem cells mediated angiogenesis by regulating NF-κB/VEGF signaling. Am. J. Transl. Res. 2017, 9, 5538–5547. [Google Scholar] [PubMed]
- Lin, Y.G.; Kunnumakkara, A.B.; Nair, A.; Merritt, W.M.; Han, L.Y.; Armaiz-Pena, G.N.; Kamat, A.A.; Spannuth, W.A.; Gershenson, D.M.; Lutgendorf, S.K.; et al. Curcumin inhibits tumor growth and angiogenesis in ovarian carcinoma by targeting the nuclear factor-κB pathway. Clin. Cancer Res. 2007, 13, 3423–3430. [Google Scholar] [CrossRef] [PubMed]
- Marquardt, J.U.; Gomez-Quiroz, L.; Arreguin Camacho, L.O.; Pinna, F.; Lee, Y.H.; Kitade, M.; Domínguez, M.P.; Castven, D.; Breuhahn, K.; Conner, E.A.; et al. Curcumin effectively inhibits oncogenic NF-kB signaling and restrains stemness features in liver cancer. J. Hepatol. 2015, 3, 661–669. [Google Scholar] [CrossRef] [PubMed]
- Tong, W.; Wang, Q.; Sun, D.; Suo, J. Curcumin suppresses colon cancer cell invasion via AMPK-induced inhibition of NF-κB, uPA activator and MMP9. Oncol. Lett. 2016, 12, 4139–4146. [Google Scholar] [CrossRef] [PubMed]
- Pan, W.; Yang, H.; Cao, C.; Song, X.; Wallin, B.; Kivlin, R.; Lu, S.; Hu, G.; Di, W.; Wan, Y. AMPK mediates curcumin-induced cell death in CaOV3 ovarian cancer cells. Oncol. Rep. 2008, 20, 1553–1559. [Google Scholar] [PubMed]
- An, H.K.; Kim, K.S.; Lee, J.W.; Park, M.H.; Moon, H.I.; Park, S.J.; Baik, J.S.; Kim, C.H.; Lee, Y.C. Mimulone-induced autophagy through p53-mediated AMPK/mTOR pathway increases caspase-mediated apoptotic cell death in A549 human lung cancer cells. PLoS ONE 2014, 9, e114607. [Google Scholar] [CrossRef] [PubMed]
- Kwon, K.M.; Chung, T.W.; Kwak, C.H.; Choi, H.J.; Kim, K.W.; Ha, S.H.; Cho, S.H.; Lee, Y.C.; Ha, K.T.; Lee, M.J.; et al. Disialyl GD2 ganglioside suppresses ICAM-1-mediated invasiveness in human breast cancer MDA-MB231 cells. Int. J. Biol. Sci. 2017, 13, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Ha, S.H.; Kang, S.K.; Choi, H.; Kwak, C.H.; Abekura, F.; Park, J.Y.; Kwon, K.M.; Chang, H.W.; Lee, Y.C.; Ha, K.T.; et al. Induction of GD3/α1-adrenergic receptor/transglutaminase 2-mediated erythroid differentiation in chronic myelogenous leukemic K562 cells. Oncotarget 2017, 8, 72205–72219. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.; Jin, U.H.; Kang, S.K.; Abekura, F.; Park, J.Y.; Kwon, K.M.; Suh, S.J.; Cho, S.H.; Ha, K.T.; Lee, Y.C.; et al. Monosialyl Ganglioside GM3 Decreases Apolipoprotein B-100 Secretion in Liver Cells. J. Cell. Biochem. 2017, 118, 2168–2181. [Google Scholar] [CrossRef] [PubMed]




© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, M.; Kim, K.-S.; Fukushi, A.; Kim, D.-H.; Kim, C.-H.; Lee, Y.-C. Transcriptional Activation of Human GD3 Synthase (hST8Sia I) Gene in Curcumin-Induced Autophagy in A549 Human Lung Carcinoma Cells. Int. J. Mol. Sci. 2018, 19, 1943. https://doi.org/10.3390/ijms19071943
Lee M, Kim K-S, Fukushi A, Kim D-H, Kim C-H, Lee Y-C. Transcriptional Activation of Human GD3 Synthase (hST8Sia I) Gene in Curcumin-Induced Autophagy in A549 Human Lung Carcinoma Cells. International Journal of Molecular Sciences. 2018; 19(7):1943. https://doi.org/10.3390/ijms19071943
Chicago/Turabian StyleLee, Miri, Kyoung-Sook Kim, Abekura Fukushi, Dong-Hyun Kim, Cheorl-Ho Kim, and Young-Choon Lee. 2018. "Transcriptional Activation of Human GD3 Synthase (hST8Sia I) Gene in Curcumin-Induced Autophagy in A549 Human Lung Carcinoma Cells" International Journal of Molecular Sciences 19, no. 7: 1943. https://doi.org/10.3390/ijms19071943
APA StyleLee, M., Kim, K.-S., Fukushi, A., Kim, D.-H., Kim, C.-H., & Lee, Y.-C. (2018). Transcriptional Activation of Human GD3 Synthase (hST8Sia I) Gene in Curcumin-Induced Autophagy in A549 Human Lung Carcinoma Cells. International Journal of Molecular Sciences, 19(7), 1943. https://doi.org/10.3390/ijms19071943




