The Small Yeast GTPase Rho5 and Its Dimeric GEF Dck1/Lmo1 Respond to Glucose Starvation
Abstract
:1. Introduction
2. Results
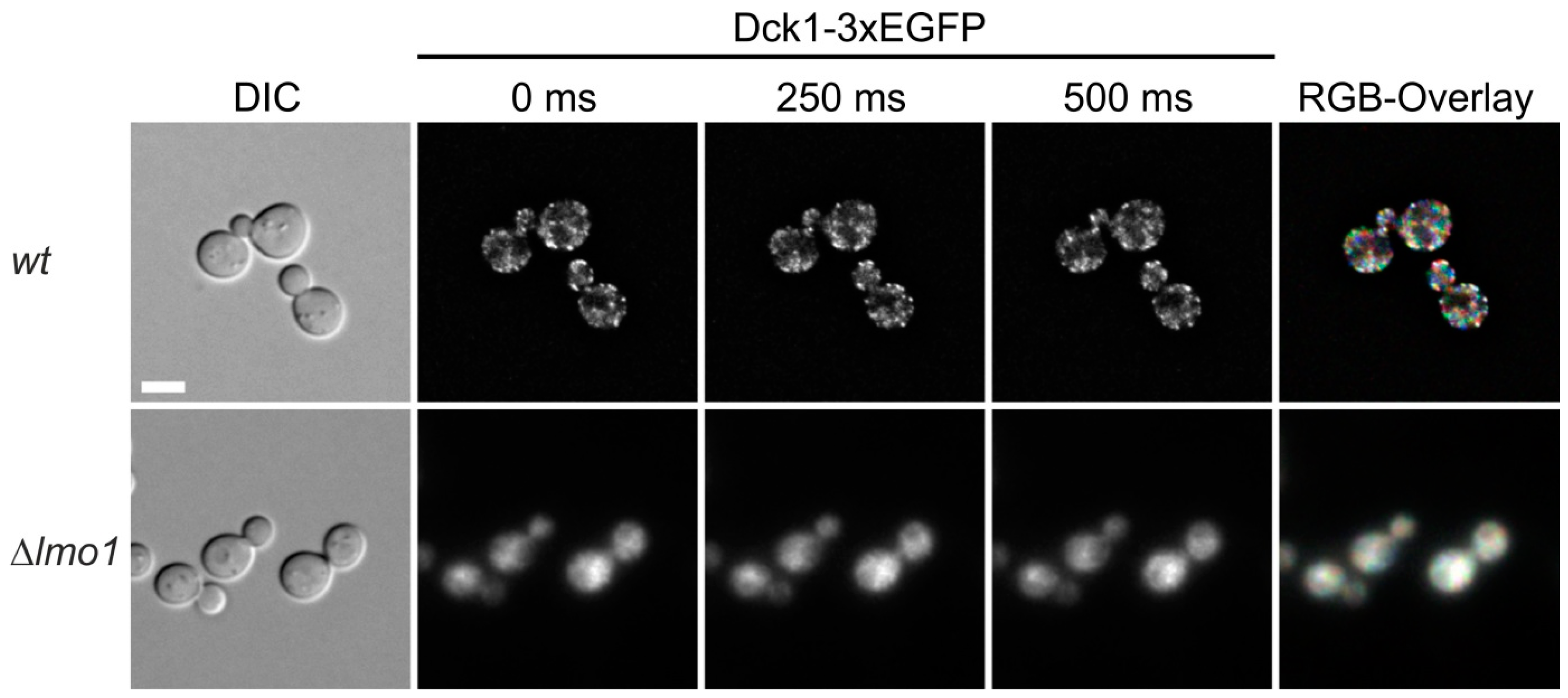
2.1. Dck1 Forms Transient Foci in the Cytoplasm and Rapidly Accumulates at Mitochondria upon Glucose Starvation
2.2. Rho5 Deletions Genetically Interact with Gpr1, Gpa2, and Sch9
2.3. Deletions of RHO5, GPR1, GPA2, or RAS2 Result in Hyper-Resistance towards Cell Wall Stress Agents
2.4. Lack of Dck1 Moderately Affects Actin Dynamics in Budding Yeast Cells
3. Discussion
4. Materials and Methods
4.1. Strains and Growth Conditions
4.2. Construction of Plasmids, Deletion Mutants and Gene Tagging
4.3. Strains and Experimental Conditions for Fluorescence Microscopy, Image Acquisition and Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AMPK | 5′ adenosine monophosphate activated kinase |
| 2-DOG | 2-deoxy glucose |
| GEF | GDP/GTP exchange factor |
| PKA | protein kinase A |
| ROS | reactive oxygen species |
| SNF | sucrose non-fermenters |
| TORC1 | target of rapamycin complex 1 |
| vATPase | vacuolar ATPase complex |
References
- Haga, R.B.; Ridley, A.J. Rho gtpases: Regulation and roles in cancer cell biology. Small GTPases 2016, 7, 207–221. [Google Scholar] [CrossRef] [PubMed]
- Zamboni, V.; Jones, R.; Umbach, A.; Ammoni, A.; Passafaro, M.; Hirsch, E.; Merlo, G.R. Rho gtpases in intellectual disability: From genetics to therapeutic opportunities. Int. J. Mol. Sci. 2018, 19, 1821. [Google Scholar] [CrossRef] [PubMed]
- Hall, A. Rho family gtpases. Biochem. Soc. Trans. 2012, 40, 1378–1382. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, H.P.; Huppert, S.; Lorberg, A.; Heinisch, J.J. Rho5p downregulates the yeast cell integrity pathway. J. Cell Sci. 2002, 115, 3139–3148. [Google Scholar] [PubMed]
- Heinisch, J.J.; Rodicio, R. Protein kinase c in fungi-more than just cell wall integrity. FEMS Microbiol. Rev. 2018, 42. [Google Scholar] [CrossRef] [PubMed]
- Annan, R.B.; Wu, C.; Waller, D.D.; Whiteway, M.; Thomas, D.Y. Rho5p is involved in mediating the osmotic stress response in saccharomyces cerevisiae, and its activity is regulated via msi1p and npr1p by phosphorylation and ubiquitination. Eukaryot. Cell 2008, 7, 1441–1449. [Google Scholar] [CrossRef] [PubMed]
- Mao, K.; Klionsky, D.J. Mapks regulate mitophagy in Saccharomyces cerevisiae. Autophagy 2011, 7, 1564–1565. [Google Scholar] [CrossRef] [PubMed]
- Mao, K.; Wang, K.; Zhao, M.; Xu, T.; Klionsky, D.J. Two mapk-signaling pathways are required for mitophagy in saccharomyces cerevisiae. J. Cell Biol. 2011, 193, 755–767. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.; Kang, P.J.; Park, H.O. The rho5 gtpase is necessary for oxidant-induced cell death in budding yeast. Proc. Natl. Acad. Sci. USA 2008, 105, 1522–1527. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, H.P.; Jendretzki, A.; Wittland, J.; Wiechert, J.; Heinisch, J.J. Identification of dck1 and lmo1 as upstream regulators of the small gtpase rho5 in Saccharomyces cerevisiae. Mol. Microbiol. 2015, 96, 306–324. [Google Scholar] [CrossRef] [PubMed]
