NLRP3-Dependent and -Independent Processing of Interleukin (IL)-1β in Active Ulcerative Colitis
Abstract
:1. Introduction
2. Results
2.1. Clinical Characteristics of IBD Cohort
2.2. NLRP3 and IL-1β Are Upregulated in Active Disease
2.3. NLRP3 is Localized to the Influx of Lamina Propria Cells in Active UC
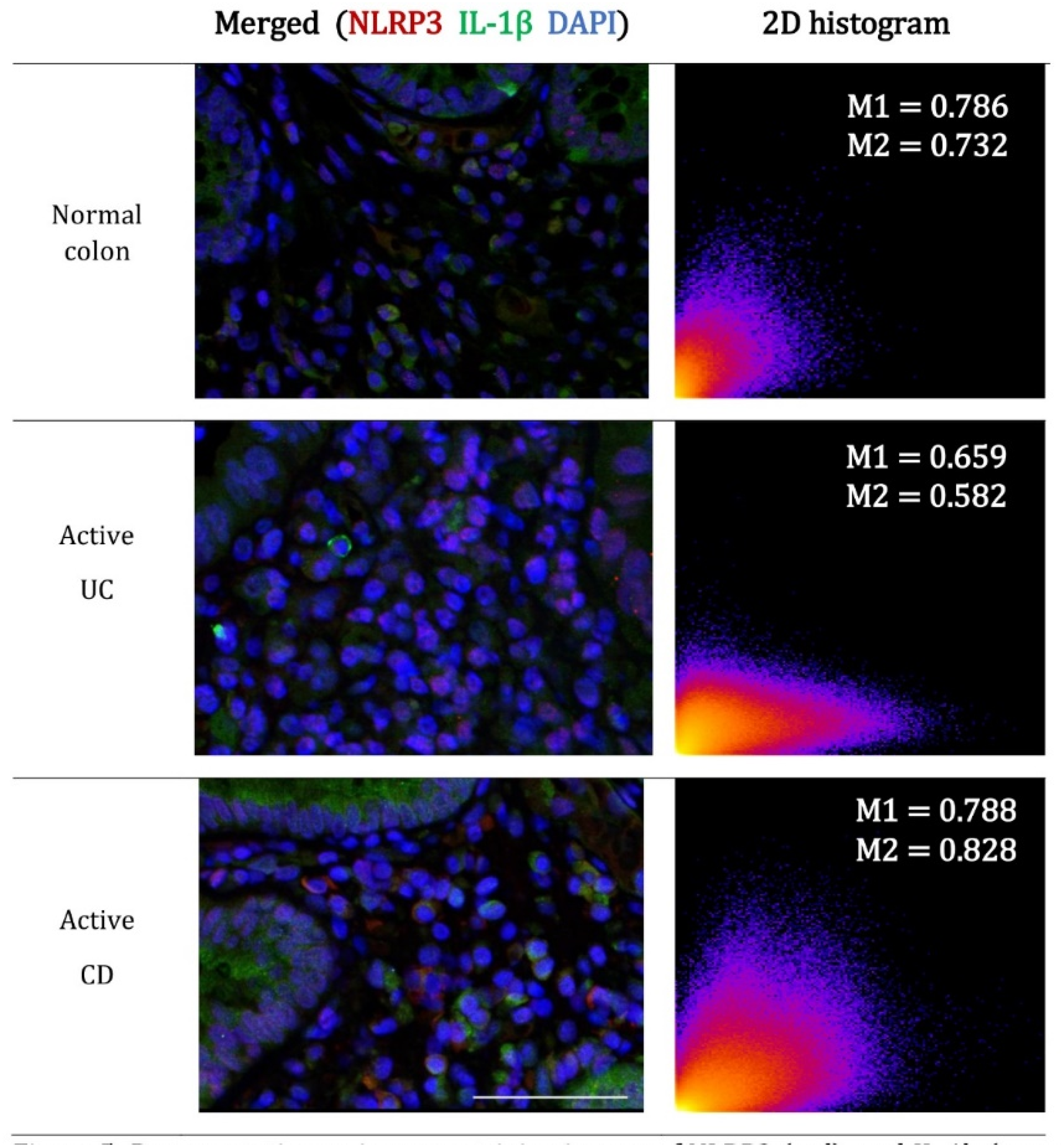
2.4. The Contribution of NLRP3 to IL-1β Is Reduced in Active UC
3. Discussion
4. Materials and Methods
4.1. Ethics
4.2. Study Participants and Biopsy Collection
4.3. Gene Expression Analysis
4.4. Immunohistochemistry Analysis
4.5. Immunofluorescence confocal Microscopy
4.6. Data Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ASC | Apoptosis-associated speck-like protein containing a CARD |
| CASP1 | Caspase-1 |
| CD | Crohn’s disease |
| IBD | Inflammatory bowel disease |
| IL-1β | Interleukin 1, beta |
| mRNA | Messenger ribonucleic acid |
| NLRP3 | Nod-like receptor family, pyrin domain containing 3 |
| UC | Ulcerative colitis |
| qRT-PCR | Quantitative real-time-polymerase chain reaction |
References
- Bickston, S.J.; Bloomfeld, R.S. Handbook of Inflammatory Bowel Disease, 1st ed.; Lippincott Williams and Wilkins: Philadelphia, PA, USA, 2010; pp. 1–175. [Google Scholar]
- Cho, J.H. The genetics and immunopathogenesis of inflammatory bowel disease. Nat. Rev. Immunol. 2008, 8, 458–466. [Google Scholar] [CrossRef] [PubMed]
- Dinarello, C.A. A clinical perspective of IL-1beta as the gatekeeper of inflammation. Eur. J. Immunol. 2011, 41, 1203–1217. [Google Scholar] [CrossRef]
- Lamkanfi, M.; Dixit, V.M. Inflammasomes: Guardians of cytosolic sanctity. Immunol. Rev. 2009, 227, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Becker, C.E.; O’Neill, L.A. Inflammasomes in inflammatory disorders: The role of TLRs and their interactions with NLRs. Semin. Immunopathol. 2007, 29, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Guarda, G.; So, A. Regulation of inflammasome activity. Immunology 2010, 130, 329–336. [Google Scholar] [CrossRef] [Green Version]
- Kanneganti, T.D. Central roles of NLRs and inflammasomes in viral infection. Nat. Rev. Immunol. 2010, 10, 688–698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sahoo, M.; Ceballos-Olvera, I.; del Barrio, L.; Re, F. Role of the inflammasome, IL-1beta, and IL-18 in bacterial infections. Sci. World J. 2011, 11, 2037–2050. [Google Scholar] [CrossRef]
- Martinon, F.; Tschopp, J. Inflammatory caspases and inflammasomes: Master switches of inflammation. Cell Death Differ. 2007, 14, 10–22. [Google Scholar] [CrossRef]
- Schroder, K.; Tschopp, J. The inflammasomes. Cell 2010, 140, 821–832. [Google Scholar] [CrossRef]
- Bauernfeind, F.G.; Horvath, G.; Stutz, A.; Alnemri, E.S.; MacDonald, K.; Speert, D.; Fernandes-Alnemri, T.; Wu, J.; Monks, B.G.; Fitzgerald, K.A.; et al. Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J. Immunol. 2009, 183, 787–791. [Google Scholar] [CrossRef]
