Understanding Ovarian Cancer: iTRAQ-Based Proteomics for Biomarker Discovery
Abstract
:1. Introduction
2. Results
3. Discussion
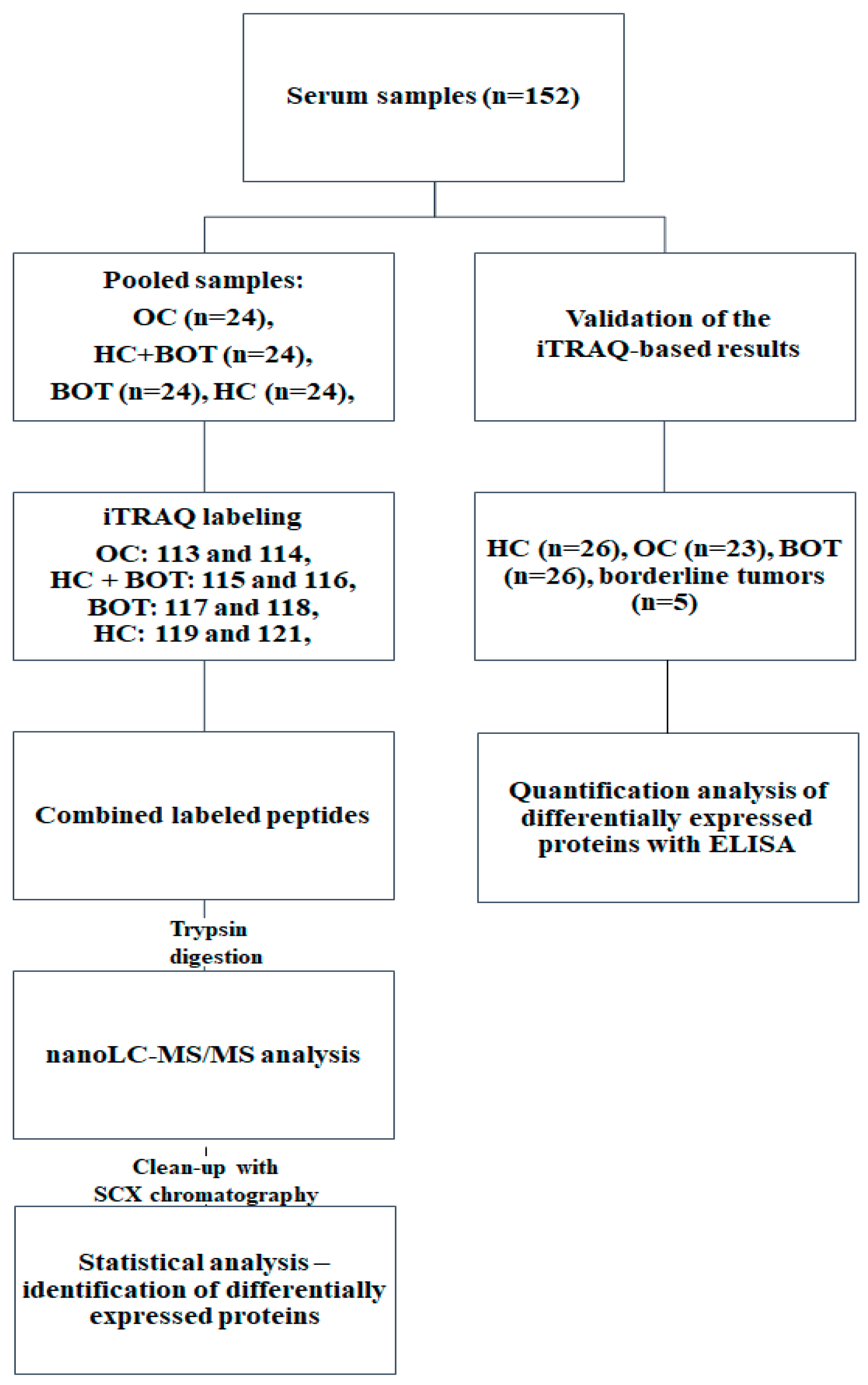
4. Materials and Methods
4.1. Samples
4.2. Protein Digestion
4.3. iTRAQ Labeling
4.4. nanoLC-MALDI-TOF/TOF MS/MS Analysis
4.5. Database Search and Data Processing
4.6. Quantification of Individual Proteins by ELISA
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| OC | ovarian cancer |
| FIGO | International Federation of Gynaecology and Obstetrics |
| CA125 | cancer antigen 125 |
| HE4 | human epididymis protein 4 |
| RMI | Risk of Malignancy Index |
| SILAC | stable isotope labelling by amino acids in cell culture |
| ICAT | Isotope-coded affinity tag |
| ITRAQ | isobaric Tags for Relative and Absolute Quantification |
| MALDI-TOF | matrix-assisted laser desorption/ionization time-of-flight |
| MS | mass spectrometry |
| BOT | benign ovarian tumors |
| HC | Healthy controls |
| ELISA | enzyme-linked immunosorbent assay |
| SC | sequences coverage |
| MW | molecular weight |
| CRP | C-reactive protein |
| FGA | Fibrinogen alpha chain |
| APOC2 | Apolipoprotein C-II |
| SAA1 | Serum amyloid A1 |
| ORM1 | Alpha-1-acid glycoprotein 1 |
| SERPINA1 | Alpha-1-antitrypsin |
| HPX | Hemopexin |
| TF | Serotransferrin |
| ALB | Serum albumin |
| CV | Coefficient of variation |
| SCX | strong cation exchange |
| LC | liquid chromatography |
| TFA | trifluoroacetic acid |
| HCCA | α-cyano-4-hydroxycinnamic acid |
| HPM | Human Proteome Map |
| ROC | receiver operating characteristic curve |
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Wolman, I. Berek and Novak’s Gynecology 15th Edition. J. Obstet. Gynecol. India 2014, 64, 150–151. [Google Scholar] [CrossRef] [Green Version]
- Horala, A.; Swiatly, A.; Matysiak, J.; Banach, P.; Nowak-Markwitz, E.; Kokot, Z. Diagnostic Value of Serum Angiogenesis Markers in Ovarian Cancer Using Multiplex Immunoassay. Int. J. Mol. Sci. 2017, 18, 123. [Google Scholar] [CrossRef] [PubMed]
- Swiatly, A.; Horala, A.; Hajduk, J.; Matysiak, J.; Nowak-Markwitz, E.; Kokot, Z.J. MALDI-TOF-MS analysis in discovery and identification of serum proteomic patterns of ovarian cancer. BMC Cancer 2017, 17, 472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plewa, S.; Horała, A.; Dereziński, P.; Klupczynska, A.; Nowak-Markwitz, E.; Matysiak, J.; Kokot, Z. Usefulness of Amino Acid Profiling in Ovarian Cancer Screening with Special Emphasis on Their Role in Cancerogenesis. Int. J. Mol. Sci. 2017, 18, 2727. [Google Scholar] [CrossRef] [PubMed]
