Characterization of Begomoviruses Sampled during Severe Epidemics in Tomato Cultivars Carrying the Ty-1 Gene
Abstract
:1. Introduction
2. Results
2.1. Aggressive Tomato Yellow Leaf Curl Virus (TYLCV) Epidemics in Resistant Tomato Crops in Murcia (Spain)
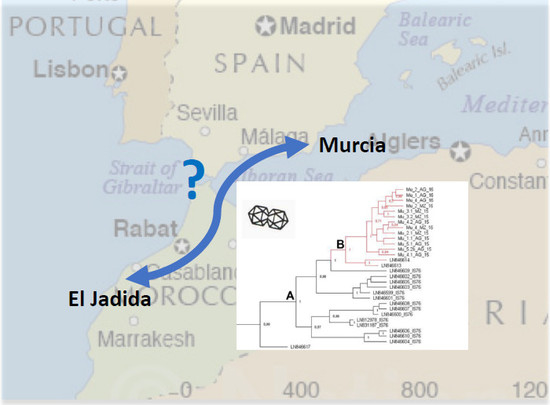
2.2. Phylogenetic Analysis of the Murcia (Spain) Isolates Sampled during 2015 to 2016
2.3. Comparing the Fitness of TYLCV-Mu 15 and TYLCV-IS76 in Susceptible and Resistant (Ty-1/Ty-3) Tomato Plants
3. Discussion
4. Materials and Methods
4.1. Samplings
4.2. DNA Extraction and Geminivirus Detection by PCR
4.3. Sequencing
4.4. Analysis of Sequences
4.5. Infectious Clone
4.6. Virus Accumulation Test
4.7. Statistical Analyses
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| TYLCD | Tomato yellow leaf curl disease |
| TYLCV | Tomato yellow leaf curl virus |
| RCA | Rolling Circle Amplification |
| HTS | High throughput Sequencing |
| NCBI | The National Center for Biotechnology Information |
| BLAST | Basic local alignment tool |
| MUSCLE | Multiple Sequence Comparison by Log-Expectation |
| MEGA | Molecular Evolutionary Genetic Analysis |
| SDT | Sequence demarcation tool |
| RDP | Recombination detection program |
| MCMC | Markov Chain Monte Carlo |
| YEB | Yeast Extract Broth |
| SIM | Simplified Induction Medium |
| dpi | Days post inoculation |
Appendix A
| Sample | Num. of Reads b | Mapped Reads | Average Coverage | % of Reference Covered c |
|---|---|---|---|---|
| Mu 1.1 MZ:15 | 20,962 | 63 | 5.03 | 97 |
| Mu 2.1 MZ:15 | 19,394 | 64 | 5.5 | 100 |
| Mu 2.2 MZ:15 | 19,822 | 7895 | 642 | 100 |
| Mu 3.1 MZ:15 | 20,322 | 26 | 1.93 | 79 |
| Mu 3.2 MZ:15 | 18,328 | 1901 | 152.44 | 100 |
| Mu 4.2 MZ:15 | 43.27 | 44 | 3.59 | 92 |
| Mu 5.2 MZ:15 | 13,452 | 358 | 358 | 100 |
| Mu 1.1 AG:15 | 12,424 | 2202 | 203.86 | 100 |
| Mu 3.1 AG:15 | 31,354 | 8560 | 693.27 | 100 |
| Mu 3.2 AG:15 | 22,558 | 133 | 10.13 | 95 |
| Mu 4.1 AG:15 | 253 | 2218 | 156.49 | 100 |
| Mu 4.2 AG:15 | 20,269 | 1775 | 128.37 | 100 |
| Mu 5.1 AG:15 | 17,316 | 1983 | 163.93 | 100 |
| Mu 5.2a AG:15 | 16,302 | 3848 | 332.42 | 100 |
| Mu 5.2b AG:15 d | 16,302 | 2471 | 199.45 | 100 |
References
- Hanssen, I.M.; Lapidot, M. Major Tomato Viruses in the Mediterranean Basin. Adv. Virus Res. 2012, 84, 31–66. [Google Scholar] [CrossRef] [PubMed]
- Picó, B.; Diez, J.; Nuez, F. Viral diseases causing the greatest economic losses to the tomato crop. II The Tomato yellow leaf curl virus-a review. Sci. Hortic. 1996, 67, 151–196. [Google Scholar] [CrossRef]
- Brown, J.K.; Zerbini, F.M.; Navas-Castillo, J.; Moriones, E.; Ramos-Sobrinho, R.; Silva, J.C.F.; Fiallo-Olivé, E.; Briddon, R.W.; Hernández-Zepeda, C.; Idris, A.; et al. Revision of Begomovirus taxonomy based on pairwise sequence comparisons. Arch. Virol. 2015, 160, 1593–1619. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Pendón, J.A.; Cañizares, M.C.; Moriones, E.; Bejarano, E.R.; Czosnek, H.; Navas-Castillo, J. Tomato yellow leaf curl viruses: Ménage à trois between the virus complex, the plant and the whitefly vector. Mol. Plant Pathol. 