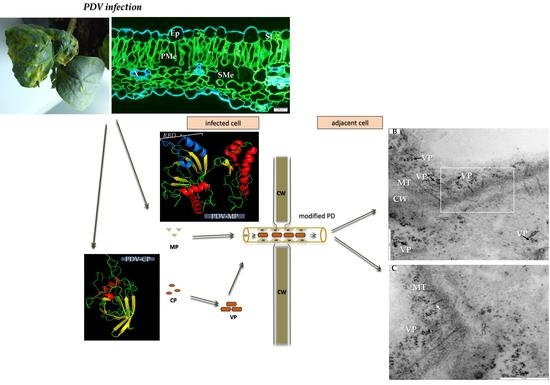
Ultrastructural Analysis of Prune Dwarf Virus Intercellular Transport and Pathogenesis
Abstract
:1. Introduction
2. Results
2.1. Symptomatology Induced by Prune Dwarf Virus (PDV) in Cucumis sativus
2.2. Immunofluorescence Localization of Coat Protein (CP) in PDV-Infected Cucumber Leaves
2.3. Ultrastructural Alterations in Cucumber Cells During PDV Infection
2.4. Modeling of 3D Structures of PDV-0599 and Alfalfa Mosaic Virus-VRU (AMV-VRU) Movement Proteins (MPs)
2.5. Localization and Quantification of PDV-CP and MP in Infected Cucumber Leaves by Double-Immunogold
3. Discussion
4. Materials and Methods
4.1. Virus Inoculation
4.2. Plant Material Preparations for Immunofluorescence and Immunofluorescence Localization of Coat Protein (CP) in Cucumber Leaves
4.3. Preparation of Plant Material for Transsmision Electron Microscopy (TEM) and Double-Immunogold Localization of CP and MP
4.4. Comparative Analyses of 3D Models of PDV-0599 and AMV-VRU
4.5. Double-Immunogold Localization and Quantification of PDV-CP and MP in Cucumber Leaves
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bujarski, J.J.; Figlerowicz, M.; Gallitelli, D.; Roossinck, M.J.; Scott, S.W. Family Bromoviridae. In Virus Taxonomy: Classification and Nomenclature of Viruses-Ninth Report of the International Committee on Taxonomy of Viruses, 1st ed.; King, A.M.Q., Adams, M.J., Carstens, E.B., Lefkowitz, E.J., Eds.; Elsevier Academic Press: San Diego, CA, USA, 2012; pp. 972–976. ISBN 978-0123846846. [Google Scholar]
- Brunt, H.A.; Crabtree, K.; Dallawitz, M.J.; Gibs, A.J.; Watson, L. Viruses of Plants, 1st ed.; CAB International UK: Wallingford, UK, 1996; ISBN 978-0851987941. [Google Scholar]
- Paduch-Cichal, E. Characterization of PNRSV and PDV. Associate Professor Thesis, Warsaw University of Life Sciences, Warsaw, Poland, 2000. [Google Scholar]
- Fulton, R.W. Prune dwarf virus. C.M.I/A.A.B. Descr. Plant Viruses 1970, 1. [Google Scholar]
- Pallas, V.; Aparicio, F.; Herranz, M.C.; Amari, K.; Sanchez-Pina, M.A.; Myrta, A.; Sanchez-Navarro, J.A. Ilarviruses of Prunus spp.: A continued concern for fruit trees. Phytopathology 2012, 102, 1108–1120. [Google Scholar] [CrossRef] [PubMed]
- Pallas, V.; Aparicio, F.; Herranz, M.C.; Sanchez-Navarro, J.A.; Scott, S.W. The molecular biology of ilarviruses. Adv. Virus Res. 2013, 87, 139–183. [Google Scholar] [CrossRef] [PubMed]
- Kalinowska, E.; Mroczkowska, K.; Paduch-Cichal, E.; Chodorska, M. Genetic variability among coat protein of Prune dwarf virus variants from different countries and different Prunus species. Eur. J. Plant Pathol. 2014, 4, 863–868. [Google Scholar] [CrossRef]
- Nemeth, M. Virus, Mycoplasma and Rikettsia Diseases of Fruit Trees, 1st ed.; Springer: Budapest, Hungary, 1986; pp. 600–650. ISBN 978-90-247-2868-8. [Google Scholar]
- Sztuba-Solińska, J.; Bujarski, J.J. Insights into the single-cell reproduction cycle of members of the family Bromoviridae: Lessons from the use of protoplast systems. J. Virol. 2008, 82, 10330–10340. [Google Scholar] [CrossRef] [PubMed]
- Bol, J.F. Replication of alfamo- and ilarviruses: Role of the coat protein. Annu. Rev. Phytopathol. 2005, 43, 39–62. [Google Scholar] [CrossRef] [PubMed]
- Kozieł, E.; Otulak, K.; Garbaczewska, G. Phylogenetic analysis of PDV movement protein compared to Bromoviridae members as justification of possible intercellular movement. Acta Biol. Crac. Ser. Bot. 2015, 57, 19–31. [Google Scholar] [CrossRef]
- Kozieł, E.; Bujarski, J.J.; Otulak, K. Molecular biology of Prune Dwarf Virus—A lesser known member of the Bromoviridae but a vital component in the dynamic virus–host cell interaction network. Int. J. Mol. Sci. 2017, 18, 2733. [Google Scholar] [CrossRef] [PubMed]
