Comparative Transcriptome Analysis Reveals an Efficient Mechanism for α -Linolenic Acid Synthesis in Tree Peony Seeds
Abstract
:1. Introduction
2. Results and Discussion
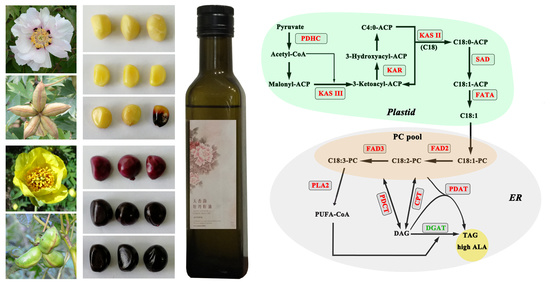
2.1. Transcriptome Data Reveals Lipid Pathways Associated with High ALA Content in Seeds
2.2. Gene Expression for Select Enzymes in TAG Metabolism Is Associated with High ALA Content
2.3. Acyl Editing and Head Group Exchange Drives PUFA Synthesis in P. rockii
2.4. An Acyl-CoA Independent Pathway May Be a Preferred Terminal Step in ALA Accumulation
2.5. Desaturases Play a Key Role in Driving the ALA Content in Seeds
2.6. Molecular Cloning and Expression Analysis of PrFAD2 and PrFAD3
2.7. Overexpression of PrFAD2 and PrFAD3 Affects the ALA/LA Ratio in Arabidopsis Seeds
3. Materials and Methods
3.1. Plant Material
3.2. RNA Extraction, Sequencing and De Novo Assembly
3.3. Normalization of Gene Expression Levels and Identification of Differentially Expressed Genes
3.4. Functional Annotation, GO and KEGG Classification
3.5. Quantitative Real-Time PCR
3.6. Cloning and in Silico Analysis of Desaturases
3.7. Overexpression and Stable Transformation
3.8. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ALA | α-linolenic acid |
| LA | linoleic acid |
| FAD | fatty acid desaturases |
| PUFAs | polyunsaturated fatty acids |
| TAG | triacylglycerol |
| PC | phosphatidylcholine |
| SAD | stearoyl-ACP desaturase |
| DEGs | differentially expressed genes |
| qRT-PCR | quantitative real-time PCR |
| ITS | internal transcribed spacer |
| RPKM | reads per kilobase per million |
References
- Zhang, Q.Y.; Yu, R.; Xie, L.H.; Rahman, M.M.; Kilaru, A.; Niu, L.X.; Zhang, Y.L. Fatty acid and associated gene expression analyses of three tree peony species reveal key genes for ALA synthesis in seeds. Front. Plant Sci. 2018, 9, 106. [Google Scholar] [CrossRef] [PubMed]
- Radovanovic, N.; Thambugala, D.; Duguid, S.; Loewen, E.; Cloutier, S. Functional characterization of flax fatty acid desaturase FAD2 and FAD3 isoforms expressed in yeast reveals a broad diversity in activity. Mol. Biotechnol. 2014, 56, 609–620. [Google Scholar] [CrossRef] [PubMed]
- Warude, D.; Joshi, K.; Harsulkar, A. Polyunsaturated fatty acids: Biotechnology. Crit. Rev. Biotechnol. 2006, 26, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Egert, S.; Baxheinrich, A.; Lee-Barkey, Y.H.; Tschoepe, D.; Wahrburg, U.; Stratmann, B. Effects of an energy-restricted diet rich in plant-derived α-linolenic acid on systemic inflammation and endothelial function in overweight-to-obese patients with metabolic syndrome traits. Br. J. Nutr. 2014, 112, 1315–1322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.J.; Go, Y.S.; Ahn, S.J.; Chung, C.H.; Suh, M.C. Functional complementation of a periila ω3 fatty acid desaturase under the seed-specific SeFAD2 promoter. J. Plant Biol. 2008, 51, 174–179. [Google Scholar] [CrossRef]
- Amjad Khan, W.; Chun-Mei, H.; Khan, N.; Iqbal, A.; Lyu, S.W.; Shah, F. Bioengineered Plants Can Be a Useful Source of Omega-3 Fatty Acids. BioMed Res. Int. 2017. [Google Scholar] [CrossRef]
- Harwood, J.L. Recent advances in the biosynthesis of plant fatty acids. Biochim. Biophys. Acta 1996, 1301, 7–56. [Google Scholar] [CrossRef]
- Shanklin, J.; Cahoon, E.B. Desaturation and related modifications of fatty acids. Annu. Rev. Plant Physiol. 1998, 49, 611–641. [Google Scholar] [CrossRef]