- Franchi, L.; Eigenbrod, T.; Nunez, G. Cutting edge: TNF-alpha mediates sensitization to ATP and silica via the NLRP3 inflammasome in the absence of microbial stimulation. J. Immunol. 2009, 183, 792–796. [Google Scholar] [CrossRef] [PubMed]
- Martinon, F.; Mayor, A.; Tschopp, J. The inflammasomes: Guardians of the body. Annu. Rev. Immunol. 2009, 27, 229–265. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Planillo, R.; Kuffa, P.; Martinez-Colon, G.; Smith, B.L.; Rajendiran, T.M.; Nunez, G. K(+) efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity 2013, 38, 1142–1153. [Google Scholar] [CrossRef] [PubMed]
- Hornung, V.; Bauernfeind, F.; Halle, A.; Samstad, E.O.; Kono, H.; Rock, K.L.; Fitzgerald, K.A.; Latz, E. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat. Immunol. 2008, 9, 847–856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimada, K.; Crother, T.R.; Karlin, J.; Dagvadorj, J.; Chiba, N.; Chen, S.; Ramanujan, V.K.; Wolf, A.J.; Vergnes, L.; Ojcius, D.M.; et al. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity 2012, 36, 401–414. [Google Scholar] [CrossRef] [PubMed]
- Nakahira, K.; Haspel, J.A.; Rathinam, V.A.; Lee, S.J.; Dolinay, T.; Lam, H.C.; Englert, J.A.; Rabinovitch, M.; Cernadas, M.; Kim, H.P.; et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat. Immunol. 2011, 12, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Iyer, S.S.; He, Q.; Janczy, J.R.; Elliott, E.I.; Zhong, Z.; Olivier, A.K.; Sadler, J.J.; Knepper-Adrian, V.; Han, R.; Qiao, L.; et al. Mitochondrial cardiolipin is required for Nlrp3 inflammasome activation. Immunity 2013, 39, 311–323. [Google Scholar] [CrossRef]
- Zhou, R.; Tardivel, A.; Thorens, B.; Choi, I.; Tschopp, J. Thioredoxin-interacting protein links oxidative stress to inflammasome activation. Nat. Immunol. 2010, 11, 136–140. [Google Scholar] [CrossRef]
- Misawa, T.; Takahama, M.; Kozaki, T.; Lee, H.; Zou, J.; Saitoh, T.; Akira, S. Microtubule-driven spatial arrangement of mitochondria promotes activation of the NLRP3 inflammasome. Nat. Immunol. 2013, 14, 454–460. [Google Scholar] [CrossRef]
- Subramanian, N.; Natarajan, K.; Clatworthy, M.R.; Wang, Z.; Germain, R.N. The adaptor MAVS promotes NLRP3 mitochondrial localization and inflammasome activation. Cell 2013, 153, 348–361. [Google Scholar] [CrossRef]
- Dinarello, C.A. Immunological and inflammatory functions of the interleukin-1 family. Annu. Rev. Immunol. 2009, 27, 519–550. [Google Scholar] [CrossRef] [PubMed]
- van de Veerdonk, F.L.; Netea, M.G.; Dinarello, C.A.; Joosten, L.A. Inflammasome activation and IL-1beta and IL-18 processing during infection. Trends Immunol. 2011, 32, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Herzog, C.; Haun, R.S.; Kaushal, V.; Mayeux, P.R.; Shah, S.V.; Kaushal, G.P. Meprin A and meprin alpha generate biologically functional IL-1beta from pro-IL-1beta. Biochem. Biophys. Res. Commun. 2009, 379, 904–908. [Google Scholar] [CrossRef] [PubMed]
