Transcriptome Analysis Reveals Multiple Hormones, Wounding and Sugar Signaling Pathways Mediate Adventitious Root Formation in Apple Rootstock
Abstract
:1. Introduction
2. Results
2.1. Morphological Observations of the Process of AR Formation in Stem Cuttings of ‘T337’ Apple Rootstocks
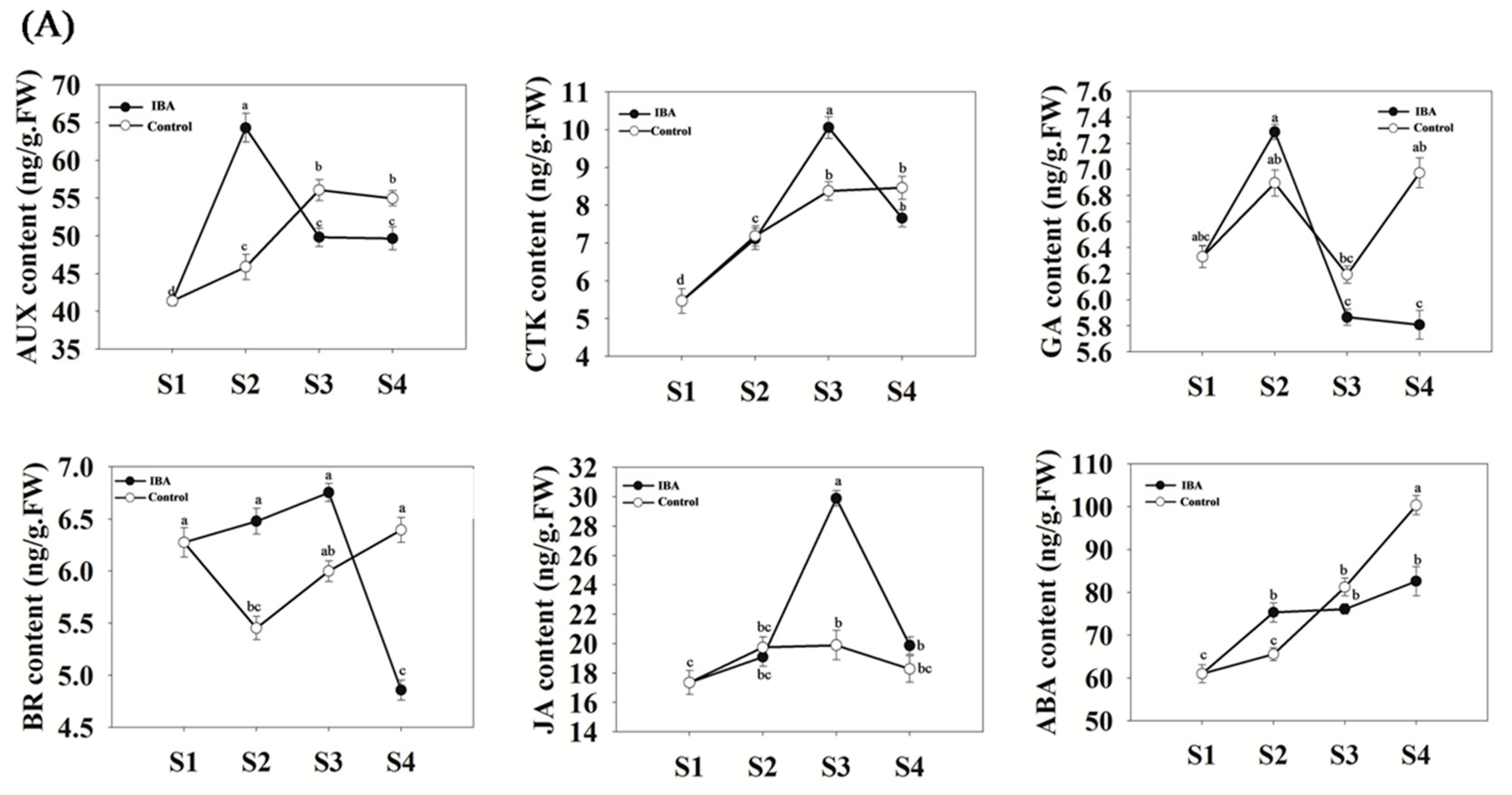
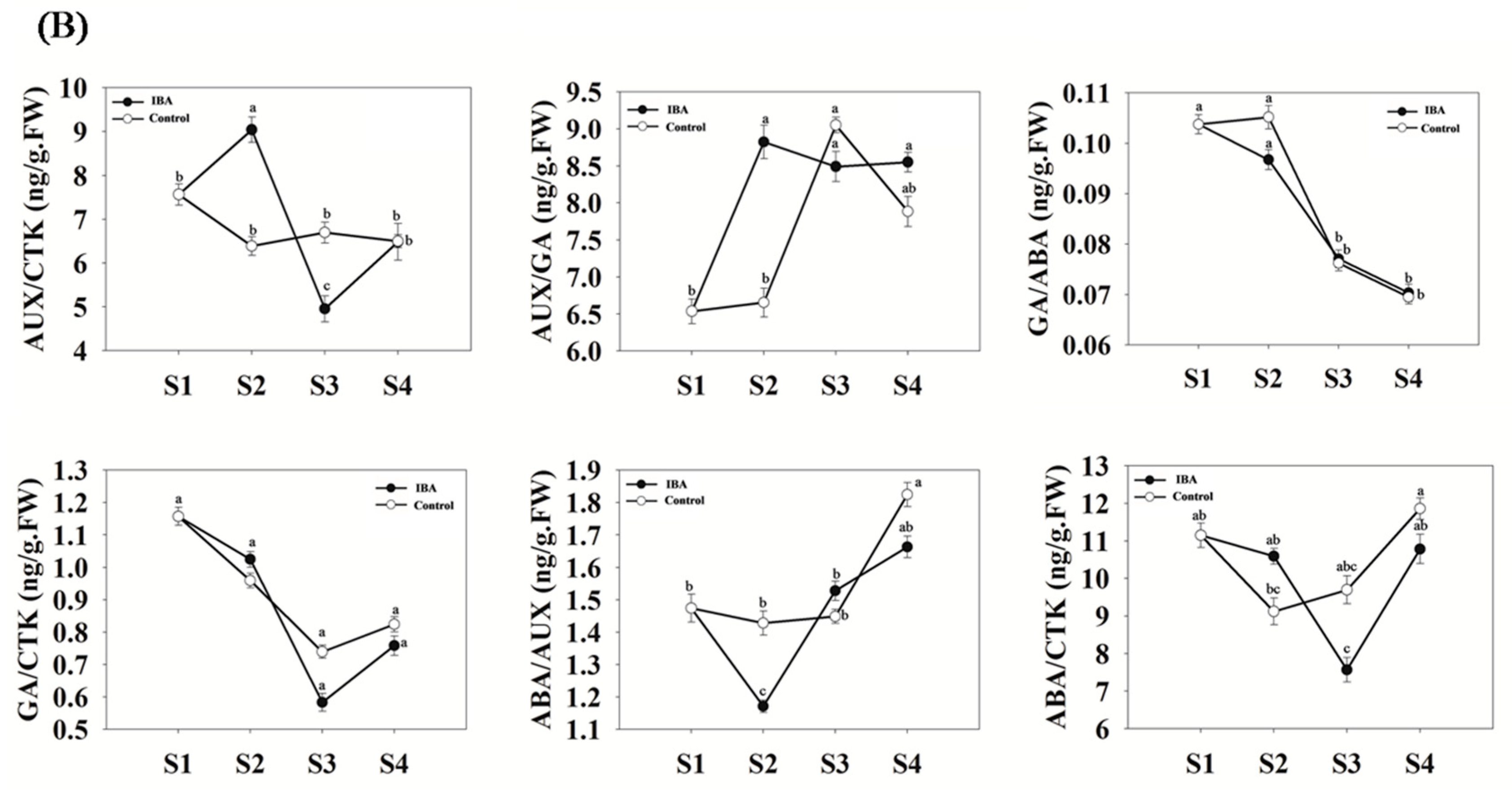
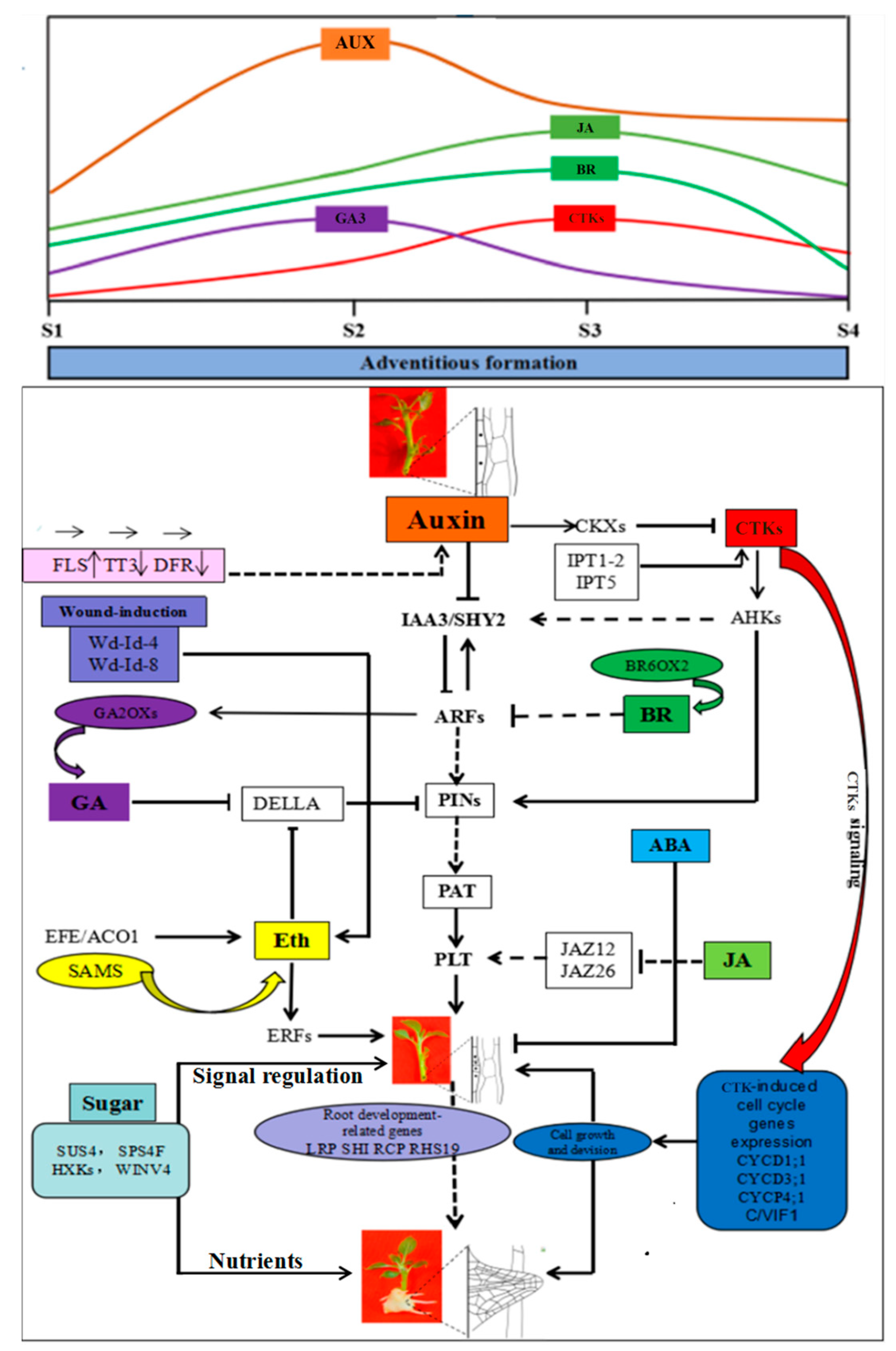
2.2. Quantitative Analyses of Hormone Levels in ‘T337’ Stem Cuttings during AR Formation
2.3. Expression Analysis of Related Genes with RNA Deep Sequencing and RT-qPCR
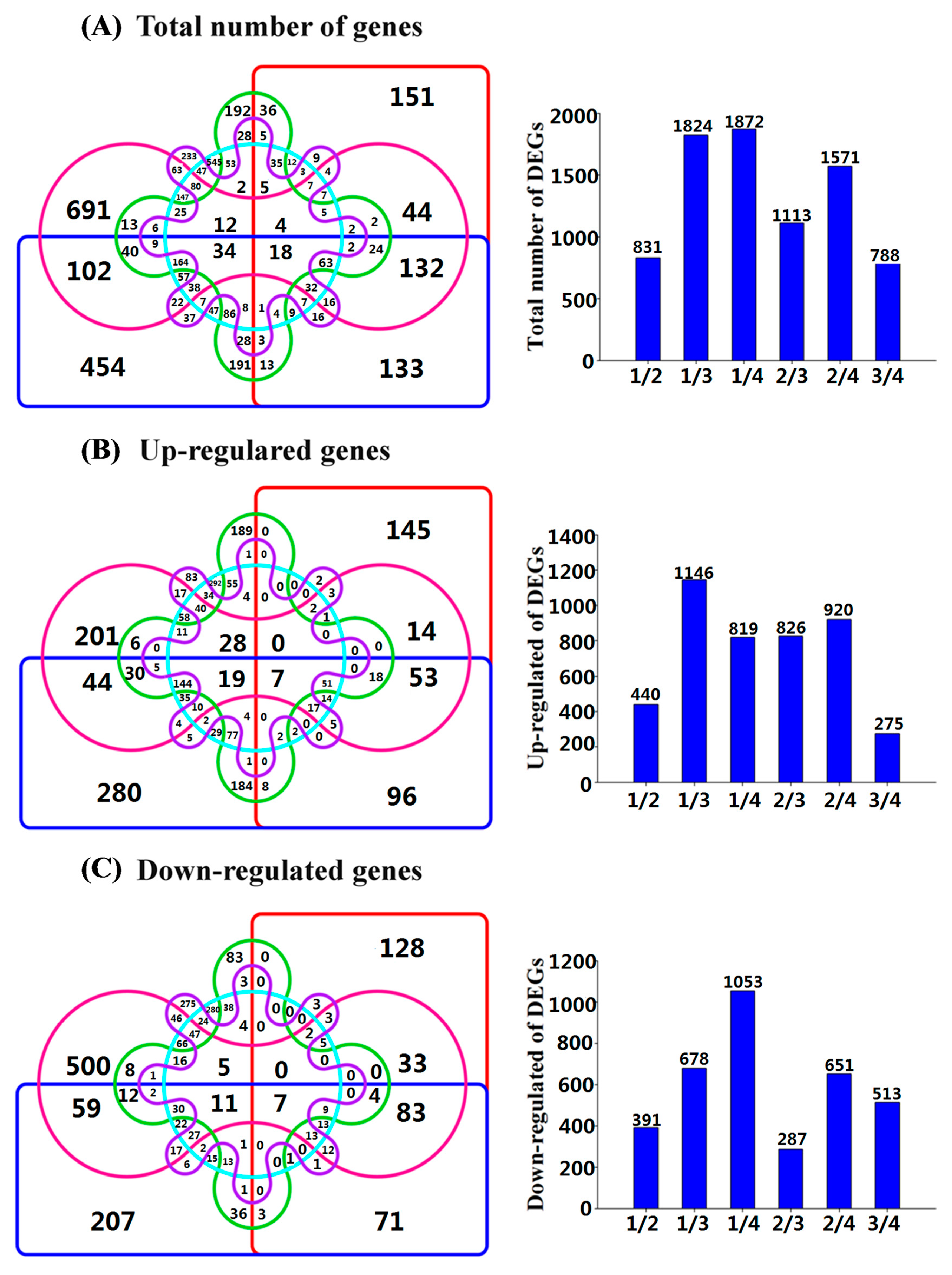
2.4. The Classification of Differentially Expressed Genes (DEGs)
2.4.1. Expression Profiles of Auxin-Related Genes
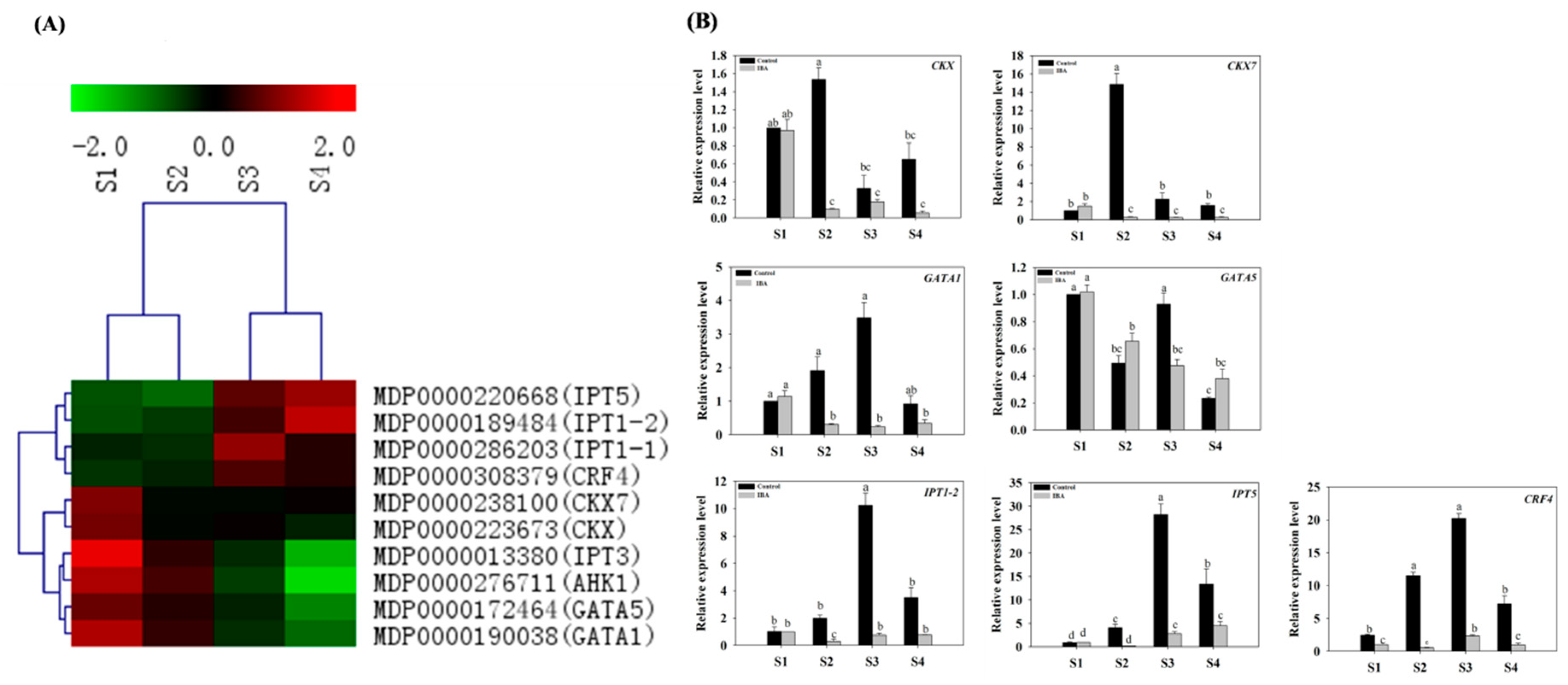
2.4.2. Expression Profiles of CTK-Related Genes
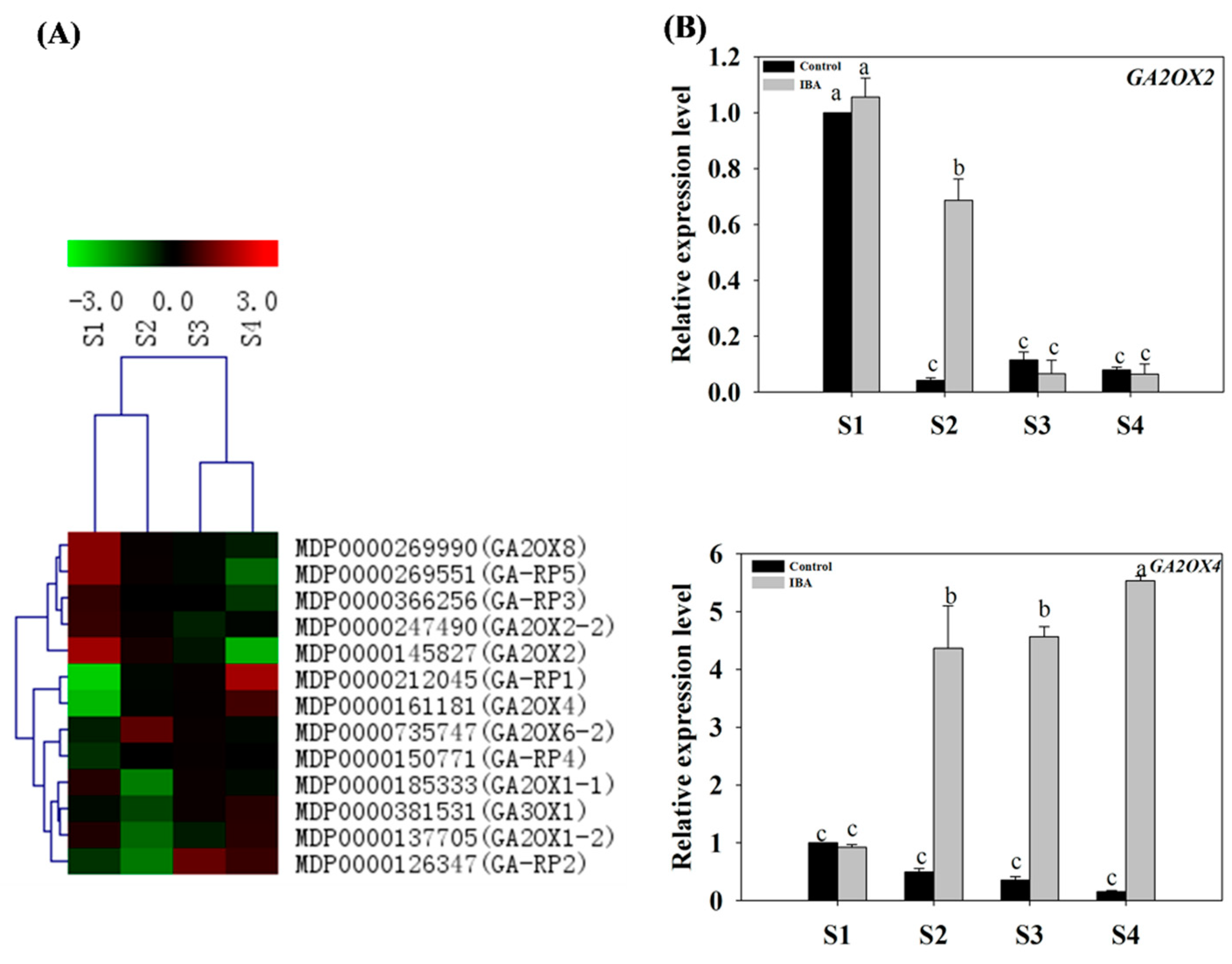
2.4.3. Expression Profiles of GA-Related Genes
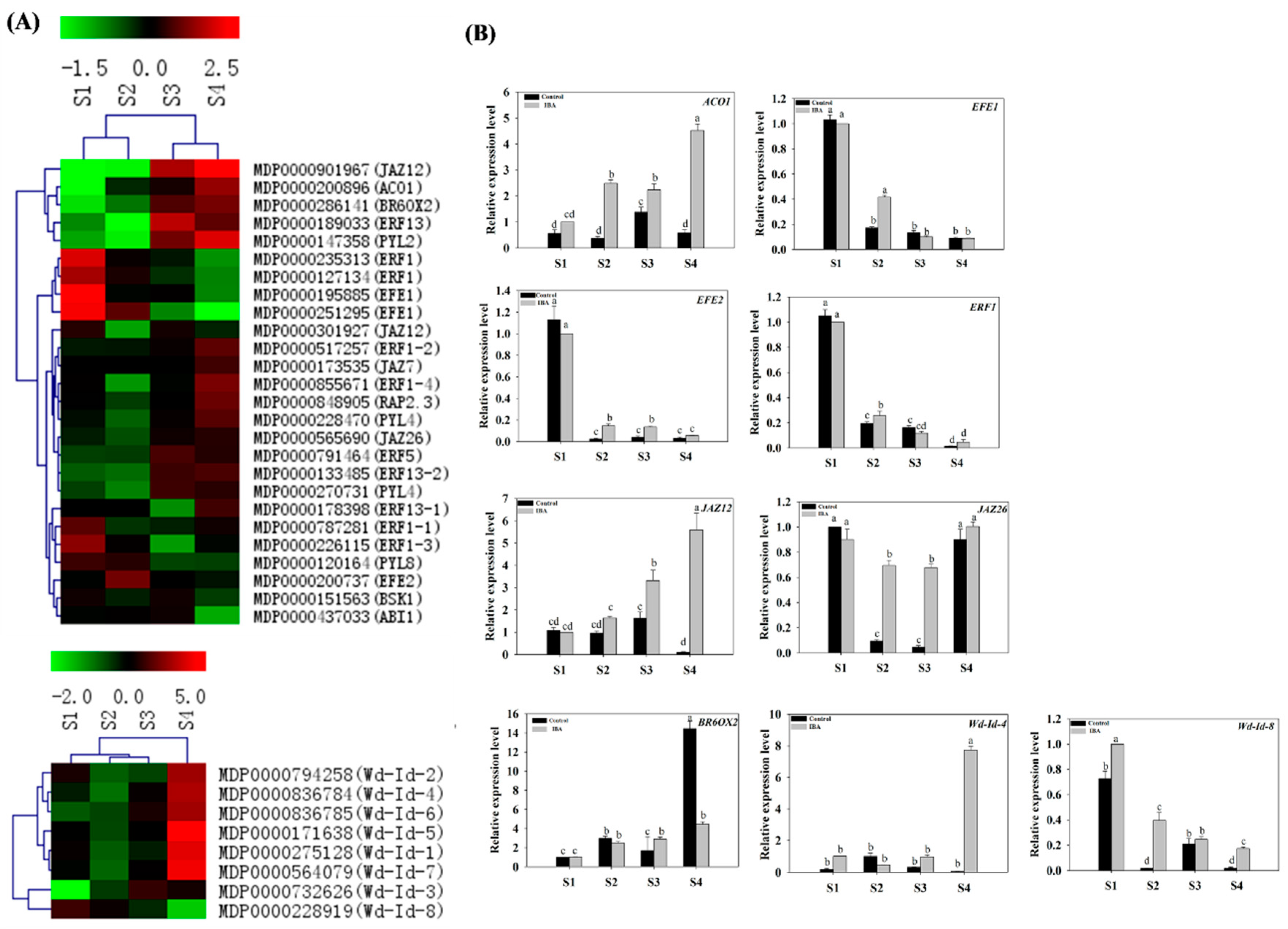
2.4.4. Expression Profiles of Eth-, JA-, and BR-Related Genes
2.4.5. Expression Profiles of Wound Induction-Related Genes
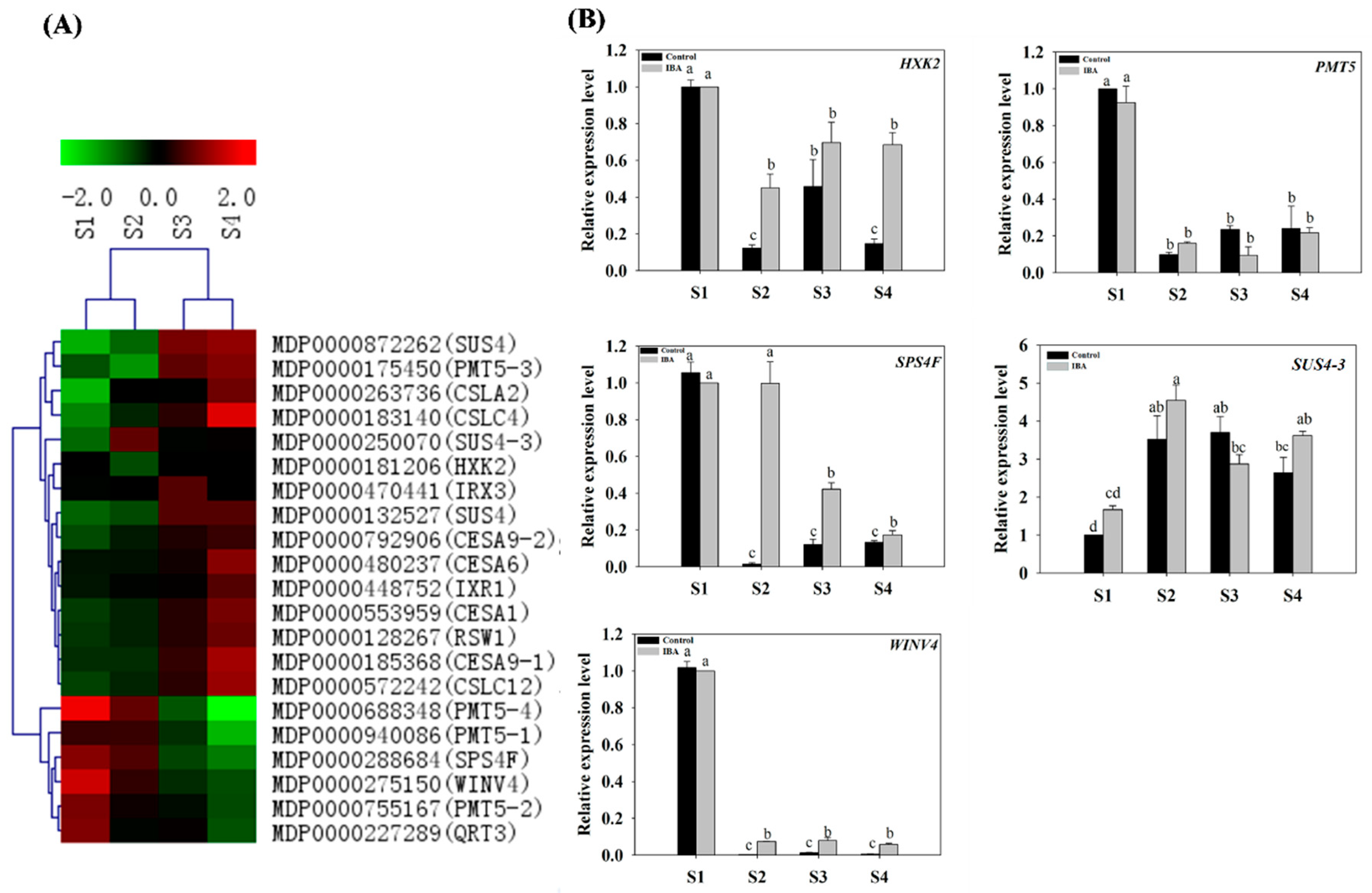
2.4.6. Expression Profiles of Sugar Metabolism-Related Genes
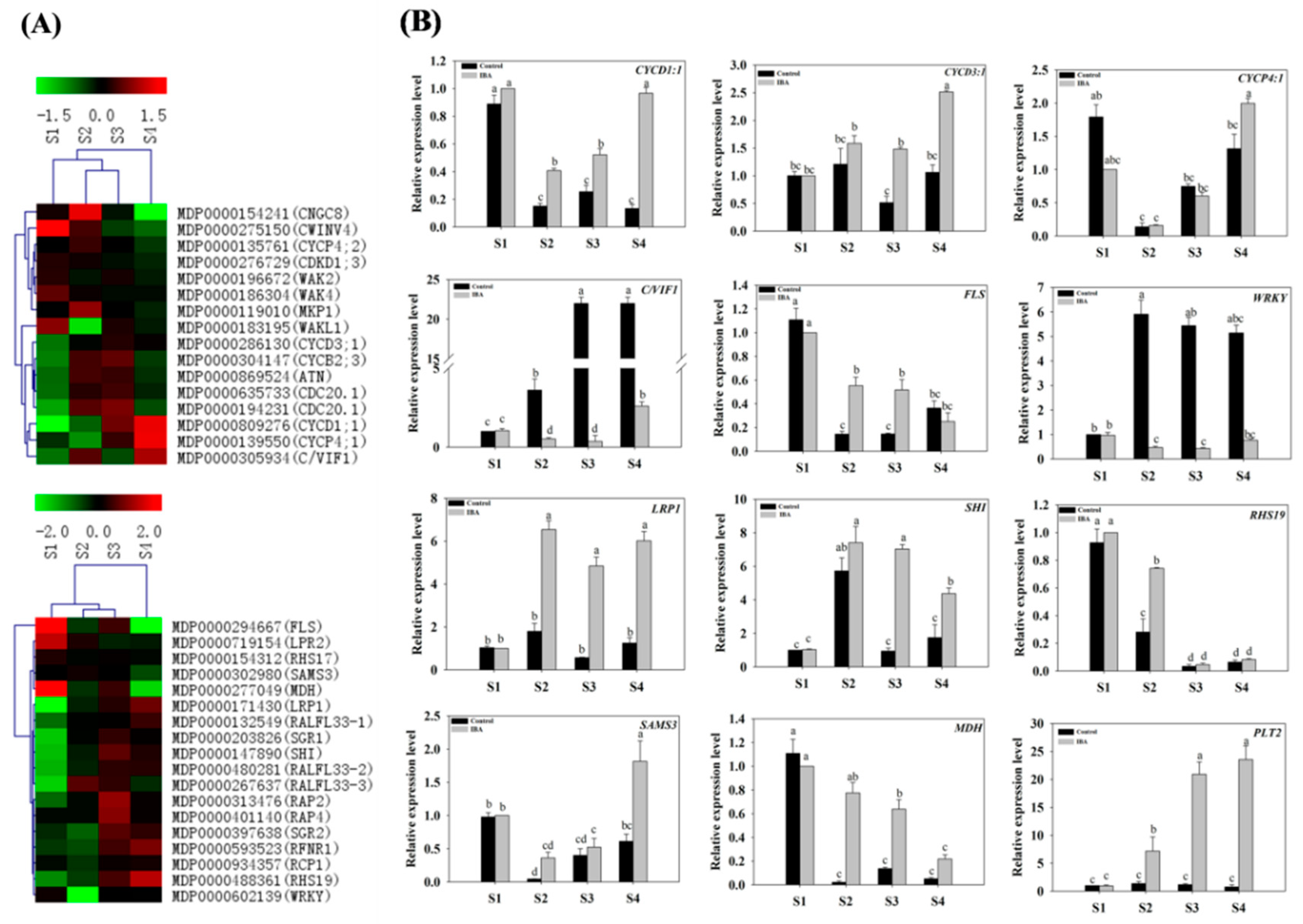
2.4.7. Expression of Genes Related to Root Development, the Cell Cycle, and Proteinase Activity
- The Expression Profiles of Root Development-Related Genes
- The Expression Profiles of Cell-Cycle-Related Genes
- The Expression Profiles of Protease-Related Genes
3. Discussion
3.1. Morphological and Anatomical Aspects of AR Formation in ‘T337’ Apple Rootstock
3.2. Multiple Hormones Signaling Pathways Interact with Auxin Signaling to Mediate IBA-Induced AR Formation in ‘T337’ Apple Rootstocks
3.3. Auxin Is Involved in the Regulation of AR Formation
3.4. Cytokinin Is Involved in the Regulation of AR Formation
3.5. Gibberellin Is Involved in the Regulation of AR Formation
3.6. Ethlene, Jasmonic Acid, and Brassinolide etc. Are Involved in the Regulation of AR Formation
3.7. Wound Signaling and AR Formation in ‘T337’ Apple Rootstocks
3.8. Sugar Signaling Mediated AR Formation in ‘T337’ Apple Rootstock
3.9. Cell Cycle- and Root Development-Related Gene Expression during AR Formation
4. Material and Methods
4.1. Plant Material, Growth Conditions, and Treatments
4.2. Paraffin Section
4.3. Extraction and Measurement of Hormones in Cuttings
4.4. RNA Extraction and cDNA Synthesis
4.5. cDNA Library Construction and RNA Deep Sequencing
- Statistical Analysis of Clean Reads
- Mapping Rate Statistics
- The Distribution of Reads in The Reference Genome
- Detection of New Transcription Sites
- Gene Expression Abundance Statistics
- Differentially Expressed Genes (DEGs)
4.6. RT-qPCR Analysis
4.7. Time-Series Cluster Analysis and Hierarchical Clustering
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AR | Adventitious root |
| LR | Lateral root |
| IBA | Indole-3-butyric acid |
| IAA | Indole-3-acetic acid |
| ELISA | enzyme linked immunosorbent assay |
| PI | primordium initiation. |
| RP | root primordium |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| FPKM | Fragment Per Kilo bases per Million |
| DEGs | Differentially Expressed Genes |