2010, 11, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Mabvakure, B.; Martin, D.P.; Kraberger, S.; Cloete, L.; van Brunschot, S.; Geering, A.D.W.; Thomas, J.E.; Bananej, K.; Lett, J.M.; Lefeuvre, P.; et al. Ongoing geographical spread of Tomato yellow leaf curl virus. Virology 2016, 498, 257–264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lefeuvre, P.; Martin, D.P.; Harkins, G.; Lemey, P.; Gray, A.J.A.; Meredith, S.; Lakay, F.; Monjane, A.; Lett, J.M.; Varsani, A.; et al. The spread of tomato yellow leaf curl virus from the middle east to the world. PLoS Pathog. 2010, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navot, N.; Pichersky, E.; Zeidan, M.; Zamir, D.; Czosnek, H. Tomato yellow leaf curl virus: A whitefly transmitted geminivirus with a single genomic component. Virology 1991, 185, 151–161. [Google Scholar] [CrossRef]
- Brown, J.K.; Fauquet, C.M.; Briddon, R.W.; Zerbini, M.; Moriones, E.; Navas-Castillo, J. Family Geminiviridae. In Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses; King, A.M., Adams, M.J., Carstens, E.B., Lefkowitz, E.J., Eds.; Elsevier: San Diego, CA, USA, 2011; pp. 351–373. [Google Scholar]
- Gronenborn, B. The Tomato Yellow Leaf Curl Virus Genome and Function of Its Proteins. In Tomato Yellow Leaf Curl Virus Disease: Management, Molecular Biology, Breeding for Resistance; Czosnek, H., Ed.; Springer: Dordrecht, The Netherlands, 2007; pp. 67–84. [Google Scholar]
- Kil, E.-J.; Kim, S.; Lee, Y.-J.; Byun, H.-S.; Park, J.; Seo, H.; Kim, C.-S.; Shim, J.-K.; Lee, J.-H.; Kim, J.-K.; et al. Tomato yellow leaf curl virus (TYLCV-IL): A seed-transmissible geminivirus in tomatoes. Sci. Rep. 2016, 6, 19013. [Google Scholar] [CrossRef] [PubMed]
- Moriones, E.; Arnó, J.; Accotto, G.P.; Noris, E.; Cavallarin, L. First report of Tomato Yellow Leaf Curl Virus in Spain. Plant Dis. 1993, 77, 953. [Google Scholar] [CrossRef]
- Noris, E.; Hidalgo, E.; Accotto, G.P.; Moriones, E. High similarity among the tomato yellow leaf curl virus isolates from the West Mediterranean Basin: The nucleotide sequence of an infectious clone from Spain. Arch. Virol. 1994, 135, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Navas-Castillo, J.; Sánchez-Campos, S.; Díaz, J.A.; Sáez-Alonso, E.; Moriones, E. First report of Tomato yellow leaf curl virus-Is in Spain: Coexistence of two different geminiviruses in the same epidemic outbreak. Plant Dis. 1997, 81, 1461. [Google Scholar] [CrossRef]
- Navas-Castillo, J.; Sánchez-Campos, S.; Noris, E.; Louro, D.; Accotto, G.P.; Moriones, E. Natural recombination between Tomato yellow leaf curl virus-Is and Tomato leaf curl virus. J. Gen. Virol. 2000, 81, 2797–2801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morilla, G.; Antúnez, C.; Bejarano, E.R.; Janssen, D.; Cuadrao, I.M. A New Tomato yellow leaf curl virus Strain in Southern Spain. Plant Dis. 2003, 87, 1004. [Google Scholar] [CrossRef]
- García-Andrés, S.; Accotto, G.P.; Navas-Castillo, J.; Moriones, E. Founder effect, plant host, and recombination shape the emergent population of begomoviruses that cause the tomato yellow leaf curl disease in the Mediterranean basin. Virology 2007, 359, 302–312. [Google Scholar] [CrossRef] [PubMed]