- Paduch-Cichal, E.; Sala-Rejczak, K. Biological properties, stability in crude sap and serological characterization of Prune dwarf virus (PDV) isolates from Prunus avium seedlings. Phytopathol. Pol. 2003, 29, 9–22. [Google Scholar]
- Boulila, M. Molecular characterization of an almond isolate of Prune dwarf virus in Tunisia: Putative recombination breakpoints in the partial sequences of the coat protein-encoding gene in isolates from different geographic origin. Phytopathol. Mediterr. 2009, 48, 411–421. [Google Scholar] [CrossRef]
- Öztürk, Y.; Çevik, B. Genetic Diversity in the coat rotein genes of Prune dwarf virus isolates from sweet cherry growing in Turkey. Plant Pathol. J. 2015, 31, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Kinoti, W.M.; Constable, F.E.; Nancarrow, N.; Plummer, K.M.; Rodoni, B. The incidence and genetic diversity of Apple mosaic virus (ApMV) and Prune dwarf virus (PDV) Prunus species in Australia. Viruses 2018, 10, 136. [Google Scholar] [CrossRef] [PubMed]
- Nemeth, M. Interferencja vizsgălatok a csonthĕjas gyümöcsfăk gyürüsfoltossăg (ringspot) virusavial. Növĕnyvĕdelem 1972, 8, 64–71. [Google Scholar]
- Myrta, A.; Savino, V. Virus and virus-like diseases of cherry in the Mediterranean region. Acta Hortic. 2008, 795, 891–896. [Google Scholar] [CrossRef]
- Çaglayan, K.; Gazel, M. Virus and virus-like diseases of stone fruits in the eastern Mediterranean area of Turkey. Acta Hortic. 1998, 472, 527–529. [Google Scholar] [CrossRef]
- Fulton, R.W. Comparative host ranges of certain mechanically transmitted viruses of Prunus. Phytopatology 1957, 47, 215–220. [Google Scholar]
- Halk, R.; Fulton, R.W. Stabilization and particle morphology of Prune dwarf virus. Virology 1978, 91, 434–443. [Google Scholar] [CrossRef]
- Fulton, R.W. Purification of Sour cherry necrotic ringspot and Prune dwarf viruses. Virology 1959, 9, 522–535. [Google Scholar] [CrossRef]
- Cropley, R.; Gilmer, R.M.; Posnette, A.F. Necrotic ring spot and Prune dwarf viruses in Prunus and in herbaceous indicators. Ann. Appl. Biol. 1964, 53, 325–332. [Google Scholar] [CrossRef]
- Malinowski, T.; Zawadzka, B. Use of ELISA method in detection of fruit trees viruses. In Proceedings of the XXXIII Poland Pomology Conference, Skierniewice, Poland, 30 August–1 September 1994; The Research Institute of Pomology: Skierniewice, Poland, 1994; pp. 9–11. [Google Scholar]
- Kozieł, E. Pathogenic Changes in Vegetative Organs of Plum Tree and Test Plants Infected by Prune Dwarf Virus (PDV). Ph.D. Thesis, Warsaw University of Life Sciences, Warsaw, Poland, 2016. [Google Scholar]
- Waterworth, H.E.; Fulton, R.W. Variation among isolates of necrotic ringspot and Prune dwarf viruses isolated from sour cherry. Phytopathology 1964, 54, 1155–1160. [Google Scholar]
- Zaumayer, W.J. Alfalfa yellow mosaic virus systemically infectious to beans. Phytopathology 1953, 43, 38–42. [Google Scholar]
- Zaumayer, W.J. Two new strains of Alfalfa mosaic virus systemically infectious to bean. Phytopathology 1963, 53, 444–449. [Google Scholar]
- Zaumayer, W.J.; Patino, G. Vein Necrosis, another systemically infectious strain of Alfalfa mosaic virus in bean. Phytopathology 1960, 50, 226–231. [Google Scholar]
- Buchanan, B.B.; Gruissem, W.; Jones, R.L. Biochemistry and Molecular Biology of Plants, 2nd ed.; Wiley-Blackwell: New York, NY, USA, 2015; pp. 984–1040. ISBN 978-0-470-71421-8. [Google Scholar]
- Schoelz, J.E.; Harries, P.A.; Nelson, R.S. Intracellular transport of plant viruses: Finding the door out of the cell. Mol. Plant 2011, 4, 813–831. [Google Scholar] [CrossRef] [PubMed]