- Qi, D.; Wang, Q.; Li, H.; Zhang, T.; Lan, R.; Kwong, D.W.J.; Wong, W.-K.; Wong, K.-L.; Li, S.; Lu, F. SILAC-based quantitative proteomics identified lysosome as a fast response target to PDT agent Gd-N induced oxidative stress in human ovarian cancer IGROV1 cells. Mol. Biosyst. 2015, 11, 3059–3067. [Google Scholar] [CrossRef] [PubMed]
- Kjellin, H.; Johansson, H.; Höög, A.; Lehtiö, J.; Jakobsson, P.-J.; Kjellman, M. Differentially expressed proteins in malignant and benign adrenocortical tumors. PLoS ONE 2014, 9, e87951. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.; Cheng, L.; White, J.T.; Lu, W.; Utleg, A.G.; Yan, X.; Urban, N.D.; Drescher, C.W.; Hood, L.; Lin, B. Quantitative Proteomics Analysis Integrated with Microarray Data Reveals That Extracellular Matrix Proteins, Catenins, and P53 Binding Protein 1 Are Important for Chemotherapy Response in Ovarian Cancers. OMI. J. Integr. Biol. 2009, 13, 345–354. [Google Scholar] [CrossRef] [PubMed]
- Russell, M.R.; Walker, M.J.; Williamson, A.J.K.; Gentry-Maharaj, A.; Ryan, A.; Kalsi, J.; Skates, S.; D’Amato, A.; Dive, C.; Pernemalm, M.; et al. Protein Z: A putative novel biomarker for early detection of ovarian cancer. Int. J. Cancer 2016, 138, 2984–2992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mi, H.; Muruganujan, A.; Casagrande, J.T.; Thomas, P.D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013, 8, 1551–1566. [Google Scholar] [CrossRef] [PubMed]
- Fan, G.; Wrzeszczynski, K.O.; Fu, C.; Su, G.; Pappin, D.J.; Lucito, R.; Tonks, N.K. A quantitative proteomics-based signature of platinum sensitivity in ovarian cancer cell lines. Biochem. J. 2015, 465, 433–442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, Y.; Tan, A.-C.; Sun, X.; Olson, M.T.; Xie, Z.; Jinawath, N.; Chan, D.W.; Shih, I.-M.; Zhang, Z.; Zhang, H. Quantitative proteomic analysis of ovarian cancer cells identified mitochondrial proteins associated with paclitaxel resistance. Proteom. Clin. Appl. 2009, 3, 1288–1295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shetty, V.; Nickens, Z.; Testa, J.; Hafner, J.; Sinnathamby, G.; Philip, R. Quantitative immunoproteomics analysis reveals novel MHC class I presented peptides in cisplatin-resistant ovarian cancer cells. J. Proteom. 2012, 75, 3270–3290. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chen, S.; Zhang, M.; Li, N.; Chen, Y.; Su, W.; Liu, Y.; Lu, D.; Li, S.; Yang, Y.; et al. Legumain: A biomarker for diagnosis and prognosis of human ovarian cancer. J. Cell. Biochem. 2012, 113, 2679–2686. [Google Scholar] [CrossRef] [PubMed]