- Chapman, K.D.; Ohlrogge, J.B. Compartmentation of triacylglycerol accumulation in plants. J. Biol. Chem. 2012, 287, 2288–2294. [Google Scholar] [CrossRef]
- Aznar-Moreno, J.A.; Venegas Calerón, M.; Martínez-Force, E.; Garcés, R.; Mullen, R.; Gidda, S.K.; Salas, J.J. Sunflower (Helianthus annuus) long-chain acyl-coenzyme A synthetases expressed at high levels in developing seeds. Physiol. Plant 2014, 150, 363–373. [Google Scholar] [CrossRef]
- Coleman, R.A.; Lee, D.P. Enzymes of triacylglycerol synthesis and their regulation. Prog. Lipid Res. 2004, 43, 134–176. [Google Scholar] [CrossRef]
- Bates, P.D.; Browse, J. The significance of different diacylgycerol synthesis pathways on plant oil composition and bioengineering. Front. Plant Sci. 2012, 3, 147. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Fan, J.; Taylor, D.C.; Ohlrogge, J.B. DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell 2009, 21, 3885–3901. [Google Scholar] [CrossRef] [PubMed]
- Okuley, J.; Lightner, J.; Feldmann, K.; Yadav, N.; Lark, E. Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant Cell 1994, 6, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Arondel, V.; Lemieux, B.; Hwang, I.; Gibson, S.; Goodman, H.M.; Somerville, C.R. Map-based cloning of a gene controlling omega-3 fatty acid desaturation in Arabidopsis. Science 1992, 258, 1353–1355. [Google Scholar] [CrossRef] [PubMed]
- Shanklin, J.; Whittle, E.; Fox, B.G. Eight histidine residues are catalytically essential in a membrane-associated iron enzyme; stearoyl-CoA desaturase; and are conserved in alkane hydroxylase and xylene monooxygenase. Biochemistry 1994, 33, 12787–12794. [Google Scholar] [CrossRef] [PubMed]
- Los, D.A.; Murata, N. Structure and expression of fatty acid desaturases. Biochim. Biophys. Acta 1998, 1394, 3–15. [Google Scholar] [CrossRef]
- Yadav, N.S.; Wierzbicki, A.; Aegerter, M.; Caster, C.S.; Pérez-Grau, L.; Kinney, A.J.; Hitz, W.D.; Booth, J.R., Jr.; Schweiger, B.; Stecca, K.L.; et al. Cloning of higher plant ω-3 fatty acid desaturases. Plant Physiol. 1993, 103, 467–476. [Google Scholar] [CrossRef] [PubMed]
- Bilyeu, K.D.; Palavalli, L.; Sleper, D.A.; Beuselinck, P.R. Three microsomal omega-3 fatty-acid desaturase genes contribute to soybean linolenic acid levels. Crop Sci. 2003, 43, 1833–1838. [Google Scholar] [CrossRef]
- Reed, D.W.; Schäfer, U.A.; Covello, P.S. Characterization of the Brassica napusextraplastidial linoleate desaturase by expression in Saccharomyces cerevisiae. Plant Physiol. 2000, 122, 715–720. [Google Scholar] [CrossRef]
- Yurchenko, O.O.; Park, S.; Ilut, D.C.; Inmon, J.J.; Millhollon, J.C.; Liechty, Z.; Page, J.T.; Jenks, M.A.; Chapman, K.D.; Udall, J.A.; et al. Genome-wide analysis of the omega-3 fatty acid desaturase gene family in Gossypium. BMC Plant Biol. 2014, 14, 312. [Google Scholar] [CrossRef] [PubMed]
- Hernández, M.L.; Mancha, M.; Martínez-Rivas, J.M. Molecular cloning and characterization of genes encoding two microsomal oleate desaturases (FAD2) from olive. Phytochemistry 2005, 66, 1417–1426. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.R.; Kim, S.H.; Go, Y.S.; Jung, S.M.; Roh, K.H.; Kim, J.B.; Suh, M.C.; Lee, S.; Kim, H.U. Molecular cloning and functional analysis of two FAD2 genes from American grape (Vitis labrusca L.). Gene 2012, 509, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Rivas, J.M.; Sperling, P.; Lühs, W.; Heinz, E. Spatial and temporal regulation of three different microsomal oleate desaturase genes (FAD2) from normal-type and high-oleic varieties of sunflower (Helianthus annuus L.). Mol. Breed. 2001, 8, 159–168. [Google Scholar] [CrossRef]
