Effect of TDP2 on the Level of TOP2-DNA Complexes and SUMOylated TOP2-DNA Complexes
Abstract
:1. Introduction
2. Results
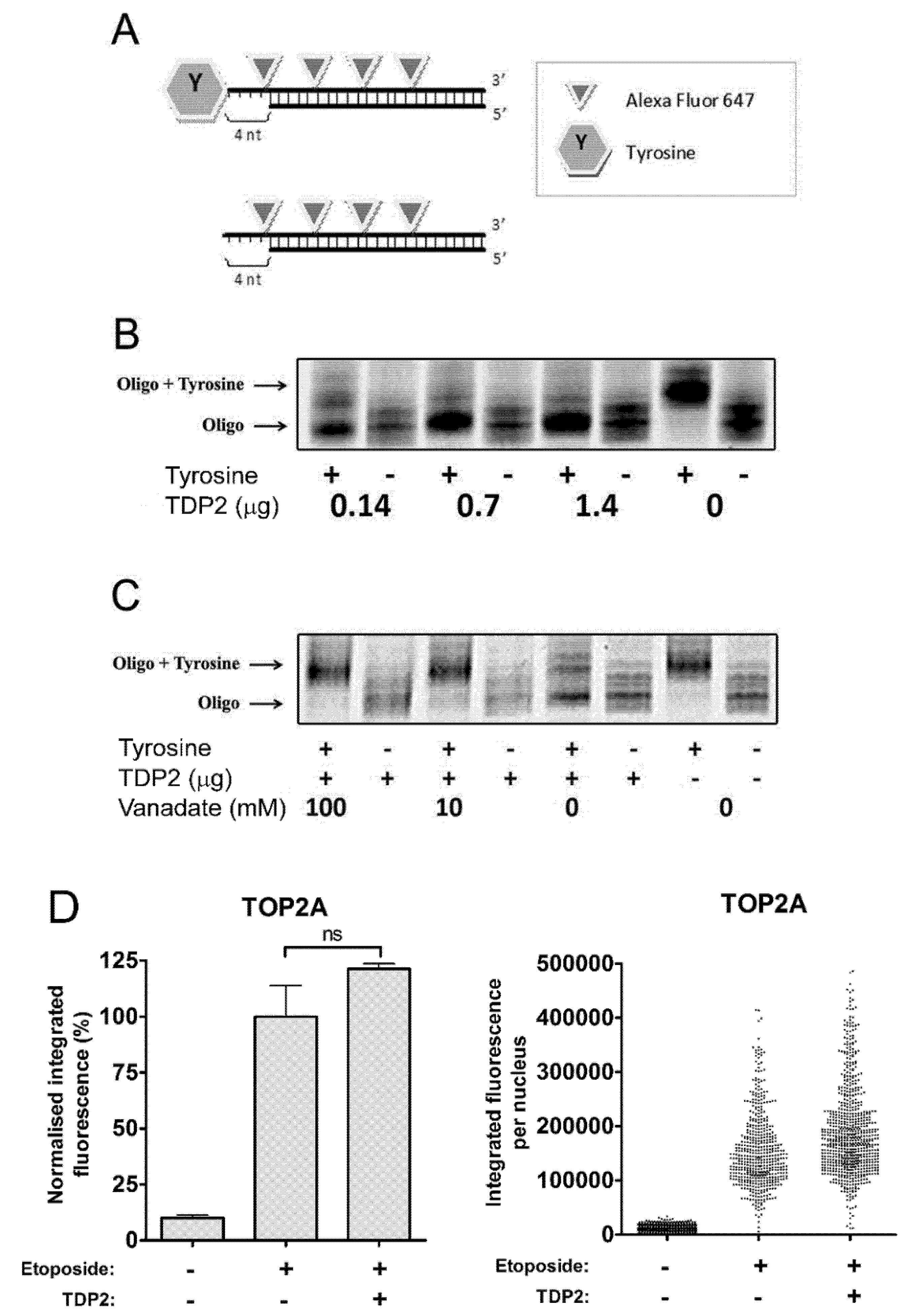
2.1. TDP2 Removes 5′-Tyrosine Adducts from a Labelled Oligonucleotide Substrate in Vitro
2.2. TDP2 Alone Does not Remove TOP2 Protein from Etoposide-Induced TOP2-DNA Covalent Complexes
2.3. TDP2 Depletion Does not Affect the Removal of Etoposide-Induced TOP2-DNA Covalent Complexes in Cells
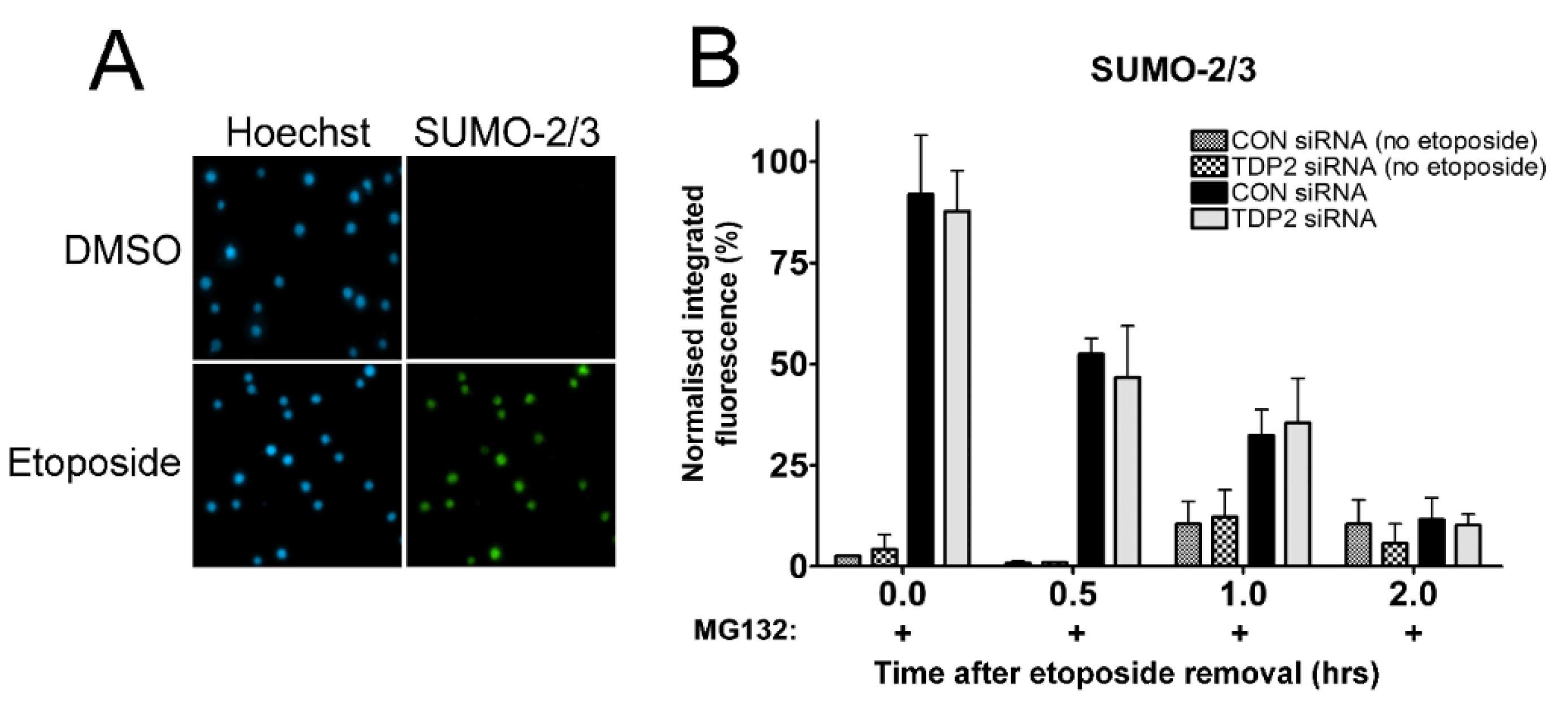
2.4. The Proteasome-Independent Removal of TOP2-DNA Complexes by TDP2 is not Detectable in K562 Cells Following TDP2 Knockdown
2.5. Investigating the Proteasome-Independent Removal of SUMOylated TOP2-DNA Complexes in TDP2 Knockdown Cells Using the TARDIS Assay
3. Discussion
4. Materials and Methods
4.1. TDP2 Activity Assay
4.2. In Vitro Trapped in Agarose Immunostaining (TARDIS)
4.3. TDP2 siRNA
4.4. Western Blotting
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ATM | Ataxia telangiectasia mutated kinase |
| ATR | Ataxia telangiectasia and Rad3 related kinase |
| ɣH2AX | Phosphorylated histone H2AX (ser 139) |
| DMSO | Dimethyl sulfoxide |
| DNA | Deoxyribonucleic acid |
| DNA-PK | DNA-dependent protein kinase |
| DSB | Double strand DNA break |
| FITC | Fluorescein isothiocyanate |
| NaOH | Sodium hydroxide |
| NHEJ | Non-homologous end joining |
| PBS | Phosphate buffered saline |
| SDS | Sodium dodecyl sulfate |
| SIM | SUMO interacting motif |
| siRNA | Small interfering RNA |
| SUMO | Small ubiquitin-like modifier |
| TOP2 | DNA Topoisomerase II |
| TDP2 | 5′-tyrosyl DNA phosphodiesterase 2 |
| TARDIS | Trapped in agarose DNA immunostaining |
| ZATT | Zinc finger protein associated with TDP2 and TOP2 |
References
- Aparicio, T.; Baer, R.; Gottesman, M.; Gautier, J. MRN, CtIP, and BRCA1 mediate repair of topoisomerase II-DNA adducts. J. Cell Biol. 2016, 212, 399–408. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, N.K.; Maizels, N. MRE11 function in response to topoisomerase poisons is independent of its function in double-strand break repair in Saccharomyces cerevisiae. PLoS ONE 2010, 5, e15387. [Google Scholar] [CrossRef] [PubMed]