| AUX | Auxin |
| CTK | Cytokinin |
| GA | Gibberellic acid |
| Eth | Ethylene |
| BR | Brassinolide |
| JA | Jasmonic acid |
| ABA | Abscisic acid |
References
- Li, S.W.; Shi, R.F.; Leng, Y. De Novo Characterization of the Mung Bean Transcriptome and Transcriptomic Analysis of Adventitious Rooting in Seedlings Using RNA-Seq. PLoS ONE 2015, 10, e132969. [Google Scholar] [CrossRef] [PubMed]
- Bellini, C.; Pacurar, D.I.; Perrone, I. Adventitious Roots and Lateral Roots: Similarities and Differences. Annu. Rev. Plant Biol. 2014, 65, 639. [Google Scholar] [CrossRef] [PubMed]
- Ludwig-Müller, J.; Vertocnik, A.; Town, C.D. Analysis of indole-3-butyric acid-induced adventitious root formation on Arabidopsis stem segments. J. Exp. Bot. 2005, 56, 2095. [Google Scholar] [CrossRef] [PubMed]
- Della, R.F.; Fattorini, L.; D’Angeli, S.; Veloccia, A.; Falasca, G.; Altamura, M.M. Auxin and cytokinin control formation of the quiescent centre in the adventitious root apex of Arabidopsis. Ann. Bot. 2013, 112, 1395–1407. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, J.A.; Rasmussen, A.; Traini, R.; Voss, U.; Sturrock, C.; Mooney, S.J.; Wells, D.M.; Bennett, M.J. Branching out in roots: Uncovering form, function, and regulation. Plant Physiol. 2014, 166, 538–550. [Google Scholar] [CrossRef] [PubMed]
- Ahkami, A.H.; Melzer, M.; Ghaffari, M.R.; Pollmann, S.; Ghorbani, J.M.; Shahinnia, F.; Hajirezaei, M.R.; Druege, U. Distribution of indole-3-acetic acid in Petunia hybrida shoot tip cuttings and relationship between auxin transport, carbohydrate metabolism and adventitious root formation. Planta 2013, 238, 499–517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garrido, G.; Ramon, G.J.; Angel, C.E.; Acosta, M.; Sanchez-Bravo, J. Origin and basipetal transport of the IAA responsible for rooting of carnation cuttings. Physiol. Plant 2002, 114, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Da, C.C.; de Almeida, M.R.; Ruedell, C.M.; Schwambach, J.; Maraschin, F.S.; Fett-Neto, A.G. When stress and development go hand in hand: Main hormonal controls of adventitious rooting in cuttings. Front. Plant Sci. 2013, 4, 133. [Google Scholar]
- Klerk, G.J.D.; Guan, H.; Huisman, P.; Marinova, S. Effects of phenolic compounds on adventitious root formation and oxidative decarboxylation of applied indoleacetic acid in Malus ‘Jork 9’. Plant Growth Regul. 2011, 63, 175–185. [Google Scholar] [CrossRef] [Green Version]
- Ramirez-Carvajal, G.A.; Morse, A.M.; Dervinis, C.; Davis, J.M. The cytokinin type-B response regulator PtRR13 is a negative regulator of adventitious root development in Populus. Plant Physiol. 2009, 150, 759–771. [Google Scholar] [CrossRef] [PubMed]
- Busov, V.; Meilan, R.; Pearce, D.W.; Rood, S.B.; Ma, C.; Tschaplinski, T.J.; Strauss, S.H. Transgenic modification of gai or rgl1 causes dwarfing and alters gibberellins, root growth, and metabolite profiles in Populus. Planta 2006, 224, 288–299. [Google Scholar] [CrossRef] [PubMed]