- Pirtle, I.L.; Kongcharoensuntorn, W.; Nampaisansuk, M.; Knesek, J.E.; Chapman, K.D.; Pirtle, R.M. Molecular cloning and functional expression of the gene for a cotton Δ-12 fatty acid desaturase (FAD2). Biochim. Biophys. Acta 2001, 1522, 122–129. [Google Scholar] [CrossRef]
- Vrinten, P.; Hu, Z.; Munchinsky, M.A.; Rowland, G.; Qiu, X. Two FAD3 desaturase genes control the level of linolenic acid in flax seed. Plant Physiol. 2005, 139, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Bourgis, F.; Kilaru, A.; Cao, X.; Ngando-Ebongue, G.F.; Drira, N.; Ohlrogge, J.B.; Arondel, V. Comparative transcriptome and metabolite analysis of oil palm and date palm mesocarp that differ dramatically in carbon partitioning. Proc. Natl. Acad. Sci. USA 2011, 108, 12527–12532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, X.D.; Pan, B.Z.; Fu, Q.; Niu, L.; Chen, M.S.; Xu, Z.F. De novo transcriptome assembly of the eight major organs of Sacha Inchi (Plukenetia volubilis) and the identification of genes involved in α-linolenic acid metabolism. BMC Genom. 2018, 19, 380. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liang, S.; Duan, J.; Wang, J.; Chen, S.; Cheng, Z.; Zhang, Q.; Liang, X.; Li, Y. De novo assembly and characterisation of the transcriptome during seed development; and generation of genic-SSR markers in peanut (Arachis hypogaea L.). BMC Genom. 2012, 13, 90. [Google Scholar] [CrossRef] [PubMed]
- Kilaru, A.; Cao, X.; Dabbs, P.B.; Sung, H.J.; Rahamn, M.M.; Thrower, N.; Zynda, G.; Podicheti, R.; Ibarra-Laclette, E.; Herrera-Estrella, L.; et al. Oil biosynthesis in a basal angiosperm: Transcriptome analysis of Persea Americana mesocarp. BMC Plant Biol. 2015, 15, 203. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.U.; Lee, K.R.; Shim, D.; Lee, J.H.; Chen, G.Q.; Hwang, S. Transcriptome analysis and identification of genes associated with ω-3 fatty acid biosynthesis in Perilla frutescens (L.) var. frutescens. BMC Genom. 2016, 17, 474. [Google Scholar] [CrossRef] [PubMed]
- Troncoso-Ponce, M.A.; Kilaru, A.; Cao, X.; Durrett, T.P.; Fan, J.; Jensen, J.K.; Thrower, N.A.; Pauly, M.; Wilkerson, C.; Ohlrogge, J.B. Comparative deep transcriptional profiling of four developing oilseeds. Plant J. 2011, 68, 1014–1027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahman, M.; McVetty, P. A review of Brassica seed color. Can. J. Plant Sci. 2011, 91, 437–446. [Google Scholar] [CrossRef]
- Qu, C.; Hasan, M.; Lu, K.; Liu, L.; Zhang, K.; Fu, F.; Wang, M.; Liu, S.; Bu, H.; Wang, R.; et al. Identification of QTL for seed coat colour and oil content in Brassica napus by association mapping using SSR markers. Can. J. Plant Sci. 2015, 95, 387–395. [Google Scholar] [CrossRef] [Green Version]
- Diederichsen, A.; Raney, J.P. Seed colour, seed weight and seed oil content in Linum usitatissimum accessions held by Plant Gene Resources of Canada. Pant Breed. 2006, 125, 372–377. [Google Scholar] [CrossRef]
- Pruitt, K.D.; Tatusova, T.; Maglott, D.R. NCBI Reference Sequence (RefSeq): A curated non-redundant sequence database of genomes; transcripts and proteins. Nucleic Acids Res. 2005, 33, D501–D504. [Google Scholar] [CrossRef] [PubMed]