- Hartsuiker, E.; Neale, M.J.; Carr, A.M. Distinct requirements for the Rad32(Mre11) nuclease and Ctp1(CtIP) in the removal of covalently bound topoisomerase I and II from DNA. Mol. Cell 2009, 33, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.C.; Padget, K.; Curtis, H.; Cowell, I.G.; Moiani, D.; Sondka, Z.; Morris, N.J.; Jackson, G.H.; Cockell, S.J.; Tainer, J.A.; et al. MRE11 facilitates the removal of human topoisomerase II complexes from genomic DNA. Biol. Open 2012, 1, 863–873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberts, S.A.; Strande, N.; Burkhalter, M.D.; Strom, C.; Havener, J.M.; Hasty, P.; Ramsden, D.A. Ku is a 5′-dRP/AP lyase that excises nucleotide damage near broken ends. Nature 2010, 464, 1214–1217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tammaro, M.; Liao, S.; Beeharry, N.; Yan, H. DNA double-strand breaks with 5′ adducts are efficiently channeled to the DNA2-mediated resection pathway. Nucleic Acids Res. 2016, 44, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Ban, Y.; Ho, C.W.; Lin, R.K.; Lyu, Y.L.; Liu, L.F. Activation of a novel ubiquitin-independent proteasome pathway when RNA polymerase II encounters a protein roadblock. Mol. Cell. Biol. 2013, 33, 4008–4016. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.R.; Peng, A.L.; Chen, H.C.; Lo, S.C.; Huang, T.H.; Li, T.K. Cellular processing pathways contribute to the activation of etoposide-induced DNA damage responses. DNA Repair 2008, 7, 452–463. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Desai, S.D.; Ting, C.Y.; Hwang, J.; Liu, L.F. 26 S proteasome-mediated degradation of topoisomerase II cleavable complexes. J. Biol. Chem. 2001, 276, 40652–40658. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Lyu, Y.L.; Lin, C.P.; Zhou, N.; Azarova, A.M.; Wood, L.M.; Liu, L.F. A protease pathway for the repair of topoisomerase II-DNA covalent complexes. J. Biol. Chem. 2006, 281, 35997–36003. [Google Scholar] [CrossRef] [PubMed]
- Cortes Ledesma, F.; El Khamisy, S.F.; Zuma, M.C.; Osborn, K.; Caldecott, K.W. A human 5′-tyrosyl DNA phosphodiesterase that repairs topoisomerase-mediated DNA damage. Nature 2009, 461, 674–678. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Cortés-Ledesma, F.; El Khamisy, S.F.; Caldecott, K.W. TDP2/TTRAP Is the Major 5′-Tyrosyl DNA Phosphodiesterase Activity in Vertebrate Cells and Is Critical for Cellular Resistance to Topoisomerase II-induced DNA Damage. J. Biol. Chem. 2011, 286, 403–409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esguerra, C.V.; Nelles, L.; Vermeire, L.; Ibrahimi, A.; Crawford, A.D.; Derua, R.; Janssens, E.; Waelkens, E.; Carmeliet, P.; Collen, D.; et al. Ttrap is an essential modulator of Smad3-dependent Nodal signaling during zebrafish gastrulation and left-right axis determination. Development 2007, 134, 4381–4393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pei, H.; Yordy, J.S.; Leng, Q.; Zhao, Q.; Watson, D.K.; Li, R. EAPII interacts with ETS1 and modulates its transcriptional function. Oncogene 2003, 22, 2699–2709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pype, S.; Declercq, W.; Ibrahimi, A.; Michiels, C.; Van Rietschoten, J.G.; Dewulf, N.; de Boer, M.; Vandenabeele, P.; Huylebroeck, D.; Remacle, J.E. TTRAP, a novel protein that associates with CD40, tumor necrosis factor (TNF) receptor-75 and TNF receptor-associated factors (TRAFs), and that inhibits nuclear factor-kappa B activation. J. Biol. Chem. 2000, 275, 18586–18593. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.-Q.; Wang, J.-J.; Li, W.-J.; Huang, L.; Tian, L.; Xue, J.-L.; Chen, J.-Z.; Jia, W. Cellular protein TTRAP interacts with HIV-1 integrase to facilitate viral integration. Biochem. Biophys. Res. Commun. 2009, 387, 256–260. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Sharma, A.; Ju, L.; Murai, J.; Umans, L.; Vermeire, L.; Pommier, Y.; Takeda, S.; Huylebroeck, D.; Caldecott, K.W.; et al. TDP2 promotes repair of topoisomerase I-mediated DNA damage in the absence of TDP1. Nucleic Acids Res. 2012, 40, 8371–8380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gómez-Herreros, F.; Romero-Granados, R.; Zeng, Z.; Álvarez-Quilón, A.; Quintero, C.; Ju, L.; Umans, L.; Vermeire, L.; Huylebroeck, D.; Caldecott, K.W.; et al. TDP2–Dependent Non-Homologous End-Joining Protects against Topoisomerase II–Induced DNA Breaks and Genome Instability in Cells and In Vivo. PLoS Genet. 2013, 9, e1003226. [Google Scholar] [CrossRef] [PubMed]