- Bergonci, T.; Ribeiro, B.; Ceciliato, P.H.; Guerrero-Abad, J.C.; Silva-Filho, M.C.; Moura, D.S. Arabidopsis thaliana RALF1 opposes brassinosteroid effects on root cell elongation and lateral root formation. J. Exp. Bot. 2014, 65, 2219–2230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.W.; Yang, Y.W.; Lur, H.S.; Tsai, Y.G.; Chang, M.C. A novel function of abscisic acid in the regulation of rice (Oryza sativa L.) root growth and development. Plant Cell Physiol. 2006, 47, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Negi, S.; Sukumar, P.; Liu, X.; Cohen, J.D.; Muday, G.K. Genetic dissection of the role of ethylene in regulating auxin-dependent lateral and adventitious root formation in tomato. Plant J. 2010, 61, 3–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Zhang, X.; Wang, Y.; Lin, M. The Dynamic Changes of Endogenous Hormones in the Process of Adventitious Root Formation for Seabuckthorn Cutting Seedling. Glob. Seabuckthorn Res. Dev. 2014, 2, 003. [Google Scholar]
- Chen, L.; Tong, J.; Xiao, L.; Ruan, Y.; Liu, J.; Zeng, M.; Huang, H.; Wang, J.W.; Xu, L. YUCCA-mediated auxin biogenesis is required for cell fate transition occurring during de novo root organogenesis in Arabidopsis. J. Exp. Bot. 2016, 67, 4273–4284. [Google Scholar] [CrossRef] [PubMed]
- Lavenus, J.; Goh, T.; Roberts, I.; Guyomarc’H, S.; Lucas, M.; De Smet, I.; Fukaki, H.; Beeckman, T.; Bennett, M.; Laplaze, L. Lateral root development in Arabidopsis: Fifty shades of auxin. Trends Plant Sci. 2013, 18, 450–458. [Google Scholar] [CrossRef] [PubMed]
- Legue, V.; Rigal, A.; Bhalerao, R.P. Adventitious root formation in tree species: Involvement of transcription factors. Physiol. Plant 2014, 151, 192–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gutierrez, L.; Bussell, J.D.; Pacurar, D.I.; Schwambach, J.; Pacurar, M.; Bellini, C. Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of AUXIN RESPONSE FACTOR transcripts and microRNA abundance. Plant Cell 2009, 21, 3119–3132. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, L.; Mongelard, G.; Flokova, K.; Pacurar, D.I.; Novak, O.; Staswick, P.; Kowalczyk, M.; Pacurar, M.; Demailly, H.; Geiss, G.; et al. Auxin controls Arabidopsis adventitious root initiation by regulating jasmonic acid homeostasis. Plant Cell 2012, 24, 2515–2527. [Google Scholar] [CrossRef] [PubMed]
- Mashiguchi, K.; Tanaka, K.; Sakai, T.; Sugawara, S.; Kawaide, H.; Natsume, M.; Hanada, A.; Yaeno, T.; Shirasu, K.; Yao, H.; et al. The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 18512–18517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.H.; Zou, M.H.; Feng, B.H.; Huang, X.; Zhang, Z.; Sun, G.M. Molecular cloning and characterization of the genes encoding an auxin efflux carrier and the auxin influx carriers associated with the adventitious root formation in mango (Mangifera indica L.) cotyledon segments. Plant Physiol. Biochem. 2012, 55, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Peret, B.; Swarup, K.; Ferguson, A.; Seth, M.; Yang, Y.; Dhondt, S.; James, N.; Casimiro, I.; Perry, P.; Syed, A.; et al. AUX/LAX genes encode a family of auxin influx transporters that perform distinct functions during Arabidopsis development. Plant Cell 2012, 24, 2874–2885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sukumar, P.; Maloney, G.S.; Muday, G.K. Localized induction of the ATP-binding cassette B19 auxin transporter enhances adventitious root formation in Arabidopsis. Plant Physiol. 2013, 162, 1392–1405. [Google Scholar] [CrossRef] [PubMed]