- UniProt Consortium. UniProt: A hub for protein information. Nucleic Acids Res. 2015, 43, D204–D212. [Google Scholar] [CrossRef]
- Beisson, F.; Koo, A.J.; Ruuska, S.; Schwender, J.; Pollard, M.; Thelen, J.J.; Paddock, T.; Salas, J.J.; Savage, L.; Milcamps, A.; et al. Arabidopsis genes involved in acyl lipid metabolism. A 2003 census of the candidates; a study of the distribution of expressed sequence tags in organs; and a web-based database. Plant Physiol. 2003, 132, 681–697. [Google Scholar] [CrossRef]
- Ke, J.; Behal, R.H.; Back, S.L.; Nikolau, B.J.; Wurtele, E.S.; Oliver, D.J. The role of pyruvate dehydrogenase and acetyl-coenzyme A synthetase in fatty acid synthesis in developing Arabidopsis seeds. Plant Physiol. 2000, 123, 497–508. [Google Scholar] [CrossRef]
- Dehesh, K.; Tai, H.; Edwards, P.; Byrne, J.; Jaworski, J.G. Overexpression of 3-ketoacyl-acyl-carrier protein synthase IIIs in plants reduces the rate of lipid synthesis. Plant Physiol. 2001, 125, 1103–1114. [Google Scholar] [CrossRef]
- Yu, N.; Xiao, W.F.; Zhu, J.; Chen, X.Y.; Peng, C.C. The Jatropha curcas KASIII gene alters fatty acid composition of seeds in Arabidopsis thaliana. Biol. Plant. 2015, 59, 773–782. [Google Scholar] [CrossRef]
- Wei, Q.; Li, J.; Zhang, L.; Wu, P.; Chen, Y.; Li, M.; Jiang, H.; Wu, G. Cloning and characterization of a β-ketoacyl-acyl carrier protein synthase II from Jatropha curcas. J. Plant Physiol. 2012, 169, 816–824. [Google Scholar] [CrossRef] [PubMed]
- Cahoon, E.B.; Clemente, T.E.; Damude, H.G.; Kinney, A.J. Modifying vegetable oils for food and non-food purposes. In Oil Crops; Vollmann, J., Rajcan, I., Eds.; Springer: New York, NY, USA, 2010; pp. 31–56. [Google Scholar]
- Salas, J.J.; Ohlrogge, J.B. Characterization of substrate specificity of plant FatA and FatB acyl-ACP thioesterases. Arch. Biochem. Biophys. 2002, 403, 25–34. [Google Scholar] [CrossRef]
- Bates, P.D.; Stymne, S.; Ohlrogge, J. Biochemical pathways in seed oil synthesis. Curr. Opin. Plant Biol. 2013, 16, 358–364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.R.; Chen, G.Q.; Kim, H.U. Current progress towards the metabolic engineering of plant seed oil for hydroxy fatty acids production. Plant Cell Rep. 2015, 34, 603–615. [Google Scholar] [CrossRef] [PubMed]
- Wallis, J.G.; Watts, J.L.; Browse, J. Polyunsaturated fatty acid synthesis: What will they think of next? Trends Biochem. Sci. 2002, 27, 467–473. [Google Scholar] [CrossRef]
- Bayon, S.; Chen, G.C.; Weselake, R. A Small Phospholipase A2-α from Castor Catalyzes the Removal of Hydroxy Fatty Acids from Phosphatidylcholine in Transgenic Arabidopsis Seeds. Plant Physiol. 2015, 167, 1259–1270. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xu, R.; Wang, R.; Liu, A. Transcriptome analysis of Sacha Inchi (Plukenetia volubilis L.) seeds at two developmental stages. BMC Genom. 2012, 13, 716. [Google Scholar] [CrossRef]
- Arroyo-Caro, J.M.; Chileh, T.; Alonso, D.L.; García-Maroto, F. Molecular characterization of a lysophosphatidylcholine acyltransferase gene belonging to the MBOAT family in Ricinus communis L. Lipids 2013, 48, 663–674. [Google Scholar] [CrossRef]