- Coeshott, C.; Ohnemus, C.; Pilyavskaya, A.; Ross, S.; Wieczorek, M.; Kroona, H.; Leimer, A.H.; Cheronis, J. Converting enzyme-independent release of tumor necrosis factor alpha and IL-1beta from a stimulated human monocytic cell line in the presence of activated neutrophils or purified proteinase 3. Proc. Nat. Acad. Sci. USA 1999, 96, 6261–6266. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, S.; Uehara, A.; Nochi, T.; Yamaguchi, T.; Ueda, H.; Sugiyama, A.; Hanzawa, K.; Kumagai, K.; Okamura, H.; Takada, H. Neutrophil proteinase 3-mediated induction of bioactive IL-18 secretion by human oral epithelial cells. J. Immunol. 2001, 167, 6568–6575. [Google Scholar] [CrossRef]
- Guma, M.; Ronacher, L.; Liu-Bryan, R.; Takai, S.; Karin, M.; Corr, M. Caspase 1-independent activation of interleukin-1beta in neutrophil-predominant inflammation. Arthritis Rheum. 2009, 60, 3642–3650. [Google Scholar] [CrossRef]
- Beausejour, A.; Grenier, D.; Goulet, J.P.; Deslauriers, N. Proteolytic activation of the interleukin-1beta precursor by Candida albicans. Infect. Immun. 1998, 66, 676–681. [Google Scholar] [PubMed]
- Netea, M.G.; van de Veerdonk, F.L.; van der Meer, J.W.; Dinarello, C.A.; Joosten, L.A. Inflammasome-independent regulation of IL-1-family cytokines. Annu. Rev. Immunol. 2015, 33, 49–77. [Google Scholar] [CrossRef]
- Nistico, R.; Florenzano, F.; Mango, D.; Ferraina, C.; Grilli, M.; Di Prisco, S.; Nobili, A.; Saccucci, S.; D’Amelio, M.; Morbin, M.; et al. Presynaptic c-Jun N-terminal Kinase 2 regulates NMDA receptor-dependent glutamate release. Sci. Rep. 2015, 5, 9035. [Google Scholar] [CrossRef] [Green Version]
- Strober, W.; Murray, P.J.; Kitani, A.; Watanabe, T. Signalling pathways and molecular interactions of NOD1 and NOD2. Nat. Rev. Immunol. 2006, 6, 9–20. [Google Scholar] [CrossRef] [PubMed]
- McAlindon, M.E.; Hawkey, C.J.; Mahida, Y.R. Expression of interleukin 1 beta and interleukin 1 beta converting enzyme by intestinal macrophages in health and inflammatory bowel disease. Gut 1998, 42, 214–219. [Google Scholar] [CrossRef] [PubMed]
- Reinecker, H.C.; Steffen, M.; Witthoeft, T.; Pflueger, I.; Schreiber, S.; MacDermott, R.P.; Raedler, A. Enhanced secretion of tumour necrosis factor-alpha, IL-6, and IL-1 beta by isolated lamina propria mononuclear cells from patients with ulcerative colitis and Crohn’s disease. Clin. Exp. Immunol. 1993, 94, 174–181. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.Y.; Ouyang, Q. Expression and significance of mucosal beta-defensin-2, TNFalpha and IL-1beta in ulcerative colitis. Zhonghua nei ke za zhi (In Chinese) 2008, 47, 11–14. [Google Scholar]
- Ligumsky, M.; Simon, P.L.; Karmeli, F.; Rachmilewitz, D. Role of interleukin 1 in inflammatory bowel disease--enhanced production during active disease. Gut 1990, 31, 686–689. [Google Scholar] [CrossRef] [PubMed]