- Waldemarson, S.; Krogh, M.; Alaiya, A.; Kirik, U.; Schedvins, K.; Auer, G.; Hansson, K.M.; Ossola, R.; Aebersold, R.; Lee, H.; et al. Protein Expression Changes in Ovarian Cancer during the Transition from Benign to Malignant. J. Proteome Res. 2012, 11, 2876–2889. [Google Scholar] [CrossRef] [PubMed]
- Kristjansdottir, B.; Levan, K.; Partheen, K.; Carlsohn, E.; Sundfeldt, K. Potential tumor biomarkers identified in ovarian cyst fluid by quantitative proteomic analysis, iTRAQ. Clin. Proteom. 2013, 10, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poersch, A.; Grassi, M.L.; de Carvalho, V.P.; Lanfredi, G.P.; de Palma, C.S.; Greene, L.J.; de Sousa, C.B.; Carrara, H.H.A.; Candido dos Reis, F.J.; Faça, V.M. A proteomic signature of ovarian cancer tumor fluid identified by highthroughput and verified by targeted proteomics. J. Proteom. 2016, 145, 226–236. [Google Scholar] [CrossRef] [PubMed]
- Boylan, K.L.; Andersen, J.D.; Anderson, L.B.; Higgins, L.; Skubitz, A.P. Quantitative proteomic analysis by iTRAQ(R) for the identification of candidate biomarkers in ovarian cancer serum. Proteome Sci. 2010, 8, 31. [Google Scholar] [CrossRef] [PubMed]
- Enakpene, C.A.; Omigbodun, A.O.; Goecke, T.W.; Odukogbe, A.-T.; Beckmann, M.W. Preoperative evaluation and triage of women with suspicious adnexal masses using risk of malignancy index. J. Obstet. Gynaecol. Res. 2009, 35, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Grenache, D.G.; Heichman, K.A.; Werner, T.L.; Vucetic, Z. Clinical performance of two multi-marker blood tests for predicting malignancy in women with an adnexal mass. Clin. Chim. Acta 2015, 438, 358–363. [Google Scholar] [CrossRef] [PubMed]
- Moshkovskii, S.A.; Serebryakova, M.V.; Kuteykin-Teplyakov, K.B.; Tikhonova, O.V.; Goufman, E.I.; Zgoda, V.G.; Taranets, I.N.; Makarov, O.V.; Archakov, A.I. Ovarian cancer marker of 11.7 kDa detected by proteomics is a serum amyloid A1. Proteomics 2005, 5, 3790–3797. [Google Scholar] [CrossRef] [PubMed]
- Toriola, A.T.; Grankvist, K.; Agborsangaya, C.B.; Lukanova, A.; Lehtinen, M.; Surcel, H.-M. Changes in pre-diagnostic serum C-reactive protein concentrations and ovarian cancer risk: A longitudinal study. Ann. Oncol. 2011, 22, 1916–1921. [Google Scholar] [CrossRef] [PubMed]
- Asher, V.; Lee, J.; Bali, A. Preoperative serum albumin is an independent prognostic predictor of survival in ovarian cancer. Med. Oncol. 2012, 29, 2005–2009. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.M.G.; Ankerst, D.P.; Andridge, R.R. Validation of biomarker-based risk prediction models. Clin. Cancer Res. 2008, 14, 5977–5983. [Google Scholar] [CrossRef] [PubMed]