- Kozieł, E.; Otulak, K.; Lockhart, B.E.L.; Garbaczewska, G. Subcelullar localization of proteins associated with Prune dwarf virus replication. Eur. J. Plant Pathol. 2017, 149, 653–668. [Google Scholar] [CrossRef]
- Mochizuki, T.; Nobuhara, S.; Nishimura, M.; Ryang, B.S.; Naoe, M.; Matsumoto, T.; Kosaka, Y.; Ohki, S.T. The entry of Cucumber mosaic virus into cucumber xylem is facilitated by co-infection with Zucchini yellow mosaic virus. Arch. Virol. 2016, 161, 2683–2692. [Google Scholar] [CrossRef] [PubMed]
- Van Pelt-Heerschap, H.; Verbeek, H.; Huisman, M.J.; Loesch-Fries, L.S.; Van Vloten-Doting, L. Non-structural proteins and RNAs of Alfalfa mosaic virus synthesized in tobacco and cowpea protoplasts. Virology 1987, 161, 190–197. [Google Scholar] [CrossRef]
- Van Pelt-Heerschap, H. Immunochemical Analysis of the Alfalfa mosaic virus Gene Products. Ph.D. Thesis, Leiden University, Leiden, The Netherlands, 1987. [Google Scholar]
- Van der Kuyl, A.C.; Neeleman, L.; Bol, J.F. Deletion analysis of cis and trans acting elements involved in the replication of alfalfa mosaic virus RNA 3 in vivo. Virology 1991, 183, 687–694. [Google Scholar] [CrossRef]
- Hull, R. Plant Virology, 5th ed.; Elsevier Academic Press: London, UK, 2013; pp. 148–603. ISBN 9780123848710. [Google Scholar]
- Matthews, R.E.F. Plant Virology, 3th ed.; Elsevier Academic Press: London, UK, 1991; pp. 143–519. ISBN 978-0-12-480553-8. [Google Scholar]
- Dangl, J.L.; Dietrich, R.A.; Richberg, M.H. Death don’t have no mercy: Cell death programs in plant-microbeinteractions. Plant Cell 1996, 8, 1793–1807. [Google Scholar] [CrossRef] [PubMed]
- Chikh Ali, M.; Karasev, A.V.; Furutani, N.; Taniguchi, M.; Kano, Y.; Sato, M.; Natsuaki, T.; Maoka, T. Occurrence of Potato virus Y strain PVY NTN in foundation seed potatoes in Japan, and screening forsymptoms in Japanese potato cultivars. Plant Pathol. 2013, 62, 1157–1165. [Google Scholar] [CrossRef]
- Otulak-Kozieł, K.; Kozieł, E.; Lockhart, B.E.L. Plant cell wall dynamics in compatible and incompatible potato response to infection caused by Potato virus Y (PVYNTN). Int. J. Mol. Sci. 2018, 19, 862. [Google Scholar] [CrossRef] [PubMed]
- Alexander, M.M.; Cilia, M. A molecular tug-of-war: Global plant proteome changes during viral infection. Curr. Plant Biol. 2016, 5, 13–24. [Google Scholar] [CrossRef]
- Di Carli, M.; Benvenuto, E.; Donini, M. Recent insights into plant–virus interactions through proteomic analysis. J. Proteome Res. 2012, 11, 4765–4780. [Google Scholar] [CrossRef] [PubMed]
- Bruknard, J.O.; Zambryski, P.C. Plasmodesmata enable multicellularity: New insights into their evolution, biogenesis, and functions in development and immunity. Curr. Opin. Plant Biol. 2017, 35, 76–83. [Google Scholar] [CrossRef]
- Bucher, G.L.; Tarina, C.; Heinlein, M.; Di Serio, F.; Meins, F., Jr.; Iglesias, V.A. Local expression of enzymatically active class I beta-1,3-glucanase enhances symptoms of TMV infection in tobacco. Plant J. 2001, 28, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Favali, M.A.; Conti, G.G. Ultrastructural observations on the chloroplasts of basil plants either infected with different viruses or treated with 3-amino-l,2,4-triazole. Protoplasma 1970, 70, 153–166. [Google Scholar] [CrossRef]
- Bamunusinghe, D.; Seo, J.K.; Rao, A.L. Subcellular localization and rearrangement of endoplasmic reticulum by Brome mosaic virus capsid protein. J. Virol. 2011, 85, 2953–2963. [Google Scholar] [CrossRef] [PubMed]
- Hull, R.; Hills, G.J.; Markham, R. Studies on Alfalfa mosaic virus: II. The structure of the virus components. Virology 1969, 37, 416–428. [Google Scholar] [CrossRef]