- Monci, F.; Sánchez-Campos, S.; Navas-Castillo, J.; Moriones, E. A natural recombinant between the geminiviruses Tomato yellow leaf curl Sardinia virus and Tomato yellow leaf curl virus exhibits a novel pathogenic phenotype and is becoming prevalent in Spanish populations. Virology 2002, 303, 317–326. [Google Scholar] [CrossRef] [PubMed]
- García-Andrés, S.; Monci, F.; Navas-Castillo, J.; Moriones, E. Begomovirus genetic diversity in the native plant reservoir Solanum nigrum: Evidence for the presence of a new virus species of recombinant nature. Virology 2006, 350, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Font, M.I.; Rubio, L.; Martínez-Culebras, P.V.; Jordá, C. Genetic structure and evolution of natural populations of viruses causing the tomato yellow leaf curl disease in Spain. Virus Res. 2007, 128, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Lapidot, M.; Friedman, M. Breeding for resistance to whitefly-transmitted geminivirus. Ann. Appl. Biol. 2002, 140, 109–127. [Google Scholar] [CrossRef]
- Butterbach, P.; Verlaan, M.G.; Dullemans, A.; Lohuis, D.; Visser, R.G.F.; Bai, Y.; Kormelink, R. Tomato yellow leaf curl virus resistance by Ty-1 involves increased cytosine methylation of viral genomes and is compromised by cucumber mosaic virus infection. Proc. Natl. Acad. Sci. USA 2014, 111, 12942–12947. [Google Scholar] [CrossRef] [PubMed]
- Zamir, D.; Ekstein-Michelson, I.; Zakay, Y.; Navot, N.; Zeidan, M.; Sarfatti, M.; Eshed, Y.; Harel, E.; Pleban, T.; van-Oss, H.; et al. Mapping and introgression of a tomato yellow leaf curl virus tolerance gene, TY-1. Theor. Appl. Genet. 1994, 88, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Schuster, D.J.; Scott, J.W. Ty-3, a begomovirus resistance locus near the Tomato yellow leaf curl virus resistance locus Ty-1 on chromosome 6 of tomato. Mol. Breed. 2007, 20, 271–284. [Google Scholar] [CrossRef]
- Verlaan, M.G.; Hutton, S.F.; Ibrahem, R.M.; Kormelink, R.; Visser, R.G.F.; Scott, J.W.; Edwards, J.D.; Bai, Y. The Tomato Yellow Leaf Curl Virus Resistance Genes Ty-1 and Ty-3 Are Allelic and Code for DFDGD-Class RNA-Dependent RNA Polymerases. PLoS Genet. 2013, 9. [Google Scholar] [CrossRef] [PubMed]
- Michelson, I.; Zamir, D.; Czosnek, H. Accumulation and translocation of tomato yellow leaf curl virus (TYLCV) in a Lycopersicon esculentum breeding line containing the L. chilense TYLCV tolerance gene Ty-1. Phytopathology 1994, 84, 928–933. [Google Scholar] [CrossRef]
- Pérez de Castro, A.; Díez, M.J.; Nuez, F. Evaluation of breeding tomato lines partially resistant to Tomato yellow leaf curl Sardinia virus and Tomato yellow leaf curl virus derived from Lycopersicon chilense. Can. J. Plant Pathol. 2005, 27, 268–275. [Google Scholar] [CrossRef]
- Belabess, Z.; Peterschmitt, M.; Granier, M.; Tahiri, A.; Blenzar, A.; Urbino, C. The non-canonical tomato yellow leaf curl virus recombinant that displaced its parental viruses in southern Morocco exhibits a high selective advantage in experimental conditions. J. Gen. Virol. 2016, 97, 3433–3445. [Google Scholar] [CrossRef] [PubMed]
- Glick, E.; Levy, Y.; Gafni, Y. The viral etiology of tomato yellow leaf curl disease-a review. Plant Prot. Sci. 2009, 45, 81–97. [Google Scholar] [CrossRef]