- Weiland, F.; Fritz, K.; Oehler, M.K.; Hoffmann, P. Methods for identification of CA125 from ovarian cancer ascites by high resolution mass spectrometry. Int. J. Mol. Sci. 2012, 13, 9942–9958. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, H.; Tan, M.; Liu, J.; Hu, Z.; Liu, D.; Gao, J.; Zhu, L.; Lin, B. Human epididymis protein 4 in association with Annexin II promotes invasion and metastasis of ovarian cancer cells. Mol. Cancer 2014, 13, 243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daniels, T.R.; Bernabeu, E.; Rodríguez, J.A.; Patel, S.; Kozman, M.; Chiappetta, D.A.; Holler, E.; Ljubimova, J.Y.; Helguera, G.; Penichet, M.L. The transferrin receptor and the targeted delivery of therapeutic agents against cancer. Biochim. Biophys. Acta 2012, 1820, 291–317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macedo, M.; Sousa, M. Transferrin and the Transferrin Receptor: Of Magic Bullets and Other Concerns. Inflamm. Allergy-Drug Targets 2008, 7, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Naoum, F.A. Iron deficiency in cancer patients. Rev. Bras. Hematol. Hemoter. 2016, 38, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Wegdam, W.; Argmann, C.A.; Kramer, G.; Vissers, J.P.; Buist, M.R.; Kenter, G.G.; Aerts, J.M.F.G.; Meijer, D.; Moerland, P.D. Label-Free LC-MSe in Tissue and Serum Reveals Protein Networks Underlying Differences between Benign and Malignant Serous Ovarian Tumors. PLoS ONE 2014, 9, e108046. [Google Scholar] [CrossRef] [PubMed]
- Rauniyar, N.; Peng, G.; Lam, T.T.; Zhao, H.; Mor, G.; Williams, K.R. Data-Independent Acquisition and Parallel Reaction Monitoring Mass Spectrometry Identification of Serum Biomarkers for Ovarian Cancer. Biomark. Insights 2017, 12, 1177271917710948. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Zhang, X.; Jiang, W.; Wang, Y.; Jin, H.; Liu, X.; Xu, C. Discovery of serum biomarkers implicated in the onset and progression of serous ovarian cancer in a rat model using iTRAQ technique. Eur. J. Obstet. Gynecol. Reprod. Biol. 2012, 165, 96–103. [Google Scholar] [CrossRef] [PubMed]
- De Buck, M.; Gouwy, M.; Wang, J.M.; Van Snick, J.; Opdenakker, G.; Struyf, S.; Van Damme, J. Structure and Expression of Different Serum Amyloid A (SAA) Variants and their Concentration-Dependent Functions During Host Insults. Curr. Med. Chem. 2016, 23, 1725–1755. [Google Scholar] [CrossRef] [PubMed]
- Malle, E.; Sodin-Semrl, S.; Kovacevic, A. Serum amyloid A: An acute-phase protein involved in tumour pathogenesis. Cell. Mol. Life Sci. 2009, 66, 9–26. [Google Scholar] [CrossRef] [PubMed]