- Pacurar, D.I.; Perrone, I.; Bellini, C. Auxin is a central player in the hormone cross-talks that control adventitious rooting. Physiol. Plant 2014, 151, 83–96. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Li, S.W.; Xue, L.G.; Xu, S.J.; Feng, H.Y.; An, L.Z. Hydrogen peroxide acts as a signal molecule in the adventitious root formation of mung bean seedlings. Environ. Exp. Bot. 2009, 65, 63–71. [Google Scholar] [CrossRef]
- Koyuncu, F.; Balta, F. Adventitious root formation in leaf-bud cuttings of tea (Camellia sinensis L.). Pak. J. Bot. 2004, 36, 763–768. [Google Scholar]
- Montain, C.R.; Haissig, B.E.; Curtis, J.D. Differentiation of adventitious root primordia in callus of Pinusbanks. Can. J. For. Res. 1983, 13, 195–200. [Google Scholar] [CrossRef]
- Goldfarb, B.; Hackett, W.P.; Furnier, G.R.; Mohn, C.A.; Plietzsch, A. Adventitious root initiation in hypocotyl and epicotyl cuttings of eastern white pine (Pinus strobus) seedlings. Physiol. Plant. 2010, 102, 513–522. [Google Scholar] [CrossRef]
- Thimann, K.V.; Went, F.W. On the chemical nature of the root forming hormone. Proc. Kon. Acad. Wetensch. Amst. 1934, 37, 456–458. [Google Scholar]
- Ludwig-Müller, J. Indole-3-butyric acid in plant growth and development. Plant Growth Regulat. 2000, 32, 219–230. [Google Scholar] [CrossRef]
- Li, Y.H.; Zhang, H.N.; Wu, Q.S.; Muday, G.K. Transcriptional sequencing and analysis of major genes involved in the adventitious root formation of mango cotyledon segments. Planta 2017, 245, 1193–1213. [Google Scholar] [CrossRef] [PubMed]
- Druege, U.; Franken, P.; Lischewski, S.; Ahkami, A.H.; Zerche, S.; Hause, B.; Hajirezaei, M.R. Transcriptomic analysis reveals ethylene as stimulator and auxin as regulator of adventitious root formation in petunia cuttings. Front. Plant Sci. 2014, 5, 494. [Google Scholar] [CrossRef] [PubMed]
- Peat, T.S.; Bottcher, C.; Newman, J.; Lucent, D.; Cowieson, N.; Davies, C. Crystal structure of an indole-3-acetic acid amido synthetase from grapevine involved in auxin homeostasis. Plant Cell 2012, 24, 4525–4538. [Google Scholar] [CrossRef] [PubMed]
- Steffens, B.; Rasmussen, A. The Physiology of Adventitious Roots. Plant Physiol. 2016, 170, 603. [Google Scholar] [CrossRef] [PubMed]
- Van Der Krieken, W.M.; Breteler, H.; Visser, M.H.; Mavridou, D. The role of the conversion of IBA into IAA on root regeneration in apple: Introduction of a test system. Plant Cell Rep. 1993, 12, 203. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Sala, C. Direct reprogramming of adult somatic cells toward adventitious root formation in forest tree species: The effect of the juvenile-adult transition. Front. Plant Sci. 2014, 5, 310. [Google Scholar] [CrossRef] [PubMed]
- BlažKová, A.; Sotta, B.; Tranvan, H.; Maldiney, R.; Bonnet, M.; Einhorn, J.; Kerhoas, L.; Miginiac, E. Auxin metabolism and rooting in young and mature clones of Sequoia sempervirens. Physiol. Plant. 2010, 99, 73–80. [Google Scholar] [CrossRef]