- Hull, R.; Rees, M.W.; Short, M.N. Studies on Alfalfa mosaic virus: I. The protein and nucleic acid. Virology 1969, 37, 404–415. [Google Scholar] [CrossRef]
- Verchot, J. Wrapping membranes around plant virus infection. Curr. Opin. Virol. 2011, 1, 388–395. [Google Scholar] [CrossRef] [PubMed]
- Bamunusinghe, D.; Chaturvedi, S.; Seo, J.K.; Rao, A.L. Mutations in the capsid protein of Brome mosaic virus affecting encapsidation eliminate vesicle induction in planta: Implications for virus cell-to-cell spread. J. Virol. 2013, 87, 8982–8992. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, A.; Hutchens, H.M.; Berg, R.H.; Loesch-Fries, S. Alfalfa mosaic virus replicase proteins, P1 and P2, localize to the tonoplast in the presence of virus RNA. Virology 2012, 433, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Rao, A.L.N. Molecular studies on Bromovirus capsid protein. III. Analysis of cell-to-cell movement competence of coat protein defective variants of cowpea chlorotic mottle virus. Virology 1997, 232, 385–395. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Navarro, J.A.; Herranz, M.C.; Pallas, V. Cell-to-cell movement of Alfalfa mosaic virus can be mediated by the movement proteins of Ilar-, bromo-, cucumo-, tobamo- and comoviruses and does not require virion formation. Virology 2006, 346, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Flasinski, S.; Dzianott, A.; Pratt, S.; Bujarski, J. Mutational analysis of coat protein gene of brome mosaic virus: Effects on replication and movement protein in barley and on Chenopodium hybridum. Mol. Plant Microbe Interact. 1995, 8, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Rao, A.L.; Grantham, G.L. Biological significance of the seven amino-terminal basic residues of brome mosaic virus coat protein. Virology 1995, 211, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Van der Vossen, E.A.; Neeleman, L.; Bol, J.F. Early and late functions of Alfalfa mosaic virus coat protein can be mutated separately. Virology 1994, 202, 891–903. [Google Scholar] [CrossRef] [PubMed]
- Van der Wel, N.N.; Goldbach, R.W.; van Lent, J. The movement protein and coat protein of Alfalfa mosaic virus accumulate in structurally modified plasmodesmata. Virology 1998, 244, 322–329. [Google Scholar] [CrossRef] [PubMed]
- Kaido, M.; Inoue, Y.; Takeda, Y.; Sugiyama, K.; Takeda, A.; Mori, M.; Tamai, A.; Meshi, T.; Okuno, T.; Mise, K. Downregulation of the NbNACa1 gene encoding a movement-protein-interacting protein reduces cell-to-cell movement of brome mosaic virus in Nicotiana benthamiana. Mol. Plant Microbe Interact. 2007, 20, 671–681. [Google Scholar] [CrossRef] [PubMed]
- Kasteel, D.T.J.; van der Wel, N.N.; Jansen, K.A.J.; Goldbach, R.W.; van Lent, J.W.M. Tubule-forming capacity of the movement proteins of Alfalfa mosaic virus and brome mosaic virus. J. Gen. Virol. 1997, 78, 2089–2093. [Google Scholar] [CrossRef] [PubMed]
- Codoñer, F.M.; Cuevas, J.M.; Sánchez-Navarro, J.A.; Pallas, V.; Elena, S.F. Molecular evolution of the plant virus family Bromoviridae based on RNA3-encoded proteins. J. Mol. Evol. 2005, 61, 697–705. [Google Scholar] [CrossRef] [PubMed]
- Codoñer, F.M.; Fares, M.A.; Elena, S.F. Adaptive covariation between the coat and movement proteins of Prunus necrotic ringspot virus. J. Virol. 2006, 80, 5833–5840. [Google Scholar] [CrossRef] [PubMed]
- International Committee on Taxonomy of Viruses (ICTV) Official Website. Available online: https://talk.ictvonline.org/ictv-reports/ictv_9th_report/positive-sense-rna-viruses-2011/w/posrna_viruses/251/bromoviridae (accessed on 16 April 2018).