- Broach, J.R. Nutritional control of growth and development in yeast. Genetics 2012, 192, 73–105. [Google Scholar] [CrossRef] [PubMed]
- Rodicio, R.; Heinisch, J. Carbohydrate Metabolism in Wine Yeast. In Biology of Microorganisms on Grapes, in must and Wine; König, H., Unden, G., Fröhlich, J., Eds.; Springer: Heidelberg/Berlin, Germany, 2017; pp. 189–213. [Google Scholar]
- Backhaus, K.; Rippert, D.; Heilmann, C.J.; Sorgo, A.G.; de Koster, C.G.; Klis, F.M.; Rodicio, R.; Heinisch, J.J. Mutations in snf1 complex genes affect yeast cell wall strength. Eur. J. Cell Biol. 2013, 92, 383–395. [Google Scholar] [CrossRef] [PubMed]
- Rippert, D.; Backhaus, K.; Rodicio, R.; Heinisch, J.J. Cell wall synthesis and central carbohydrate metabolism are interconnected by the snf1/mig1 pathway in Kluyveromyces lactis. Eur. J. Cell Biol. 2017, 96, 70–81. [Google Scholar] [CrossRef] [PubMed]
- Park, J.I.; Collinson, E.J.; Grant, C.M.; Dawes, I.W. Rom2p, the rho1 gtp/gdp exchange factor of Saccharomyces cerevisiae, can mediate stress responses via the ras-camp pathway. J. Biol. Chem. 2005, 280, 2529–2535. [Google Scholar] [CrossRef] [PubMed]
- Garcia, R.; Bravo, E.; Diez-Muniz, S.; Nombela, C.; Rodriguez-Pena, J.M.; Arroyo, J. A novel connection between the cell wall integrity and the pka pathways regulates cell wall stress response in yeast. Sci. Rep. 2017, 7, 5703. [Google Scholar] [CrossRef] [PubMed]
- Ho, J.; Bretscher, A. Ras regulates the polarity of the yeast actin cytoskeleton through the stress response pathway. Mol. Biol. Cell 2001, 12, 1541–1555. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, N.; Wu, D.; Bi, Q.; Meng, S. The longevity of tor1delta, sch9delta, and ras2delta mutants depends on actin dynamics in Saccharomyces cerevisiae. Cell Biosci. 2015, 5, 18. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Lopez, J.L.; Laboy, R.; Jaimes-Miranda, F.; Garay, E.; DeLuna, A.; Funes, S. Slm35 links mitochondrial stress response and longevity through tor signaling pathway. Aging 2016, 8, 3255–3271. [Google Scholar] [CrossRef] [PubMed]
- Dechant, R.; Binda, M.; Lee, S.S.; Pelet, S.; Winderickx, J.; Peter, M. Cytosolic ph is a second messenger for glucose and regulates the pka pathway through v-atpase. EMBO J. 2010, 29, 2515–2526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gourlay, C.W.; Ayscough, K.R. Actin-induced hyperactivation of the ras signaling pathway leads to apoptosis in saccharomyces cerevisiae. Mol. Cell Biol. 2006, 26, 6487–6501. [Google Scholar] [CrossRef] [PubMed]