- Tonon, G.; Kevers, C.; Gaspar, T. Changes in polyamines, auxins and peroxidase activity during in vitro rooting of Fraxinus angustifolia shoots: An auxin-independent rooting model. Tree Physiol. 2001, 21, 655–663. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Chen, S.; Wang, S.; Liu, G.; Li, H.; Huang, H.; Jiang, J. BpGH3.5, an early auxin-response gene, regulates root elongation in Betula platyphylla × Betula pendula. Plant Cell Tissue Organ Cult. 2015, 120, 239–250. [Google Scholar] [CrossRef]
- Cano, A.; Sánchez-García, A.B.; Albacete, A.; González-Bayón, R.; Justamante, M.S.; Ibáñez, S.; Acosta, M.; Pérez-Pérez, J.M. Enhanced Conjugation of Auxin by GH3 Enzymes Leads to Poor Adventitious Rooting in Carnation Stem Cuttings. Front. Plant Sci. 2018, 9, 566. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Kaur, N.; Tyagi, A.K.; Khurana, J.P. The auxin-responsive GH3 gene family in rice (Oryza sativa). Funct. Integr. Genom. 2006, 6, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Bollmark, M.; Kubát, B.; Eliasson, L. Variation in Endogenous Cytokinin Content during Adventitious Root Formation in Pea Cuttings. J. Plant Physiol. 1988, 132, 262–265. [Google Scholar] [CrossRef]
- Gonzalez-Rizzo, S.; Crespi, M.; Frugier, F. The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti. Plant Cell 2006, 18, 2680–2693. [Google Scholar] [CrossRef] [PubMed]
- Laplaze, L.; Benkova, E.; Casimiro, I.; Maes, L.; Vanneste, S.; Swarup, R.; Weijers, D.; Calvo, V.; Parizot, B.; Herrera-Rodriguez, M.B.; et al. Cytokinins act directly on lateral root founder cells to inhibit root initiation. Plant Cell 2007, 19, 3889–3900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rani, D.B.; Taketa, S.; Ichii, M. Cytokinin inhibits lateral root initiation but stimulates lateral root elongation in rice (Oryza sativa). J. Plant Physiol. 2005, 162, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Werner, T.; Motyka, V.; Laucou, V.; Smets, R.; Van Onckelen, H.; Schmulling, T. Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 2003, 15, 2532–2550. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Mo, X.; Shou, H.; Wu, P. Cytokinin-mediated cell cycling arrest of pericycle founder cells in lateral root initiation of Arabidopsis. Plant Cell Physiol. 2006, 47, 1112–1123. [Google Scholar] [CrossRef] [PubMed]
- Lo, S.F.; Yang, S.Y.; Chen, K.T.; Hsing, Y.I.; Zeevaart, J.A.; Chen, L.J.; Yu, S.M. A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell 2008, 20, 2603–2618. [Google Scholar] [CrossRef] [PubMed]
- Lombardi-Crestana, S.; Da, S.A.M.; e Silva, G.F.; Pino, L.E.; Appezzato-da-Gloria, B.; Figueira, A.; Nogueira, F.T.; Peres, L.E. The tomato (Solanum lycopersicum cv. Micro-Tom) natural genetic variation Rg1 and the DELLA mutant procera control the competence necessary to form adventitious roots and shoots. J. Exp. Bot. 2012, 63, 5689–5703. [Google Scholar] [CrossRef] [PubMed]
- Mauriat, M.; Petterle, A.; Bellini, C.; Moritz, T. Gibberellins inhibit adventitious rooting in hybrid aspen and Arabidopsis by affecting auxin transport. Plant J. 2014, 78, 372–384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steffens, B.; Sauter, M. Epidermal cell death in rice is regulated by ethylene, gibberellin, and abscisic acid. Plant Physiol. 2005, 139, 713–721. [Google Scholar] [CrossRef] [PubMed]