- Tomenius, K.; Clapham, D.; Meshi, T. Localization by immunogold cytochemistry of the virus coded 30K protein in plasmodesmata of leaves infected with tobacco mosaic virus. Virology 1987, 160, 363–371. [Google Scholar] [CrossRef]
- Lucas, W.J.; Ding, B.; Van der Schoot, C. Plasmodesmata and the supracellular nature of plants. New Phytol. 1993, 125, 435–476. [Google Scholar] [CrossRef] [Green Version]
- Herranz, M.C.; Sanchez-Navarro, J.A.; Sauri, A.; Mingarro, I.; Pallas, V. Mutational analysis of the RNA-binding domain of the prunus necrotic ringspot virus (PNRSV) movement protein reveals its requirement for cell-to-cell movement. Virology 2005, 339, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, I.B.; Zhang, L.; Palukatis, P. Characterization of Cucumber mosaic virus: Cell to cell movement requires capsid protein but not virions. Virology 1998, 246, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, I.; Rao, A.L.N. Deletions in the conserved amino-terminal basic arm of cucumber mosaic virus coat protein disrupt virion assembly but do not abolish infectivity and cell-to-cell movement. Virology 1998, 248, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Kuwata, S.; Kataoka, J.; Masuta, C.; Nitta, N.; Takanami, Y. Functional analysis of deletion mutants of Cucumber mosaic virus RNA3 using an in vitro transcription system. Virology 1991, 183, 106–113. [Google Scholar] [CrossRef]
- Canto, T.; Prior, D.A.M.; Hellwald, K.H.; Oparka, K.J.; Palukaitis, P. Characterization of Cucumber mosaic virus. IV. Movement protein and coat protein are both essential for cell-to-cell movement of CMV. Virology 1997, 237, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, I.B.; Gal-On, A.; Palukaitis, P. Characterization of Cucumber mosaic virus. III. Localization of sequences in the move- ment protein controlling systemic infection in cucurbits. Virology 1997, 230, 343–349. [Google Scholar] [CrossRef] [PubMed]
- Canto, T.; Palukaitis, P. Are tubules generated by the 3a protein necessary for cucumber mosaic virus movement? Mol. Plant Microbe Interact. 1999, 12, 985–993. [Google Scholar] [CrossRef]
- Lucas, W. Plant viral movement proteins: Agents for cell-to-cell trafficking of viral genomes. Virology 2006, 344, 169–184. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.B.; Palukaitis, P. Comparison of the nucleic acid- and NTP-binding properties of the movement protein of cucumber mosaic cucumovirus and tobacco mosaic tobamovirus. Virology 1996, 216, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Okinaka, Y.; Mise, K.; Suzuki, E.; Okuno, T.; Furusawa, I. The C terminus of brome mosaic virus coat protein controls viral cell-to-cell and long-distance movement. J. Virol. 2001, 75, 5385–5390. [Google Scholar] [CrossRef] [PubMed]
- Otulak, K.; Kozieł, E.; Lockhart, B.E.L.; Garbaczewska, G. Ultrastructural effects of PVYNTN infection of Capsicum annuum L. cv. Yolo Wonder generative organs; a first step in describing seed transmission. Phytopathol. Mediterr. 2017, 56, 379–391. [Google Scholar] [CrossRef]
- Otulak, K.; Garbaczewska, G. Ultrastructural events during hypersensitive response of potato cv. Rywal infected with necrotic strains of potato virus Y. Acta Physiol. Plant. 2010, 32, 635–644. [Google Scholar] [CrossRef]
- Hayat, M. Basic Techniques for Transmission Electron Microscopy, 1st ed.; Elsevier Academic Press International: San Diego, CA, USA, 1986; pp. 285–370. ISBN 978-0-12-333926-3. [Google Scholar]
- NCBI Protein Database Official Website. Available online: https://www.ncbi.nlm.nih.gov/protein/ (accessed on 28 April 2018).