- Kummer, J.A.; Broekhuizen, R.; Everett, H.; Agostini, L.; Kuijk, L.; Martinon, F.; van Bruggen, R.; Tschopp, J. Inflammasome components NALP 1 and 3 show distinct but separate expression profiles in human tissues suggesting a site-specific role in the inflammatory response. J. Histochem. Cytochem. 2007, 55, 443–452. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, D.; Balsitis, M.; Gallivan, S.; Dixon, M.F.; Gilmour, H.M.; Shepherd, N.A.; Theodossi, A.; Williams, G.T. Guidelines for the initial biopsy diagnosis of suspected chronic idiopathic inflammatory bowel disease. The British Society of Gastroenterology Initiative. J. Clin. Pathol. 1997, 50, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Keren, D.F.; Appelman, H.D.; Dobbins, W.O., 3rd; Wells, J.J.; Whisenant, B.; Foley, J.; Dieterle, R.; Geisinger, K. Correlation of histopathologic evidence of disease activity with the presence of immunoglobulin-containing cells in the colons of patients with inflammatory bowel disease. Hum. Pathol. 1984, 15, 757–763. [Google Scholar] [CrossRef]
- Riley, S.A.; Mani, V.; Goodman, M.J.; Dutt, S.; Herd, M.E. Microscopic activity in ulcerative colitis: What does it mean? Gut 1991, 32, 174–178. [Google Scholar] [CrossRef]
- Bryant, R.V.; Burger, D.C.; Delo, J.; Walsh, A.J.; Thomas, S.; von Herbay, A.; Buchel, O.C.; White, L.; Brain, O.; Keshav, S.; et al. Beyond endoscopic mucosal healing in UC: Histological remission better predicts corticosteroid use and hospitalisation over 6 years of follow-up. Gut 2016, 65, 408–414. [Google Scholar] [CrossRef]
- Geboes, K.; Riddell, R.; Ost, A.; Jensfelt, B.; Persson, T.; Lofberg, R. A reproducible grading scale for histological assessment of inflammation in ulcerative colitis. Gut 2000, 47, 404–409. [Google Scholar] [CrossRef] [Green Version]
- Stahlberg, D.; Veress, B.; Mare, K.; Granqvist, S.; Agren, B.; Richter, S.; Lofberg, R. Leukocyte migration in acute colonic inflammatory bowel disease: Comparison of histological assessment and Tc-99m-HMPAO labeled leukocyte scan. Am. J. Gastroenterol. 1997, 92, 283–288. [Google Scholar] [PubMed]
- Weldon, M.J.; Masoomi, A.M.; Britten, A.J.; Gane, J.; Finlayson, C.J.; Joseph, A.E.; Maxwell, J.D. Quantification of inflammatory bowel disease activity using technetium-99m HMPAO labelled leucocyte single photon emission computerised tomography (SPECT). Gut 1995, 36, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Seldenrijk, C.A.; Morson, B.C.; Meuwissen, S.G.; Schipper, N.W.; Lindeman, J.; Meijer, C.J. Histopathological evaluation of colonic mucosal biopsy specimens in chronic inflammatory bowel disease: Diagnostic implications. Gut 1991, 32, 1514–1520. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Saito, H.; Fukuda, S.; Sasaki, Y.; Munakata, A.; Kudo, H. Simple mucosal biopsy criteria differentiating among Crohn disease, ulcerative colitis, and other forms of colitis: Measurement of validity. Scand. J. Gastroenterol. 2000, 35, 281–286. [Google Scholar] [PubMed]