- Maede, Y.; Shimizu, H.; Fukushima, T.; Kogame, T.; Nakamura, T.; Miki, T.; Takeda, S.; Pommier, Y.; Murai, J. Differential and common DNA repair pathways for topoisomerase I- and II-targeted drugs in a genetic DT40 repair cell screen panel. Mol. Cancer Ther. 2014, 13, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Schellenberg, M.J.; Appel, C.D.; Adhikari, S.; Robertson, P.D.; Ramsden, D.A.; Williams, R.S. Mechanism of repair of 5′-topoisomerase II-DNA adducts by mammalian tyrosyl-DNA phosphodiesterase 2. Nat. Struct. Mol. Biol. 2012, 19, 1363–1371. [Google Scholar] [CrossRef] [PubMed]
- Shi, K.; Kurahashi, K.; Gao, R.; Tsutakawa, S.E.; Tainer, J.A.; Pommier, Y.; Aihara, H. Structural basis for recognition of 5′-phosphotyrosine adducts by Tdp2. Nat. Struct. Mol. Biol. 2012, 19, 1372–1377. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Schellenberg, M.J.; Huang, S.Y.; Abdelmalak, M.; Marchand, C.; Nitiss, K.C.; Nitiss, J.L.; Williams, R.S.; Pommier, Y. Proteolytic degradation of topoisomerase II (Top2) enables the processing of Top2.DNA and Top2.RNA covalent complexes by tyrosyl-DNA-phosphodiesterase 2 (TDP2). J. Biol. Chem. 2014, 289, 17960–17969. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Huang, S.Y.; Marchand, C.; Pommier, Y. Biochemical characterization of human tyrosyl-DNA phosphodiesterase 2 (TDP2/TTRAP): A Mg2+/Mn2+-dependent phosphodiesterase specific for the repair of topoisomerase cleavage complexes. J. Biol. Chem. 2012, 287, 30842–30852. [Google Scholar] [CrossRef] [PubMed]
- Schellenberg, M.J.; Lieberman, J.A.; Herrero-Ruiz, A.; Butler, L.R.; Williams, J.G.; Munoz-Cabello, A.M.; Mueller, G.A.; London, R.E.; Cortes-Ledesma, F.; Williams, R.S. ZATT (ZNF451)-mediated resolution of topoisomerase 2 DNA-protein cross-links. Science 2017, 357, 1412–1416. [Google Scholar] [CrossRef] [PubMed]
- Zagnoli-Vieira, G.; Caldecott, K.W. TDP2, TOP2, and SUMO: What is ZATT about? Cell Res. 2017, 27, 1405–1406. [Google Scholar] [CrossRef] [PubMed]
- Uuskula-Reimand, L.; Hou, H.; Samavarchi-Tehrani, P.; Rudan, M.V.; Liang, M.; Medina-Rivera, A.; Mohammed, H.; Schmidt, D.; Schwalie, P.; Young, E.J.; et al. Topoisomerase II beta interacts with cohesin and CTCF at topological domain borders. Genome Biol. 2016, 17, 182. [Google Scholar] [PubMed]