- Deprez, M.A.; Eskes, E.; Winderickx, J.; Wilms, T. The torc1-sch9 pathway as a crucial mediator of chronological lifespan in the yeast saccharomyces cerevisiae. FEMS Yeast Res. 2018, 18. [Google Scholar] [CrossRef] [PubMed]
- Deprez, M.A.; Eskes, E.; Wilms, T.; Ludovico, P.; Winderickx, J. Ph homeostasis links the nutrient sensing pka/torc1/sch9 menage-a-trois to stress tolerance and longevity. Microb. Cell 2018, 5, 119–136. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Tang, Y.; Quan, Z.; Zhang, Z.; Oliver, S.G.; Zhang, N. Chronological lifespan in yeast is dependent on the accumulation of storage carbohydrates mediated by yak1, mck1 and rim15 kinases. PLoS Genet. 2016, 12, e1006458. [Google Scholar] [CrossRef] [PubMed]
- Patel, M.; Pelletier, A.; Cote, J.F. Opening up on elmo regulation: New insights into the control of rac signaling by the dock180/elmo complex. Small GTPases 2011, 2, 268–275. [Google Scholar] [CrossRef] [PubMed]
- Grimsley, C.M.; Kinchen, J.M.; Tosello-Trampont, A.C.; Brugnera, E.; Haney, L.B.; Lu, M.; Chen, Q.; Klingele, D.; Hengartner, M.O.; Ravichandran, K.S. Dock180 and elmo1 proteins cooperate to promote evolutionarily conserved rac-dependent cell migration. J. Biol. Chem. 2004, 279, 6087–6097. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, H.; Kiyokawa, E.; Tanaka, S.; Nagashima, K.; Gotoh, N.; Shibuya, M.; Kurata, T.; Matsuda, M. Dock180, a major crk-binding protein, alters cell morphology upon translocation to the cell membrane. Mol. Cell Biol. 1996, 16, 1770–1776. [Google Scholar] [CrossRef] [PubMed]
- Peppler, W.T.; MacPherson, R.E. Rac1 is a novel regulator of exercise-induced glucose uptake. J. Physiol. 2016, 594, 7155–7156. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.R.; Rajgor, D.; Hanley, J.G. Differential regulation of the rac1 gtpase-activating protein (gap) bcr during oxygen/glucose deprivation in hippocampal and cortical neurons. J. Biol. Chem. 2017, 292, 20173–20183. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, B.J.; Zhu, Y.; Lu, Q. Rho gtpases as therapeutic targets in alzheimer′s disease. Alzheimers Res. Ther. 2017, 9, 97. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Zhou, M.; Xie, J.; Chao, P.; Feng, Q.; Wu, J. High glucose levels promote the proliferation of breast cancer cells through gtpases. Breast Cancer (Dove Med. Press) 2017, 9, 429–436. [Google Scholar] [CrossRef] [PubMed]
- Sylow, L.; Moller, L.L.V.; Kleinert, M.; D′Hulst, G.; De Groote, E.; Schjerling, P.; Steinberg, G.R.; Jensen, T.E.; Richter, E.A. Rac1 and ampk account for the majority of muscle glucose uptake stimulated by ex vivo contraction but not in vivo exercise. Diabetes 2017, 66, 1548–1559. [Google Scholar] [CrossRef] [PubMed]