- García-Arenal, F.; Fraile, A.; Malpica, J.M. Variation and evolution of plant virus populations. Int. Microbiol. 2003, 6, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Lefeuvre, P.; Moriones, E. Recombination as a motor of host switches and virus emergence: Geminiviruses as case studies. Curr. Opin. Virol. 2015, 10, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Belabess, Z.; Dallot, S.; El-Montaser, S.; Granier, M.; Majde, M.; Tahiri, A.; Blenzar, A.; Urbino, C.; Peterschmitt, M. Monitoring the dynamics of emergence of a non-canonical recombinant of Tomato yellow leaf curl virus and displacement of its parental viruses in tomato. Virology 2015, 486, 291–306. [Google Scholar] [CrossRef] [PubMed]
- Muhire, B.M.; Varsani, A.; Martin, D.P. SDT: A virus classification tool based on pairwise sequence alignment and identity calculation. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Nei, M. Genetic Distance. In Encyclopedia of Genetics; Brenner, S., Miller, J., Eds.; Elsevier: San Diego, CA, USA, 2001; pp. 828–832. [Google Scholar]
- Duffy, S.; Holmes, E.C. Phylogenetic Evidence for Rapid Rates of Molecular Evolution in the Single-Stranded DNA Begomovirus Tomato Yellow Leaf Curl Virus. J. Virol. 2008, 82, 957–965. [Google Scholar] [CrossRef] [PubMed]
- Pamilo, P.; Bianchi, N.O. Evolution of the Zfx and Zfy genes: Rates and interdependence between the genes. Mol. Biol. Evol. 1993, 10, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Li, W.H. Unbiased estimation of the rates of synonymous and nonsynonymous substitution. J. Mol. Evol. 1993, 36, 96–99. [Google Scholar] [CrossRef] [PubMed]
- Hagen, C.; Frizzi, A.; Gabriels, S.; Huang, M.; Salati, R.; Gabor, B.; Huang, S. Accurate and sensitive diagnosis of geminiviruses through enrichment, high-throughput sequencing and automated sequence identification. Arch. Virol. 2012, 157, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Idris, A.; Al-Saleh, M.; Piatek, M.J.; Al-Shahwan, I.; Ali, S.; Brown, J.K. Viral metagenomics: Analysis of begomoviruses by illumina high-throughput sequencing. Viruses 2014, 6, 1219–1236. [Google Scholar] [CrossRef] [PubMed]
- Juarez, M.; Tovar, R.; Fiallo-Olive, E.; Aranda, M.A.; Gosalvez, B.; Castillo, P.; Moriones, E.; Navas-Castillo, J. First Detection of Tomato leaf curl New Delhi virus Infecting Zucchini in Spain. Plant Dis. 2013, 98, 857. [Google Scholar] [CrossRef]
- Ruiz, M.L.; Simon, A.; Velasco, L.; Garcia, M.C.; Janssen, D. First Report of Tomato leaf curl New Delhi virus Infecting Tomato in Spain. Plant Dis. 2015, 99, 894. [Google Scholar] [CrossRef]
- Zhou, X. Advances in understanding begomovirus satellites. Annu. Rev. Phytopathol. 2013, 51, 357–381. [Google Scholar] [CrossRef] [PubMed]
- Belabess, Z.; Urbino, C.; Granier, M.; Tahiri, A.; Blenzar, A.; Peterschmitt, M. The typical RB76 recombination breakpoint of the invasive recombinant tomato yellow leaf curl virus of Morocco can be generated experimentally but is not positively selected in tomato. Virus Res. 2018, 243, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Navas-Castillo, J.; Fiallo-Olivé, E.; Sánchez-Campos, S. Emerging Virus Diseases Transmitted by Whiteflies. Annu. Rev. Phytopathol. 2011, 49, 219–248. [Google Scholar] [CrossRef] [PubMed]