- Cowell, I.G.; Tilby, M.J.; Austin, C.A. An overview of the visualisation and quantitation of low and high MW DNA adducts using the trapped in agarose DNA immunostaining (TARDIS) assay. Mutagenesis 2011, 26, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Willmore, E.; Frank, A.J.; Padget, K.; Tilby, M.J.; Austin, C.A. Etoposide targets topoisomerase IIa and IIb in leukemic cells: Isoform-specific cleavable complexes visualized and quantified in situ by a novel immunofluorescence technique. Mol. Pharmacol. 1998, 54, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Frank, A.J.; Proctor, S.J.; Tilby, M.J. Detection and quantification of melphalan-DNA adducts at the single cell level in hematopoietic tumor cells. Blood 1996, 88, 977–984. [Google Scholar] [PubMed]
- Lee, K.C.; Bramley, R.L.; Cowell, I.G.; Jackson, G.H.; Austin, C.A. Proteasomal inhibition potentiates drugs targeting DNA topoisomerase II. Biochem. Pharmacol. 2016, 103, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Errington, F.; Willmore, E.; Leontiou, C.; Tilby, M.J.; Austin, C.A. Differences in the longevity of topo IIa and topo IIb drug-stabilized cleavable complexes and the relationship to drug sensitivity. Cancer Chemother. Pharmacol. 2004, 53, 155–162. [Google Scholar] [PubMed]
- Alchanati, I.; Teicher, C.; Cohen, G.; Shemesh, V.; Barr, H.M.; Nakache, P.; Ben-Avraham, D.; Idelevich, A.; Angel, I.; Livnah, N.; et al. The E3 ubiquitin-ligase Bmi1/Ring1A controls the proteasomal degradation of Top2alpha cleavage complex—A potentially new drug target. PLoS ONE 2009, 4, e8104. [Google Scholar] [CrossRef] [PubMed]
- Martensson, S.; Nygren, J.; Osheroff, N.; Hammarsten, O. Activation of the DNA-dependent protein kinase by drug-induced and radiation-induced DNA strand breaks. Radiat. Res. 2003, 160, 291–301. [Google Scholar] [CrossRef]
- Paull, T.T.; Rogakou, E.P.; Yamazaki, V.; Kirchgessner, C.U.; Gellert, M.; Bonner, W.M. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. CB 2000, 10, 886–895. [Google Scholar] [CrossRef]
- Revet, I.; Feeney, L.; Bruguera, S.; Wilson, W.; Dong, T.K.; Oh, D.H.; Dankort, D.; Cleaver, J.E. Functional relevance of the histone gammaH2Ax in the response to DNA damaging agents. Proc. Natl. Acad. Sci. USA 2011, 108, 8663–8667. [Google Scholar] [CrossRef] [PubMed]
- Rogakou, E.P.; Pilch, D.R.; Orr, A.H.; Ivanova, V.S.; Bonner, W.M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998, 273, 5858–5868. [Google Scholar] [CrossRef] [PubMed]