- Nazarko, V.Y.; Thevelein, J.M.; Sibirny, A.A. G-protein-coupled receptor gpr1 and g-protein gpa2 of camp-dependent signaling pathway are involved in glucose-induced pexophagy in the yeast saccharomyces cerevisiae. Cell Biol. Int. 2008, 32, 502–504. [Google Scholar] [CrossRef] [PubMed]
- Shi, L. Dock protein family in brain development and neurological disease. Commun. Integr. Biol. 2013, 6, e26839. [Google Scholar] [CrossRef] [PubMed]
- Ayscough, K.R. Endocytosis and the development of cell polarity in yeast require a dynamic f-actin cytoskeleton. Curr. Biol. 2000, 10, 1587–1590. [Google Scholar] [CrossRef]
- Goode, B.L.; Eskin, J.A.; Wendland, B. Actin and endocytosis in budding yeast. Genetics 2015, 199, 315–358. [Google Scholar] [CrossRef] [PubMed]
- Ayscough, K.R.; Stryker, J.; Pokala, N.; Sanders, M.; Crews, P.; Drubin, D.G. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-a. J. Cell Biol. 1997, 137, 399–416. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.A.; Hall, A.E.; Rose, M.D. Membrane curvature directs the localization of cdc42p to novel foci required for cell-cell fusion. J. Cell Biol. 2017, 216, 3971–3980. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, D.; Lickfeld, M.; Warnsmann, V.; Wiechert, J.; Jendretzki, A.; Schmitz, H.P. The small gtp-binding proteins agrho2 and agrho5 regulate tip-branching, maintenance of the growth axis and actin-ring-integrity in the filamentous fungus Ashbya gossypii. PLoS ONE 2014, 9, e106236. [Google Scholar] [CrossRef] [PubMed]
- Conrad, M.; Schothorst, J.; Kankipati, H.N.; Van Zeebroeck, G.; Rubio-Texeira, M.; Thevelein, J.M. Nutrient sensing and signaling in the yeast saccharomyces cerevisiae. FEMS Microbiol. Rev. 2014, 38, 254–299. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, A.; Hall, M.N. Nutrient sensing and tor signaling in yeast and mammals. EMBO J. 2017, 36, 397–408. [Google Scholar] [CrossRef] [PubMed]
- Morano, K.A.; Grant, C.M.; Moye-Rowley, W.S. The response to heat shock and oxidative stress in saccharomyces cerevisiae. Genetics 2012, 190, 1157–1195. [Google Scholar] [CrossRef] [PubMed]
- Thevelein, J.M.; de Winde, J.H. Novel sensing mechanisms and targets for the camp-protein kinase a pathway in the yeast saccharomyces cerevisiae. Mol. Microbiol. 1999, 33, 904–918. [Google Scholar] [CrossRef] [PubMed]
- Heinisch, J.J.; Brandt, R. Signaling pathways and posttranslational modifications of tau in alzheimer′s disease: The humanization of yeast cells. Microb. Cell 2016, 3, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Rodicio, R.; Heinisch, J.J. Yeast on the milky way: Genetics, physiology and biotechnology of kluyveromyces lactis. Yeast 2013, 30, 165–177. [Google Scholar] [CrossRef] [PubMed]
- Arvanitidis, A.; Heinisch, J.J. Studies on the function of yeast phosphofructokinase subunits by in vitro mutagenesis. J. Biol. Chem. 1994, 269, 8911–8918. [Google Scholar] [PubMed]