- Biaoxue, R.; Hua, L.; Wenlong, G.; Shuanying, Y. Increased serum amyloid A as potential diagnostic marker for lung cancer: A meta-analysis based on nine studies. BMC Cancer 2016, 16, 836. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Liu, F.; Higuchi, K.; Sawashita, J.; Fu, X.; Zhang, L.; Zhang, L.; Fu, L.; Tong, Z.; Higuchi, K. Serum amyloid A expression in the breast cancer tissue is associated with poor prognosis. Oncotarget 2016, 7, 35843–35852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urieli-Shoval, S.; Finci-Yeheskel, Z.; Dishon, S.; Galinsky, D.; Linke, R.P.; Ariel, I.; Levin, M.; Ben-Shachar, I.; Prus, D. Expression of serum amyloid a in human ovarian epithelial tumors: Implication for a role in ovarian tumorigenesis. J. Histochem. Cytochem. 2010, 58, 1015–1023. [Google Scholar] [CrossRef] [PubMed]
- Moshkovskii, S.A.; Vlasova, M.A.; Pyatnitskiy, M.A.; Tikhonova, O.V.; Safarova, M.R.; Makarov, O.V.; Archakov, A.I. Acute phase serum amyloid A in ovarian cancer as an important component of proteome diagnostic profiling. Proteom. Clin. Appl. 2007, 1, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Dossus, L.; Jimenez-Corona, A.; Romieu, I.; Boutron-Ruault, M.-C.; Boutten, A.; Dupré, T.; Fagherazzi, G.; Clavel-Chapelon, F.; Mesrine, S. C-reactive protein and postmenopausal breast cancer risk: Results from the E3N cohort study. Cancer Causes Control 2014, 25, 533–539. [Google Scholar] [CrossRef] [PubMed]
- Babic, A.; Schnure, N.; Neupane, N.P.; Zaman, M.M.; Rifai, N.; Welch, M.W.; Brais, L.K.; Rubinson, D.A.; Morales-Oyarvide, V.; Yuan, C.; et al. Plasma inflammatory cytokines and survival of pancreatic cancer patients. Clin. Transl. Gastroenterol. 2018, 9, 145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.; Choe, J.-W.; Kim, H.-K.; Sung, J. High-sensitivity C-reactive protein and cancer. J. Epidemiol. 2011, 21, 161–168. [Google Scholar] [CrossRef] [PubMed]
- McSorley, M.A.; Alberg, A.J.; Allen, D.S.; Allen, N.E.; Brinton, L.A.; Dorgan, J.F.; Pollak, M.; Tao, Y.; Helzlsouer, K.J. C-Reactive Protein Concentrations and Subsequent Ovarian Cancer Risk. Obstet. Gynecol. 2007, 109, 933–941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Jiao, X.; Yuan, Z.; Qiu, H.; Guo, R. C-reactive protein and risk of ovarian cancer: A systematic review and meta-analysis. Medicine 2017, 96, e7822. [Google Scholar] [CrossRef] [PubMed]
- Merlot, A.M.; Kalinowski, D.S.; Richardson, D.R. Unraveling the mysteries of serum albumin-more than just a serum protein. Front. Physiol. 2014, 5, 299. [Google Scholar] [CrossRef] [PubMed]