- Seal, S.E.; Jeger, M.J.; Van den Bosch, F. Begomovirus Evolution and Disease Management. Adv. Virus Res. 2006, 67, 297–316. [Google Scholar] [CrossRef] [PubMed]
- Delatte, H.; Dalmon, A.; Rist, D.; Soustrade, I.; Wuster, G.; Lett, J.M.; Goldbach, R.W.; Peterschmitt, M.; Reynaud, B. Tomato yellow leaf curl virus Can Be Acquired and Transmitted by Bemisia tabaci (Gennadius) from Tomato Fruit. Plant Dis. 2003, 87, 1297–1300. [Google Scholar] [CrossRef]
- Lapidot, M.; Polston, J.E. Resistance to Tomato yellow leaf curl virus in Tomato\n. Nat. Resist. Mech. Plants Viruses 2006, 503–520. [Google Scholar] [CrossRef]
- Seal, S.E.; Van den Bosch, F.; Jeger, M.J. Factors influencing begomovirus evolution and their increasing global significance: Implications for sustainable control. Crit. Rev. Plant Sci. 2006, 25, 23–46. [Google Scholar] [CrossRef]
- Puchades, A.V.; Carpino, C.; Alfaro-Fernandez, A.; Font-San-Ambrosio, M.I.; Davino, S.; Guerri, J.; Rubio, L.; Galipienso, L. Detection of Southern tomato virus by molecular hybridisation. Ann. Appl. Biol. 2017. [Google Scholar] [CrossRef]
- Doyle, J. DNA Protocols for Plants. In Molecular Techniques in Taxonomy; Hewitt, G.M., Johnston, A.W.B., Young, J.P.W., Eds.; Springer: Berlin, Germany, 1991; pp. 283–293. [Google Scholar]
- Accotto, G.P.; Navas-Castillo, J.; Noris, E.; Moriones, E.; Louro, D. Typing of tomato yellow leaf curl viruses in Europe. Eur. J. Plant Pathol. 2000, 106, 179–186. [Google Scholar] [CrossRef]
- Haible, D.; Kober, S.; Jeske, H. Rolling circle amplification revolutionizes diagnosis and genomics of geminiviruses. J. Virol. Methods 2006, 135, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.; Gish, W.; Miller, W.; Myers, E.; Lipman, D. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef] [PubMed]
- Stöver, B.C.; Müller, K.F. TreeGraph 2: Combining and visualizing evidence from different phylogenetic analyses. BMC Bioinform. 2010, 11, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Mol. Biol. Evol. 1992, 9, 678–687. [Google Scholar]
- Bouckaert, R.; Heled, J.; Kühnert, D.; Vaughan, T.G.; Wu, C.-H.; Xie, D.; Suchard, M.A.; Rambaut, A.; Drummond, A.J. Beast2: A software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2013. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 1–3. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015, 1, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Dellaporta, S.L.; Wood, J.; Hicks, J.B. A plant DNA minipreparation: Version II. Plant Mol. Biol. Report. 1983, 1, 19–21. [Google Scholar] [CrossRef]
- Urbino, C.; Gutiérrez, S.; Antolik, A.; Bouazza, N.; Doumayrou, J.; Granier, M.; Martin, D.P.; Peterschmitt, M. Within-Host Dynamics of the Emergence of Tomato Yellow Leaf Curl Virus Recombinants. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruijter, J.M.; Ramakers, C.; Hoogaars, W.M.H.; Karlen, Y.; Bakker, O.; van den hoff, M.J.B.; Moorman, A.F.M. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009, 37. [Google Scholar] [CrossRef] [PubMed]








| Tomato Cultivar a | Number of Samples | Sequence Name b | Year | Location | Field |
|---|---|---|---|---|---|
| ND | 2 | Mu 1.1 MZ:15 | 2015 | Mazarrón | 1 |
| Boludo | 2 | Mu 2.1 MZ:15/Mu 2.2 MZ:15 | 2015 | Mazarrón | 2 |