- Kirchrath, L.; Lorberg, A.; Schmitz, H.P.; Gengenbacher, U.; Heinisch, J.J. Comparative genetic and physiological studies of the map kinase mpk1p from kluyveromyces lactis and saccharomyces cerevisiae. J. Mol. Biol. 2000, 300, 743–758. [Google Scholar] [CrossRef] [PubMed]
- Schacherer, J.; Ruderfer, D.M.; Gresham, D.; Dolinski, K.; Botstein, D.; Kruglyak, L. Genome-wide analysis of nucleotide-level variation in commonly used Saccharomyces cerevisiae strains. PLoS ONE 2007, 2, e322. [Google Scholar] [CrossRef] [PubMed]
- Rose, M.D.; Winston, F.; Hieter, P. Methods in Yeast Genetics; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 1990. [Google Scholar]
- Jendretzki, A.; Ciklic, I.; Rodicio, R.; Schmitz, H.P.; Heinisch, J.J. Cyk3 acts in actomyosin ring independent cytokinesis by recruiting inn1 to the yeast bud neck. Mol. Genet. Genom. 2009, 282, 437–451. [Google Scholar] [CrossRef] [PubMed]
- Straede, A.; Corran, A.; Bundy, J.; Heinisch, J.J. The effect of tea tree oil and antifungal agents on a reporter for yeast cell integrity signalling. Yeast 2007, 24, 321–334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rothstein, R. Targeting, disruption, replacement, and allele rescue: Integrative DNA transformation in yeast. Methods Enzymol. 1991, 194, 281–301. [Google Scholar] [PubMed]
- Longtine, M.S.; McKenzie, A., 3rd; Demarini, D.J.; Shah, N.G.; Wach, A.; Brachat, A.; Philippsen, P.; Pringle, J.R. Additional modules for versatile and economical pcr-based gene deletion and modification in saccharomyces cerevisiae. Yeast 1998, 14, 953–961. [Google Scholar] [CrossRef]
- Gueldener, U.; Heinisch, J.; Koehler, G.J.; Voss, D.; Hegemann, J.H. A second set of loxp marker cassettes for cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 2002, 30, e23. [Google Scholar] [CrossRef] [PubMed]
- Maeder, C.I.; Hink, M.A.; Kinkhabwala, A.; Mayr, R.; Bastiaens, P.I.; Knop, M. Spatial regulation of fus3 map kinase activity through a reaction-diffusion mechanism in yeast pheromone signalling. Nat. Cell Biol. 2007, 9, 1319–1326. [Google Scholar] [CrossRef] [PubMed]
- Rueden, C.T.; Schindelin, J.; Hiner, M.C.; DeZonia, B.E.; Walter, A.E.; Arena, E.T.; Eliceiri, K.W. Imagej2: Imagej for the next generation of scientific image data. BMC Bioinform. 2017, 18, 529. [Google Scholar] [CrossRef] [PubMed]
- Ihaka, R.; Gentleman, R. A language for data analysis and graphics. J. Comput. Graph. Stat. 1996, 5, 299–314. [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. 2011. Available online: http://www.R-project.org/ (accessed on 10 June 2018).
- R Development Core Team. Rstudio: Integrated Development for R. 2016. Available online: http://www.R-project.org/ (accessed on 10 June 2018).