- Gupta, D.; Lis, C.G. Pretreatment serum albumin as a predictor of cancer survival: A systematic review of the epidemiological literature. Nutr. J. 2010, 9, 69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parker, D.; Bradley, C.; Bogle, S.M.; Lay, J.; Masood, M.; Hancock, A.K.; Naylor, B.; Price, J.J. Serum albumin and CA125 are powerful predictors of survival in epithelial ovarian cancer. Br. J. Obstet. Gynaecol. 1994, 101, 888–893. [Google Scholar] [CrossRef] [PubMed]
- Chiang, J.M.; Chang, C.J.; Jiang, S.F.; Yeh, C.Y.; You, J.F.; Hsieh, P.S.; Huang, H.Y. Pre-operative serum albumin level substantially predicts post-operative morbidity and mortality among patients with colorectal cancer who undergo elective colectomy. Eur. J. Cancer Care 2017, 26, e12403. [Google Scholar] [CrossRef] [PubMed]
- Moujaess, E.; Fakhoury, M.; Assi, T.; Elias, H.; El Karak, F.; Ghosn, M.; Kattan, J. The Therapeutic use of human albumin in cancer patients’ management. Crit. Rev. Oncol. Hematol. 2017, 120, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Awadallah, S. Protein Antioxidants in Thalassemia. Adv. Clin. Chem. 2013, 60, 85–128. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Xie, X.; Nie, S.; Buckanovich, R.J.; Lubman, D.M. Altered Expression of Sialylated Glycoproteins in Ovarian Cancer Sera Using Lectin-based ELISA Assay and Quantitative Glycoproteomics Analysis. J. Proteome Res. 2013, 12, 3342–3352. [Google Scholar] [CrossRef] [PubMed]
- Dowling, P.; O’Driscoll, L.; Meleady, P.; Henry, M.; Roy, S.; Ballot, J.; Moriarty, M.; Crown, J.; Clynes, M. 2-D difference gel electrophoresis of the lung squamous cell carcinomaversus normal sera demonstrates consistent alterations in the levels of ten specific proteins. Electrophoresis 2007, 28, 4302–4310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyamoto, S.; Ruhaak, L.R.; Stroble, C.; Salemi, M.R.; Phinney, B.; Lebrilla, C.B.; Leiserowitz, G.S. Glycoproteomic Analysis of Malignant Ovarian Cancer Ascites Fluid Identifies Unusual Glycopeptides. J. Proteome Res. 2016, 15, 3358–3376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cine, N.; Baykal, A.T.; Sunnetci, D.; Canturk, Z.; Serhatli, M.; Savli, H. Identification of ApoA1, HPX and POTEE genes by omic analysis in breast cancer. Oncol. Rep. 2014, 32, 1078–1086. [Google Scholar] [CrossRef] [PubMed]
- Ang, C.-S.; Rothacker, J.; Patsiouras, H.; Burgess, A.W.; Nice, E.C. Murine fecal proteomics: A model system for the detection of potential biomarkers for colorectal cancer. J. Chromatogr. A 2010, 1217, 3330–3340. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, S.; Stroble, C.D.; Taylor, S.; Hong, Q.; Lebrilla, C.B.; Leiserowitz, G.S.; Kim, K.; Ruhaak, L.R. Multiple Reaction Monitoring for the Quantitation of Serum Protein Glycosylation Profiles: Application to Ovarian Cancer. J. Proteome Res. 2018, 17, 222–233. [Google Scholar] [CrossRef] [PubMed]