- Agullo-Anton, M.A.; Ferrandez-Ayela, A.; Fernandez-Garcia, N.; Nicolas, C.; Albacete, A.; Perez-Alfocea, F.; Sanchez-Bravo, J.; Perez-Perez, J.M.; Acosta, M. Early steps of adventitious rooting: Morphology, hormonal profiling and carbohydrate turnover in carnation stem cuttings. Physiol. Plant 2014, 150, 446–462. [Google Scholar] [CrossRef] [PubMed]
- Vidoz, M.L.; Loreti, E.; Mensuali, A.; Alpi, A.; Perata, P. Hormonal interplay during adventitious root formation in flooded tomato plants. Plant J. 2010, 63, 551–562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geiss, G.; Gutierrez, L.; Bellini, C. Adventitious Root Formation: New Insights and Perspectives; Wiley-Blackwell: Hoboken, NJ, USA, 2009. [Google Scholar]
- Kurepin, L.; Haslam, T.; Lopez-Villalobos, A.; Oinam, G.; Yeung, E. Adventitious root formation in ornamental plants: II. The role of plant growth regulators. Propag. Ornam. Plants 2011, 11, 161–171. [Google Scholar]
- Müssig, C.; Shin, G.H.; Altmann, T. Brassinosteroids Promote Root Growth in Arabidopsis. Plant Physiol. 2003, 133, 1261–1271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bao, F.; Shen, J.; Brady, S.R.; Muday, G.K.; Asami, T.; Yang, Z. Brassinosteroids interact with auxin to promote lateral root development in Arabidopsis. Plant Physiol. 2004, 134, 1624–1631. [Google Scholar] [CrossRef] [PubMed]
- Mouchel, C.F.; Briggs, G.C.; Hardtke, C.S. Natural genetic variation in Arabidopsis identifies BREVIS RADIX, a novel regulator of cell proliferation and elongation in the root. Genes Dev. 2004, 18, 700–714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nemhauser, J.L.; Mockler, T.C.; Chory, J. Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol. 2004, 2, e258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yong, H.C.; Chang, H.S.; Gupta, R.; Wang, X.; Zhu, T.; Luan, S. Transcriptional Profiling Reveals Novel Interactions between Wounding, Pathogen, Abiotic Stress, and Hormonal Responses in Arabidopsis. Plant Physiol. 2002, 129, 661. [Google Scholar]
- Yang, L.; Conway, S.R.; Poethig, R.S. Vegetative phase change is mediated by a leaf-derived signal that represses the transcription of miR156. Development 2011, 138, 245–249. [Google Scholar] [CrossRef] [PubMed]
- Emons, A.M.C. Somatic embryogenesis: Cell biological aspects. Plant Biol. 1994, 43, 1–14. [Google Scholar] [CrossRef]
- Klerk, G.J.D.; Keppel, M.; Brugge, J.T.; Meekes, H. Timing of the phases in adventitious root formation in apple microcuttings. J. Exp. Bot. 1995, 46, 965–972. [Google Scholar] [CrossRef]
- Pawlicki, N.; Welander, M. Influence of carbohydrate source, auxin concentration and time of exposure on adventitious rooting of the apple rootstock Jork 9. Plant Sci. 1995, 106, 167–176. [Google Scholar] [CrossRef]
- Calamar, A.; Klerk, G.J.D. Effect of sucrose on adventitious root regeneration in apple. Plant Cell Tissue Organ Cult. 2002, 70, 207–212. [Google Scholar] [CrossRef]
- Koch, K. Sucrose metabolism: Regulatory mechanisms and pivotal roles in sugar sensing and plant development. Curr. Opin. Plant Biol. 2004, 7, 235–246. [Google Scholar] [CrossRef] [PubMed]