- Collins, A.R.; Duthie, S.J.; Dobson, V.L. Direct enzymic detection of endogenous oxidative base damage in human lymphocyte DNA. Carcinogenesis 1993, 14, 1733–1735. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Crawford, G.E. DNase-seq: A high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harb. Protoc. 2010, 2010. [Google Scholar] [CrossRef]
- Gomez-Herreros, F.; Zagnoli-Vieira, G. TDP2 suppresses chromosomal translocations induced by DNA topoisomerase II during gene transcription. Nat. Commun. 2017, 8, 233. [Google Scholar] [CrossRef] [PubMed]
- Adachi, N.; Suzuki, H.; Iiizumi, S.; Koyama, H. Hypersensitivity of nonhomologous DNA end-joining mutants to VP-16 and ICRF-193: Implications for the repair of topoisomerase II-mediated DNA damage. J. Biol. Chem. 2003, 278, 35897–35902. [Google Scholar] [CrossRef] [PubMed]
- Neale, M.J.; Pan, J.; Keeney, S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature 2005, 436, 1053–1057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ayene, I.S.; Ford, L.P.; Koch, C.J. Ku protein targeting by Ku70 small interfering RNA enhances human cancer cell response to topoisomerase II inhibitor and gamma radiation. Mol. Cancer Ther. 2005, 4, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Cowell, I.G.; Austin, C.A. Visualization and Quantification of Topoisomerase-DNA Covalent Complexes Using the Trapped in Agarose Immunostaining (TARDIS) Assay. Methods Mol. Biol. 2018, 1703, 301–316. [Google Scholar] [PubMed]




© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, K.C.; Swan, R.L.; Sondka, Z.; Padget, K.; Cowell, I.G.; Austin, C.A. Effect of TDP2 on the Level of TOP2-DNA Complexes and SUMOylated TOP2-DNA Complexes. Int. J. Mol. Sci. 2018, 19, 2056. https://doi.org/10.3390/ijms19072056
Lee KC, Swan RL, Sondka Z, Padget K, Cowell IG, Austin CA. Effect of TDP2 on the Level of TOP2-DNA Complexes and SUMOylated TOP2-DNA Complexes. International Journal of Molecular Sciences. 2018; 19(7):2056. https://doi.org/10.3390/ijms19072056
Chicago/Turabian StyleLee, Ka Cheong, Rebecca L. Swan, Zbyslaw Sondka, Kay Padget, Ian G. Cowell, and Caroline A. Austin. 2018. "Effect of TDP2 on the Level of TOP2-DNA Complexes and SUMOylated TOP2-DNA Complexes" International Journal of Molecular Sciences 19, no. 7: 2056. https://doi.org/10.3390/ijms19072056