- Timms, J.F.; Arslan-Low, E.; Kabir, M.; Worthington, J.; Camuzeaux, S.; Sinclair, J.; Szaub, J.; Afrough, B.; Podust, V.N.; Fourkala, E.-O.; et al. Discovery of serum biomarkers of ovarian cancer using complementary proteomic profiling strategies. Proteom. Clin. Appl. 2014, 8, 982–993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yip, P.; Chen, T.-H.; Seshaiah, P.; Stephen, L.L.; Michael-Ballard, K.L.; Mapes, J.P.; Mansfield, B.C.; Bertenshaw, G.P. Comprehensive Serum Profiling for the Discovery of Epithelial Ovarian Cancer Biomarkers. PLoS ONE 2011, 6, e29533. [Google Scholar] [CrossRef] [PubMed]
- Harima, Y.; Ikeda, K.; Utsunomiya, K.; Komemushi, A.; Kanno, S.; Shiga, T.; Tanigawa, N. Apolipoprotein C-II Is a Potential Serum Biomarker as a Prognostic Factor of Locally Advanced Cervical Cancer After Chemoradiation Therapy. Int. J. Radiat. Oncol. 2013, 87, 1155–1161. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-B.; Su, Y.-C.; Huang, T.-T.; Ho, H.-C.; Chang, Y.-T.; Tung, Y.-T.; Lee, W.-C. Differentially expressed serum haptoglobin alpha chain isoforms with potential application for diagnosis of head and neck cancer. Clin. Chim. Acta 2008, 398, 48–52. [Google Scholar] [CrossRef] [PubMed]
- Seebacher, V.; Aust, S.; D’Andrea, D.; Grimm, C.; Reiser, E.; Tiringer, D.; Von Mersi, H.; Polterauer, S.; Reinthaller, A.; Helmy-Bader, S. Development of a tool for prediction of ovarian cancer in patients with adnexal masses: Value of plasma fibrinogen. PLoS ONE 2017, 12, e0182383. [Google Scholar] [CrossRef] [PubMed]
- Xie, N.; Li, Z.; Zuo, R.; Qi, S.; Zhu, T.; Liu, L.; Wan, L.; Yuan, J. Serum SAA1 and APOE are novel indicators for human cytomegalovirus infection. Sci. Rep. 2017, 7, 13407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.-S.; Pinto, S.M.; Getnet, D.; Nirujogi, R.S.; Manda, S.S.; Chaerkady, R.; Madugundu, A.K.; Kelkar, D.S.; Isserlin, R.; Jain, S.; et al. A draft map of the human proteome. Nature 2014, 509, 575–581. [Google Scholar] [CrossRef] [PubMed] [Green Version]


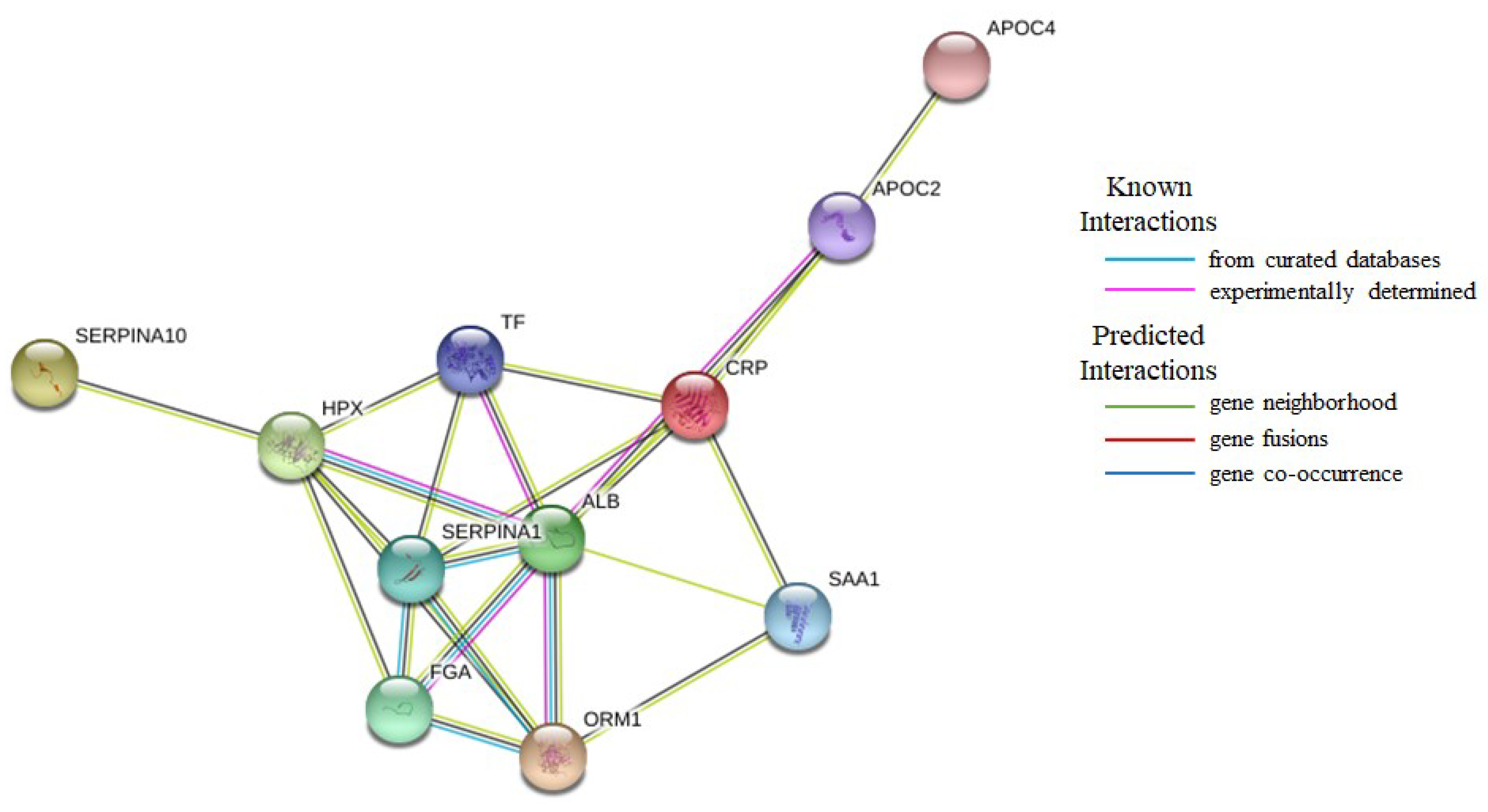
| Accession | Protein Name | Protein Class | iTRAQ Ratios | |||
|---|---|---|---|---|---|---|
| OC/HC + BOT | OC/HC | OC/BOT | BOT/HC | |||
| ORM1 | α-1-acid glycoprotein 1 | - | 0.56 | 0.81 | 0.64 | 1.28 |
| SERPINA1 | α-1-antitrypsin | Enzyme modulator | 0.75 | 0.48 | 0.89 | 0.54 |
| APOC2 | Apolipoprotein C-II | - | 1.24 | 0.96 | 1.67 | 0.59 |