- Jalview Official website. Available online: http://www.jalview.org/ (accessed on 28 April 2018).
- AIDA Server Official Website. Available online: http://aida.godziklab.org/AIDA/ (accessed on 28 April 2018).
- PyMOL Official Website. Available online: https://pymol.org/2/ (accessed on 28 April 2018).
- Mayhew, T.M. Quantifying immunogold localization on electron microscopic thin sections: A compendium of new approaches for plant cell biologists. J. Exp. Bot. 2011, 62, 4101–4113. [Google Scholar] [CrossRef] [PubMed]
- GraphPad Software Official Website. Available online: https://www.graphpad.com/quickcalcs/contingency1.cfm (accessed on 28 April 2018).
- Mayhew, T.M.; Lucocq, J.M. Multiple-labelling immunoEM using different sizes of colloidal gold: Alternative approaches to test for differential distribution and colocalization in subcellular structures. Histochem. Cell Biol. 2011, 135, 317–326. [Google Scholar] [CrossRef] [PubMed]









| Double-Immunolocalization Parameters | ||||
|---|---|---|---|---|
| (A) PDV-infected mesophyll cells: | ||||
| -plasmodesmata and tubular structures | ||||
| Protein | MPg10+ | MPg10− | Row totals | Ratio MPg10+/MPg10− |
| CPg20+ | 47 | 6 | 53 | 7.83 |
| CPg20− | 13 | 20 | 14 | 0.65 |
| Column totals | 60 | 27 | 87 | OR = 12.04 |
| -cytoplasm | ||||
| Protein | MPg10+ | MPg10− | Row totals | Ratio MPg10+/MPg10− |
| CPg20+ | 41 | 24 | 65 | 1.7 |
| CPg20− | 9 | 30 | 39 | 0.3 |
| Column totals | 50 | 54 | 104 | OR = 5.66 |
| -vacuole | ||||
| Protein | MPg10+ | MPg10− | Row totals | Ratio MPg10+/MPg10− |
| CPg20+ | 20 | 16 | 36 | 1.25 |
| CPg20− | 12 | 23 | 34 | 0.52 |
| Column totals | 32 | 38 | 70 | OR = 2.40 |
| (B) Mock inoculated mesophyll cells: | ||||
| -Plasmodesmata, tubular structures, cytoplasm and vacuole | ||||
| Protein | MPg10+ | MPg10− | Row totals | Ratio MPg10+/MPg10− |
| CPg20+ | 0 | 0 | 0 | 0 |
| CPg20− | 0 | 0 | 0 | 0 |
| Column totals | 0 | 0 | 0 | 0 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kozieł, E.; Otulak-Kozieł, K.; Bujarski, J.J. Ultrastructural Analysis of Prune Dwarf Virus Intercellular Transport and Pathogenesis. Int. J. Mol. Sci. 2018, 19, 2570. https://doi.org/10.3390/ijms19092570
Kozieł E, Otulak-Kozieł K, Bujarski JJ. Ultrastructural Analysis of Prune Dwarf Virus Intercellular Transport and Pathogenesis. International Journal of Molecular Sciences. 2018; 19(9):2570. https://doi.org/10.3390/ijms19092570
Chicago/Turabian StyleKozieł, Edmund, Katarzyna Otulak-Kozieł, and Józef J. Bujarski. 2018. "Ultrastructural Analysis of Prune Dwarf Virus Intercellular Transport and Pathogenesis" International Journal of Molecular Sciences 19, no. 9: 2570. https://doi.org/10.3390/ijms19092570