- Zheng, Q.; Li, J.Q.; Kazachkov, M.; Liu, K.; Zou, J. Identification of Brassica napus lysophosphatidylcholine acyltransferase genes through yeast functional screening. Phytochemistry 2012, 75, 21–31. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Jasieniecka-Gazarkiewicz, K.; Wan, X.; Luo, L.; Zhang, Y.; Banas, A.; Jiang, M.; Gong, Y. Molecular characterization of two lysophospholipid: Acyl-CoA acyltransferases belonging to the MBOAT family in nicotiana benthamiana. PLoS ONE 2015, 10, e0144653. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Xin, Z.; Ren, Z.; Miquel, M.; Browse, J. An enzyme regulating triacylglycerol composition is encoded by the ROD1 gene of Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 18837–18842. [Google Scholar] [CrossRef] [PubMed]
- Slack, C.R.; Roughan, P.G.; Browse, J.A.; Gardiner, S.E. Some properties of cholinephosphotransferase from developing safflower cotyledons. Biochim. Biophys. Acta 1985, 833, 438–448. [Google Scholar] [CrossRef]
- Dahlqvist, A.; Ståhl, U.; Lenman, M.; Banas, A.; Lee, M.; Sandager, L.; Ronne, H.; Stymne, S. Phospholipid: Diacylglycerol acyltransferase: An enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc. Natl. Acad. Sci. USA 2000, 97, 6487–6492. [Google Scholar] [CrossRef] [PubMed]
- Marmon, S.K.; Sturtevant, D.; Herrfurth, C.; Chapman, K.D.; Stymne, S.; Feussner, I. Two acyltransferases contribute differently to linolenic acid levels in seed oil. Plant Physiol. 2017, 173, 2081–2095. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Carlsson, A.S.; Francis, T.; Zhang, M.; Hoffman, T.; Giblin, M.E.; Taylor, D.C. Triacylglycerol synthesis by PDAT1 in the absence of DGAT1 activity is dependent on re-acylation of LPC by LPCAT2. BMC Plant. Biol. 2012, 12, 4. [Google Scholar] [CrossRef]
- Ståhl, U.; Carlsson, A.S.; Lenman, M.; Dahlqvist, A.; Huang, B.; Banaś, W.; Banaś, A.; Stymne, S. Cloning and functional characterization of a phospholipid: Diacylglycerol acyltransferase from Arabidopsis. Plant Physiol. 2004, 135, 1324–1335. [Google Scholar] [CrossRef]
- Katavic, V.; Reed, D.W.; Taylor, D.C.; Giblin, E.M.; Barton, D.L.; Zou, J.; Mackenzie, S.L.; Covello, P.S.; Kunst, L. Alteration of seed fatty acid composition by an ethyl methanesulfonate-induced mutation in Arabidopsis thaliana affecting diacylglycerol acyltransferase activity. Plant Physiol. 1995, 108, 399–409. [Google Scholar] [CrossRef]
- Pan, X.; Siloto, R.M.; Wickramarathna, A.D.; Mietkiewska, E.; Weselake, R.J. Identification of a pair of phospholipid: Diacylglycerol acyltransferases from developing flax (Linum usitatissimum L.) seed catalyzing the selective production of trilinolenin. J. Biol. Chem. 2013, 288, 24173–24188. [Google Scholar] [CrossRef]
- Yuan, L.; Mao, X.; Zhao, K.; Ji, X.; Ji, C.; Xue, J.; Li, R. Characterisation of phospholipid: Diacylglycerol acyltransferases (PDATs) from Camelina sativa and their roles in stress responses. Biol. Open 2017, 6, 1024–1034. [Google Scholar] [CrossRef]
- Bhunia, R.K.; Chakraborty, A.; Kaur, R.; Maiti, M.K.; Sen, S.K. Enhancement of α-linolenic acid content in transgenic tobacco seeds by targeting a plastidial ω-3 fatty acid desaturase (fad7) gene of Sesamum indicum to ER. Plant Cell Rep. 2016, 35, 213–226. [Google Scholar] [CrossRef]
- Murphy, D.J. Production of novel oils in plants. Curr. Opin. Biotechnol. 1999, 10, 175–180. [Google Scholar] [CrossRef]
- Tao, F.; Zhu, S.W.; Fan, J.; Cheng, B.J. Cloning and sequence analysis of maize FAD2 gene. J. Plant Physiol. Mol. Biol. 2006, 32, 649–656. [Google Scholar]