| APOC4 | Apolipoprotein C-IV | - | 1.04 | 0.74 | 1.28 | 0.59 |
| CRP | C-reactive protein | - | 2.47 | 2.37 | 3.07 | 0.78 |
| FGA | Fibrinogen alpha chain | - | 2.31 | 1.97 | 1.83 | 1.09 |
| HPX | Hemopexin | Hydrolase | 0.44 | 0.97 | 0.67 | 1.55 |
| SERPINA10 | Protein Z-dependent protease inhibitor | Enzyme modulator | 0.98 | 0.76 | 1.20 | 0.60 |
| TF | Serotransferrin | Transfer/carrier protein; Receptor; Hydrolase; Defense/immunity protein | 0.64 | 0.91 | 0.65 | 1.39 |
| ALB | Serum albumin | Transfer/carrier protein | 0.60 | 0.75 | 0.58 | 1.33 |
| SAA1 | Serum amyloid A protein | Transporter; Transfer/carrier protein; Defense/immunity protein | 3.35 | 3.59 | 4.33 | 0.83 |
| Protein | OC vs. HC + BOT | OC vs. HC | OC vs. BOT | BOT vs. HC | ||||
|---|---|---|---|---|---|---|---|---|
| AUC | p-Values | AUC | p-Values | AUC | p-Values | AUC | p-Values | |
| ORM1 | 0.518 | 0.4022 | 0.505 | 0.5679 | 0.540 | 0.4580 | 0.536 | 0.6967 |
| SERPINA1 | 0.622 | 0.0949 | 0.616 | 0.1091 | 0.627 | 0.2083 | 0.515 | 0.7274 |
| APOC2 | 0.621 | 0.1187 | 0.635 | 0.1377 | 0.607 | 0.2330 | 0.523 | 0.6806 |
| APOC4 | 0.595 | 0.256 | 0.620 | 0.1769 | 0.569 | 0.5362 | 0.561 | 0.4416 |
| CRP | 0.777 | <0.0001 | 0.761 | <0.0001 | 0.793 | <0.0001 | 0.544 | 0.3409 |
| HPX | 0.779 | <0.0001 | 0.762 | 0.0011 | 0.796 | <0.0001 | 0.540 | 0.4167 |
| SERPINA10 | 0.602 | 0.2756 | 0.559 | 0.5114 | 0.645 | 0.1834 | 0.580 | 0.6550 |
| TF | 0.851 | <0.0001 | 0.845 | <0.0001 | 0.858 | <0.0001 | 0.531 | 0.6127 |
| ALB | 0.787 | <0.0001 | 0.758 | <0.0001 | 0.875 | <0.0001 | 0.596 | 0.3449 |
| SAA1 | 0.825 | <0.0001 | 0.799 | <0.0001 | 0.851 | <0.0001 | 0.593 | 0.1719 |
| CA125 * | - | - | - | - | 0.965 | <0.0001 | - | - |
| HE4 * | - | - | - | - | 1.0 | <0.0001 | - | - |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Swiatly, A.; Horala, A.; Matysiak, J.; Hajduk, J.; Nowak-Markwitz, E.; Kokot, Z.J. Understanding Ovarian Cancer: iTRAQ-Based Proteomics for Biomarker Discovery. Int. J. Mol. Sci. 2018, 19, 2240. https://doi.org/10.3390/ijms19082240
Swiatly A, Horala A, Matysiak J, Hajduk J, Nowak-Markwitz E, Kokot ZJ. Understanding Ovarian Cancer: iTRAQ-Based Proteomics for Biomarker Discovery. International Journal of Molecular Sciences. 2018; 19(8):2240. https://doi.org/10.3390/ijms19082240
Chicago/Turabian StyleSwiatly, Agata, Agnieszka Horala, Jan Matysiak, Joanna Hajduk, Ewa Nowak-Markwitz, and Zenon J. Kokot. 2018. "Understanding Ovarian Cancer: iTRAQ-Based Proteomics for Biomarker Discovery" International Journal of Molecular Sciences 19, no. 8: 2240. https://doi.org/10.3390/ijms19082240