- Rolland, F.; Baena-Gonzalez, E.; Sheen, J. Sugar sensing and signaling in plants: Conserved and novel mechanisms. Annu. Rev. Plant Biol. 2006, 57, 675–709. [Google Scholar] [CrossRef] [PubMed]
- Appeldoorn, N.J.; Sergeeva, L.; Vreugdenhil, D.; van Der Plas, L.H.; Visser, R.G. In situ analysis of enzymes involved in sucrose to hexose-phosphate conversion during stolon-to-tuber transition of potato. Physiol. Plant 2002, 115, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Kromer, K.; Gamian, A. Analysis of soluble carbohydrates, proteins and lipids in shoots of M7 apple rootstock cultured in vitro during regeneration of adventitious roots. J. Plant Physiol. 2000, 156, 775–782. [Google Scholar] [CrossRef]
- Smeekens, S.; Ma, J.; Hanson, J.; Rolland, F. Sugar signals and molecular networks controlling plant growth. Curr. Opin. Plant Biol. 2010, 13, 274–279. [Google Scholar] [CrossRef] [PubMed]
- Moreno, F.; Ahuatzi, D.; Riera, A.; Palomino, C.A.; Herrero, P. Glucose sensing through the Hxk2-dependent signalling pathway. Biochem. Soc. Trans. 2005, 33, 265–268. [Google Scholar] [CrossRef] [PubMed]
- Zhong, C.; Kai, G.; Su, X.; Rao, P.; An, X. Genome-Wide Identification of the Invertase Gene Family in Populus. PLoS ONE 2015, 10, e138540. [Google Scholar]
- Dello, I.R.; Linhares, F.S.; Scacchi, E.; Casamitjana-Martinez, E.; Heidstra, R.; Costantino, P.; Sabatini, S. Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr. Biol. 2007, 17, 678–682. [Google Scholar] [CrossRef] [PubMed]
- De Smet, I.; Tetsumura, T.; De Rybel, B.; Frei, D.F.N.; Laplaze, L.; Casimiro, I.; Swarup, R.; Naudts, M.; Vanneste, S.; Audenaert, D.; et al. Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development 2007, 134, 681–690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Himanen, K.; Boucheron, E.; Vanneste, S.; de Almeida, E.J.; Inze, D.; Beeckman, T. Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell 2002, 14, 2339–2351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolters, H.; Jurgens, G. Survival of the flexible: Hormonal growth control and adaptation in plant development. Nat. Rev. Genet. 2009, 10, 305–317. [Google Scholar] [CrossRef] [PubMed]
- Link, M.; Rausch, T.; Greiner, S. In Arabidopsis thaliana, the invertase inhibitors AtC/VIF1 and 2 exhibit distinct target enzyme specificities and expression profiles. FEBS Lett. 2004, 573, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.L.; Fedoroff, N.V. LRP1, a gene expressed in lateral and adventitious root primordia of arabidopsis. Plant Cell 1995, 7, 735–745. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Wang, J.; Ren, X.; Bao, L.; Zhang, L.; Zhang, L.; Han, M.; Zhan, D. Root growth, yield and fruit quality of ‘Red Fuji’ apple trees in relation to planting depth of dwarfing interstock on the Loess Plateau. Eur. J. Horticult. Sci. 2015, 80, 109–116. [Google Scholar] [CrossRef]
- Cvrckova, F.; Bezvoda, R.; Zarsky, V. Computational identification of root hair-specific genes in Arabidopsis. Plant Signal. Behav. 2010, 5, 1407–1418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Q.; Chai, F.; An, X.; Han, S. Production Method for Paraffin Section of Invasive Species of Bemisia tabaci. Plant Dis. Pests 2012, 3, 46–48. [Google Scholar]