- Cook, M.G.; Dixon, M.F. An analysis of the reliability of detection and diagnostic value of various pathological features in Crohn’s disease and ulcerative colitis. Gut 1973, 14, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Vonk, A.G.; Netea, M.G.; van der Meer, J.W.; Kullberg, B.J. Host defence against disseminated Candida albicans infection and implications for antifungal immunotherapy. Expert. Opin. Biol. Ther. 2006, 6, 891–903. [Google Scholar] [CrossRef] [PubMed]
- Kullberg, B.J.; Van’T Wout, J.W.; Van Furth, R. Role of Granulocytes in Increased Host Resistance to Candida albicans Induced by Recombinant Interleukin-1. Infect. Immun. 1990, 58, 3319–3324. [Google Scholar]
- Joosten, L.A.; Netea, M.G.; Fantuzzi, G.; Koenders, M.I.; Helsen, M.M.; Sparrer, H.; Pham, C.T.; van der Meer, J.W.; Dinarello, C.A.; van den Berg, W.B. Inflammatory arthritis in caspase 1 gene-deficient mice: Contribution of proteinase 3 to caspase 1-independent production of bioactive interleukin-1beta. Arthritis Rheum. 2009, 60, 3651–3662. [Google Scholar] [CrossRef] [PubMed]
- Cassel, S.L.; Janczy, J.R.; Bing, X.; Wilson, S.P.; Olivier, A.K.; Otero, J.E.; Iwakura, Y.; Shayakhmetov, D.M.; Bassuk, A.G.; Abu-Amer, Y.; et al. Inflammasome-independent IL-1beta mediates autoinflammatory disease in Pstpip2-deficient mice. Proc. Nat. Acad. Sci. USA 2014, 111, 1072–1077. [Google Scholar] [CrossRef] [PubMed]
- Lukens, J.R.; Gross, J.M.; Calabrese, C.; Iwakura, Y.; Lamkanfi, M.; Vogel, P.; Kanneganti, T.D. Critical role for inflammasome-independent IL-1beta production in osteomyelitis. Proc. Nat. Acad. Sci. USA 2014, 111, 1066–1071. [Google Scholar] [CrossRef] [PubMed]
- Coll, R.C.; Robertson, A.A.; Chae, J.J.; Higgins, S.C.; Munoz-Planillo, R.; Inserra, M.C.; Vetter, I.; Dungan, L.S.; Monks, B.G.; Stutz, A.; et al. A small-molecule inhibitor of the NLRP3 inflammasome for the treatment of inflammatory diseases. Nat. Med. 2015, 21, 248–255. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Variable | Patients with Active Disease | Patients in Remission | |
|---|---|---|---|
| Biopsies Taken from both Active and Inactive Regions | Biopsies Taken from Active Regions only | Biopsies Taken from Inactive Regions | |
| Patients, n | 30 | 4 | 10 |
| Age (years) | 50 ± 18 | 39 ± 21 | 49 ± 11 |
| Gender | |||
| female | 16 | 2 | 6 |
| male | 14 | 2 | 4 |
| Smoker, n (%) | 1 (3) | 0 | 1 (10) |
| Disease extent * | |||
| Proctitis only | 7 (23) | - | - |
| Proctosigmoiditis | 14 (47) | - | - |
| Left-sided colitis | 8 (27) | - | - |
| Pancolitis | 1 (3) | 4 (100) | 4 (100) |
| Current Medication for UC | |||
| Untreated | 12 (40) | 2 (50) | 3 (30) |
| Aminosalicylates (5-ASA) | 16 (53) | 1 (25) | 5 (50) |
| Corticosteroids | 6 (20) | 2 (50) | 3 (30) |
| Immunomodulator therapy | 4 (13) | - | - |
| Topical therapy | 0 | - | - |