| ND | 2 | Mu 3.1 MZ:15/Mu 3.2 MZ:15 | 2015 | Mazarrón | 3 |
| ND | 2 | Mu 4.2 MZ:15 | 2015 | Mazarrón | 4 |
| ND | 2 | Mu 5.2 MZ:15 | 2015 | Mazarrón | 5 |
| Patriarca | 2 | Mu 1.1 AG:15 | 2015 | Águilas | 1 |
| Boludo | 2 | − | 2015 | Águilas | 2 |
| Cecilio | 2 | Mu 3.1 AG:15/Mu 3.2 AG:15 | 2015 | Águilas | 3 |
| Boludo | 2 | Mu 4.1 AG:15/Mu 4.2 AG:15 | 2015 | Águilas | 4 |
| Patriarca | 2 | Mu 5.1 AG:15 Mu 5.2a AG:15/Mu 5.2b AG:15 | 2015 | Águilas | 5 |
| Jawara | 5 | Mu 1 MZ:16 | 2016 | Mazarrón | 1 |
| Boludo | 5 | Mu 2 MZ:16 | 2016 | Mazarrón | 2 |
| Boludo, Duratom | 5 | Mu 3 MZ:16 | 2016 | Mazarrón | 3 |
| Ramyle, Boludo | 5 | Mu 4 MZ:16 | 2016 | Mazarrón | 4 |
| Jawara, Patriarca | 5 | Mu 1 AG:16 | 2016 | Águilas | 1 |
| Boludo, Jawara | 5 | Mu 2 AG:16 | 2016 | Águilas | 2 |
| Myla | 5 | Mu 3 AG:16 | 2016 | Águilas | 3 |
| Grandoly | 5 | Mu 4 AG:16 | 2016 | Águilas | 4 |
| Gene | dn | ds | dns − ds |
|---|---|---|---|
| V1 | 0.006 ± 0.002 | 0.057 ± 0.014 | −0.051 |
| V2 | 0012 ± 0.004 | 0.03 ± 0.013 | −0.018 |
| C1 | 0.005 ± 0.001 | 0.014 ± 0.004 | −0.009 |
| C4 | 0.005 ± 0.003 | 0.009 ± 0.008 | −0.004 |
| C2 | 0.006 ± 0.002 | 0.012 ± 0.006 | −0.006 |
| C3 | 0.013 ± 0.004 | 0.005 ± 0.004 | 0.008 |
| Cultivar | Treatment a | Num. Infected Plants |
|---|---|---|
| Pristyla (Ty-1/ty-1) | Mu15 | 11 |
| IL | 6 | |
| IS76 | 6 | |
| Mu15 + IS76 | 12 | |
| IL + IS76 | 6 | |
| Mock b | 3 | |
| Susceptible (ty-1/ty-1) | Mu15 | 11 |
| IL | 7 | |
| IS76 | 7 | |
| Mu15 + IS76 | 8 | |
| IL + IS76 | 12 | |
| Mock b | 3 |
| Clone (GenBank acc.) | Primer | Sequence (5′-3′) | Amplicon Size (bp) | Temperature (°C) | Reference |
|---|---|---|---|---|---|
| TYLCV-IL (AM409201) | Il-fw | AATGGCTATTTGGTAATTTCG | 146 | 63 | [27] |
| Il-rev | CGTCTGTGGAACCCTCG | ||||
| TYLCV-IS76 (LN812978) | IS76-fw | CCGATAAAGTAGTAGGCCCTACGCA | 135 | 60 | [27] |
| IS76-rev | AGTGGGTCCCACATATTGCAAGAC | ||||
| pTYLCV-Mu15 a | Mu-fw | AATGGCTATTTGGTAATTTCG | 146 | 63 | [27] |
| Mu-rev | CGTCTGTGGAACCCTAG | ||||
| Tomato gene 25S RNA b | 25S-fw | AGAACTGGCGATGCGGGATG | 161 | 60 | [27] |
| 25S-rev | GTTGATTCGGCAGGTGAGTTGT |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Torre, C.; Donaire, L.; Gómez-Aix, C.; Juárez, M.; Peterschmitt, M.; Urbino, C.; Hernando, Y.; Agüero, J.; Aranda, M.A. Characterization of Begomoviruses Sampled during Severe Epidemics in Tomato Cultivars Carrying the Ty-1 Gene. Int. J. Mol. Sci. 2018, 19, 2614. https://doi.org/10.3390/ijms19092614
Torre C, Donaire L, Gómez-Aix C, Juárez M, Peterschmitt M, Urbino C, Hernando Y, Agüero J, Aranda MA. Characterization of Begomoviruses Sampled during Severe Epidemics in Tomato Cultivars Carrying the Ty-1 Gene. International Journal of Molecular Sciences. 2018; 19(9):2614. https://doi.org/10.3390/ijms19092614
Chicago/Turabian StyleTorre, Covadonga, Livia Donaire, Cristina Gómez-Aix, Miguel Juárez, Michel Peterschmitt, Cica Urbino, Yolanda Hernando, Jesús Agüero, and Miguel A. Aranda. 2018. "Characterization of Begomoviruses Sampled during Severe Epidemics in Tomato Cultivars Carrying the Ty-1 Gene" International Journal of Molecular Sciences 19, no. 9: 2614. https://doi.org/10.3390/ijms19092614