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009. [Google Scholar]
- Wickham, H. The split-apply-combine strategy for data analysis. J. Stat. Softw. 2011, 40, 1–29. [Google Scholar] [CrossRef]






| Strain | Genotype | Source |
|---|---|---|
| DHD5 | MATa/α ura3-52/ura3-52 leu2-3,112/leu2-3,112 his3-11,15/his3-11,15 | [47] |
| HAJ6-A | MATa segregant from DHD5 | [50] |
| HAJ6-B | MATα segregant from DHD5 | [50] |
| DAJ119 | as DHD5 except dck1::kanMX/DCK1 sch9::SkHIS3/SCH9 | This study |
| DAJ128 | as DHD5 except lmo1::kanMX/LMO1 sch9::SkHIS3/SCH9 | This study |
| DAJ138 | as DHD5 except rho5::KanMX/RHO5 sch9::SkHIS3/SCH9 | This study |
| DAJ139 | as DHD5 except ras1::KanMX /RAS1 sch9::SkHIS3/SCH9 | This study |
| DAJ140 | as DHD5 except ras2::KanMX /RAS2 sch9::SkHIS3/SCH9 | This study |
| DAJ144 | as DHD5 except rho5::KanMX/RHO5 gpa2::KanMX/GPA2 | This study |
| DAJ145 | as DHD5 except rho5::KanMX/RHO5 gpr1::KanMX/GPR1 | This study |
| HAJ152-A | as HAJ6-A except DCK1-3xmyEGFP::SpHIS5 | [10] |
| HAJ187-A | as HAJ6-A except gpr1::KanMX | This study |
| HAJ188-A | as HAJ6-A except gpa2::KanMX | This study |
| HAJ201-A | as HAJ6-A except lmo1::KanMX | This study |
| HAJ201-B | as HAJ6-B except lmo1::KanMX | This study |
| HAJ204-A | as HAJ6-A except gpr1::KanMX DCK1-3xEGFP::SpHIS5 | This study |
| HAJ205-A | as HAJ6-A except gpa2::KanMX DCK1-3xEGFP::SpHIS5 | This study |
| HAJ206-A | as HAJ6-A except sch9::KanMX DCK1-3xEGFP::SpHIS5 | This study |
| HAJ207-A | as HAJ6-A except lmo1::KanMX DCK1-3xEGFP::SpHIS5 | This study |
| HAJ216-A | as HAJ6-B except rho5::KanMX | [10] |
| HAJ217-A | as HAJ6-A except ras1::KanMX | This study |
| HAJ218-A | as HAJ6-A except ras2::KanMX | This study |
| HCS076-A | as HAJ6-A except rho5::KanMX IDP1-mCherry::SkHIS3 | This study |
| HMZ18-A | as HAJ6-A except lmo1::SkHIS3 | This study |
| HOD294.2 | as DHD5 except rho5::kanMX/RHO5 snf1::SpHIS5/SNF1 | This study |
| HOD320 | as DHD5 except rho5::kanMX/RHO5 ras2::SkHIS3/RAS2 | This study |
| HOD294.2-1A | as HAJ6-A | This study |
| HOD294.2-1B | as HAJ6-B | This study |
| HOD294.2-2B | as HAJ6-B except rho5::KanMX | This study |
| HOD294.2-3B | as HAJ6-A except rho5::KanMX | This study |
| HOD309-1D | as HAJ6-A except rho5::KanMX CAP2-EGFP::SpHIS5 | This study |
| HOD310-1B | as HAJ6-A except CAP2-EGFP::SpHIS5 | This study |
| HOD310-4A | as HAJ6-A except lmo1::SkHIS3 CAP2-EGFP::SpHIS5 | This study |
| HOD314-2C | as HAJ6-A except dck1::KlURA3 CAP2-EGFP::SpHIS5 | This study |
| HOD314-5A | as HAJ6-A except dck1::KlURA3 | This study |
| HOD314-8A | as HAJ6-B except dck1::KlURA3 | This study |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schmitz, H.-P.; Jendretzki, A.; Sterk, C.; Heinisch, J.J. The Small Yeast GTPase Rho5 and Its Dimeric GEF Dck1/Lmo1 Respond to Glucose Starvation. Int. J. Mol. Sci. 2018, 19, 2186. https://doi.org/10.3390/ijms19082186
Schmitz H-P, Jendretzki A, Sterk C, Heinisch JJ. The Small Yeast GTPase Rho5 and Its Dimeric GEF Dck1/Lmo1 Respond to Glucose Starvation. International Journal of Molecular Sciences. 2018; 19(8):2186. https://doi.org/10.3390/ijms19082186
Chicago/Turabian StyleSchmitz, Hans-Peter, Arne Jendretzki, Carolin Sterk, and Jürgen J. Heinisch. 2018. "The Small Yeast GTPase Rho5 and Its Dimeric GEF Dck1/Lmo1 Respond to Glucose Starvation" International Journal of Molecular Sciences 19, no. 8: 2186. https://doi.org/10.3390/ijms19082186