- Zhang, D.; Pirtle, I.L.; Park, S.J.; Nampaisansuk, M.; Neogi, P.; Wanjie, S.W.; Pirtle, R.M.; Chapman, K.D. Identification and expression of a new delta-12 fatty acid desaturase (FAD2-4) gene in upland cotton and its functional expression in yeast and Arabidopsis thaliana plants. Plant Physiol. Biochem. 2009, 47, 462–471. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Chen, B.; Win, A.N.; Fu, C.; Lian, J.; Liu, X.; Wang, R.; Zhang, X.; Chai, Y. Omega-3 fatty acid desaturase gene family from two ω-3 sources; Salvia hispanica and Perilla frutescens: Cloning; characterization and expression. PLoS ONE 2018, 13, e0191432. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Cui, Q.; Xu, Y.; Yang, S.; Gao, M.; Wang, Y. Effects of tung oilseed FAD2 and DGAT2 genes on unsaturated fatty acid accumulation in Rhodotorula glutinis and Arabidopsis thaliana. Mol. Genet. Genom. 2015, 290, 1605–1613. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Gao, L.; Yu, X.; Zheng, Y.; Li, D.; Wang, X. Identification of a Δ12 fatty acid desaturase from oil palm (Elaeis guineensis Jacq.) involved in the biosynthesis of linoleic acid by heterologous expression in Saccharomyces cerevisiae. Gene 2016, 591, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644. [Google Scholar] [CrossRef]
- Rajkumar, A.P.; Qvist, P.; Lazarus, R.; Lescai, F.; Ju, J.; Nyegaard, M.; Mors, O.; Børglum, A.D.; Li, Q.; Christensen, J.H. Experimental validation of methods for differential gene expression analysis and sample pooling in RNA-seq. BMC Genom. 2015, 16, 548. [Google Scholar] [CrossRef]
- Audic, S.; Claverie, J.M. The significance of digital gene expression profiles. Genome Res. 1997, 7, 986–995. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation; visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Fang, L.; Zheng, H.; Zhang, Y.; Chen, J.; Zhang, Z.; Wang, J.; Li, S.; Li, R.; Bolund, L.; et al. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34, W293–W297. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Sternberg, M.J. Protein structure prediction on the Web: A case study using the Phyre server. Nat. Protoc. 2009, 4, 363. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood; evolutionary distance; and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Zhang, Q.Y.; Yu, R.; Sun, D.Y.; Bai, Z.Z.; Li, H.; Xue, L.; Zhang, Y.L.; Niu, L. PrLPAAT4, a putative lysophosphatidic acid acyltransferase from Paeonia rockii, plays an important role in seed fatty acid biosynthesis. Molecules 2017, 22, 1694. [Google Scholar] [CrossRef]










© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Q.; Yu, R.; Sun, D.; Rahman, M.M.; Xie, L.; Hu, J.; He, L.; Kilaru, A.; Niu, L.; Zhang, Y. Comparative Transcriptome Analysis Reveals an Efficient Mechanism for α -Linolenic Acid Synthesis in Tree Peony Seeds. Int. J. Mol. Sci. 2019, 20, 65. https://doi.org/10.3390/ijms20010065
Zhang Q, Yu R, Sun D, Rahman MM, Xie L, Hu J, He L, Kilaru A, Niu L, Zhang Y. Comparative Transcriptome Analysis Reveals an Efficient Mechanism for α -Linolenic Acid Synthesis in Tree Peony Seeds. International Journal of Molecular Sciences. 2019; 20(1):65. https://doi.org/10.3390/ijms20010065
Chicago/Turabian StyleZhang, Qingyu, Rui Yu, Daoyang Sun, Md Mahbubur Rahman, Lihang Xie, Jiayuan Hu, Lixia He, Aruna Kilaru, Lixin Niu, and Yanlong Zhang. 2019. "Comparative Transcriptome Analysis Reveals an Efficient Mechanism for α -Linolenic Acid Synthesis in Tree Peony Seeds" International Journal of Molecular Sciences 20, no. 1: 65. https://doi.org/10.3390/ijms20010065