- Yang, J.P. Improvement of traditional paraffin section preparation methods. J. Biol. 2006, 23, 45–46. [Google Scholar]
- Weiler, E.W.; Jourdan, P.S.; Conrad, W. Levels of indole-3-acetic acid in intact and decapitated coleoptiles as determined by a specific and highly sensitive solid-phase enzyme immunoassay. Planta 1981, 153, 561–571. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Zhang, D.; Lei, C.; Chen, H.F.; Xing, L.B.; Ma, J.J.; Zhao, C.P.; Han, M.Y. Proteome analyses using itraq labeling reveals critical mechanisms in alternate bearing malus prunifolia. J. Proteome Res. 2016, 15, 3602–3616. [Google Scholar] [CrossRef] [PubMed]
- Gambino, G.; Perrone, I.; Gribaudo, I. A Rapid and effective method for RNA extraction from different tissues of grapevine and other woody plants. Phytochem. Anal. 2008, 19, 520–525. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.; Liu, Y.; Qi, Y.; Jiao, S.; Tian, F.; Jiang, L.; Wang, Y. Transcriptome sequencing and metabolite analysis reveals the role of delphinidin metabolism in flower colour in grape hyacinth. J. Exp. Bot. 2014, 65, 3157. [Google Scholar]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Zhang, Y.; Ye, Z.Q.; Liu, X.Q.; Zhao, S.Q.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, W345–W349. [Google Scholar] [CrossRef] [PubMed]
- Frazee, A.C.; Pertea, G.; Jaffe, A.E.; Langmead, B.; Salzberg, S.L.; Leek, J.T. Flexible isoform-level differential expression analysis with Ballgown. Biorxiv 2014. [Google Scholar] [CrossRef]
- Alexa, A.; Rahnenfuhrer, J. topGO: Enrichment Analysis for Gene Ontology; R Package Version; Elsevier: New York, NY, USA, 2006. [Google Scholar]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Shalom, L.; Samuels, S.; Zur, N.; Shlizerman, L.; Doron-Faigenboim, A.; Blumwald, E.; Sadka, A. Fruit load induces changes in global gene expression and in abscisic acid (ABA) and indole acetic acid (IAA) homeostasis in citrus buds. J. Exp. Bot. 2014, 65, 3029–3044. [Google Scholar] [CrossRef] [PubMed] [Green Version]


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, K.; Liang, Y.; Xing, L.; Mao, J.; Liu, Z.; Dong, F.; Meng, Y.; Han, M.; Zhao, C.; Bao, L.; et al. Transcriptome Analysis Reveals Multiple Hormones, Wounding and Sugar Signaling Pathways Mediate Adventitious Root Formation in Apple Rootstock. Int. J. Mol. Sci. 2018, 19, 2201. https://doi.org/10.3390/ijms19082201
Li K, Liang Y, Xing L, Mao J, Liu Z, Dong F, Meng Y, Han M, Zhao C, Bao L, et al. Transcriptome Analysis Reveals Multiple Hormones, Wounding and Sugar Signaling Pathways Mediate Adventitious Root Formation in Apple Rootstock. International Journal of Molecular Sciences. 2018; 19(8):2201. https://doi.org/10.3390/ijms19082201
Chicago/Turabian StyleLi, Ke, Yongqi Liang, Libo Xing, Jiangping Mao, Zhen Liu, Feng Dong, Yuan Meng, Mingyu Han, Caiping Zhao, Lu Bao, and et al. 2018. "Transcriptome Analysis Reveals Multiple Hormones, Wounding and Sugar Signaling Pathways Mediate Adventitious Root Formation in Apple Rootstock" International Journal of Molecular Sciences 19, no. 8: 2201. https://doi.org/10.3390/ijms19082201