| Biologic therapy | 0 | - | - |
| Antibiotics/probiotics/vitamin supplements | 6 | - | 1 |
| Variable | Patients with Active Disease | Patients in Remission | |
|---|---|---|---|
| Biopsies Taken from both Active and Inactive Regions | Biopsies Taken from Active Regions only | Biopsies Taken from Inactive Regions | |
| Patients, n | 15 | 2 | 4 |
| Age (years) | 44 ± 16 | 37 ± 13 | 20 ± 7 |
| Gender | |||
| female | 7 | 1 | 3 |
| male | 8 | 1 | 1 |
| Smoker, n (%) | 4 (27) | 0 | 1 (25) |
| Disease extent * | |||
| Ileum only (L1) | 4 (27) | 1 | - |
| Ileum and colon (L3) | 2 (13) | - | - |
| Colon only (L2) | 9 (60) | 1 | - |
| Extra-intestinal manifestations (e.g., ileocecal/perianal/proctitis) | 4 (27) | - | - |
| Previous surgical resection | 4 (27) | - | 1 |
| Current Medication for CD | |||
| Untreated | 4 (27) | - | 1 (25) |
| Aminosalicylates (5-ASA) | 5 (33) | - | 1 |
| Corticosteroids | 2 (13) | - | - |
| Immunomodulator therapy | 6 (40) | 1 | 1 |
| Topical therapy | - | - | - |
| Biologic therapy | 2 (13) | 1 | 1 |
| Antibiotics/probiotics/vitamin supplements | - | - | - |
| TargetGene | Disease Phenotype | Quiescent Disease | Active Disease | p | ||
|---|---|---|---|---|---|---|
| Median R.E | IQR | Median R.E | IQR | |||
| NLRP3 | UC # | 1.5 | 1.0–2.3 | 3.7 | 2.5–5.7 | <0.001 |
| CD ^ | 1.6 | 0.9–2.8 | 5.2 | 2.9–15.1 | <0.001 | |
| IL-1β | UC # | 2.2 | 0.8–5.9 | 28.1 | 13.2–78.7 | <0.001 |
| CD ^ | 4.1 | 1.5–7.0 | 55.1 | 17.2–215.7 | <0.001 | |
| CASP1 | UC # | 1.3 | 0.8–2.3 | 5.1 | 2.9–9.3 | <0.001 |
| CD ^ | 2.5 | 0.9–3.9 | 6.9 | 4.5–10.2 | <0.001 | |
| ASC | UC # | 1.0 | 0.5–1.7 | 4.8 | 2.3–9.2 | <0.001 |
| CD ^ | 1.3 | 0.7–2.8 | 3.5 | 1.9–16.9 | 0.001 | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ranson, N.; Veldhuis, M.; Mitchell, B.; Fanning, S.; Cook, A.L.; Kunde, D.; Eri, R. NLRP3-Dependent and -Independent Processing of Interleukin (IL)-1β in Active Ulcerative Colitis. Int. J. Mol. Sci. 2019, 20, 57. https://doi.org/10.3390/ijms20010057
Ranson N, Veldhuis M, Mitchell B, Fanning S, Cook AL, Kunde D, Eri R. NLRP3-Dependent and -Independent Processing of Interleukin (IL)-1β in Active Ulcerative Colitis. International Journal of Molecular Sciences. 2019; 20(1):57. https://doi.org/10.3390/ijms20010057
Chicago/Turabian StyleRanson, Nicole, Mark Veldhuis, Brent Mitchell, Scott Fanning, Anthony L. Cook, Dale Kunde, and Rajaraman Eri. 2019. "NLRP3-Dependent and -Independent Processing of Interleukin (IL)-1β in Active Ulcerative Colitis" International Journal of Molecular Sciences 20, no. 1: 57. https://doi.org/10.3390/ijms20010057
APA StyleRanson, N., Veldhuis, M., Mitchell, B., Fanning, S., Cook, A. L., Kunde, D., & Eri, R. (2019). NLRP3-Dependent and -Independent Processing of Interleukin (IL)-1β in Active Ulcerative Colitis. International Journal of Molecular Sciences, 20(1), 57. https://doi.org/10.3390/ijms20010057





