MiR-150-5p May Contribute to Pathogenesis of Human Leiomyoma via Regulation of the Akt/p27Kip1 Pathway In Vitro
Abstract
:1. Introduction
2. Results
2.1. Clinical Characteristics and miR Profiles of Uterine Leiomyoma
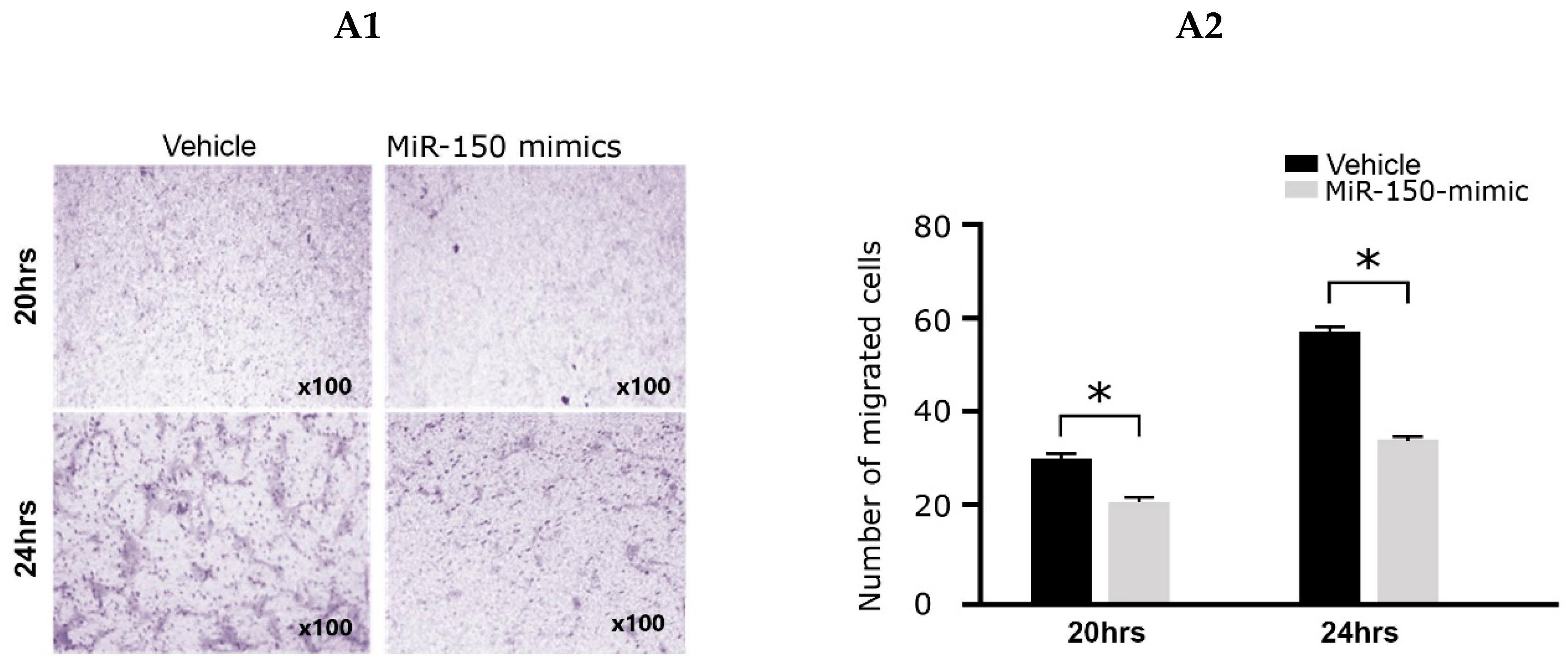
2.2. Effects of the miR-150 Mimic on Cell Migration and Collagen Gel Contraction
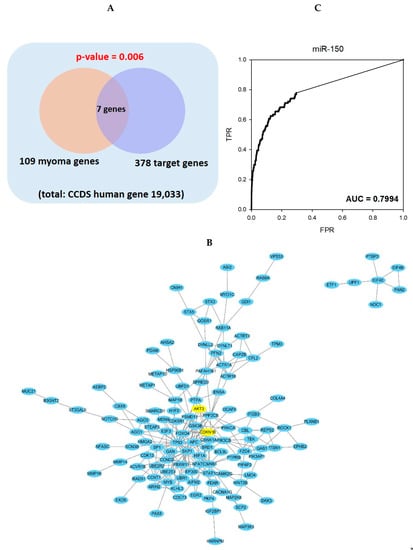
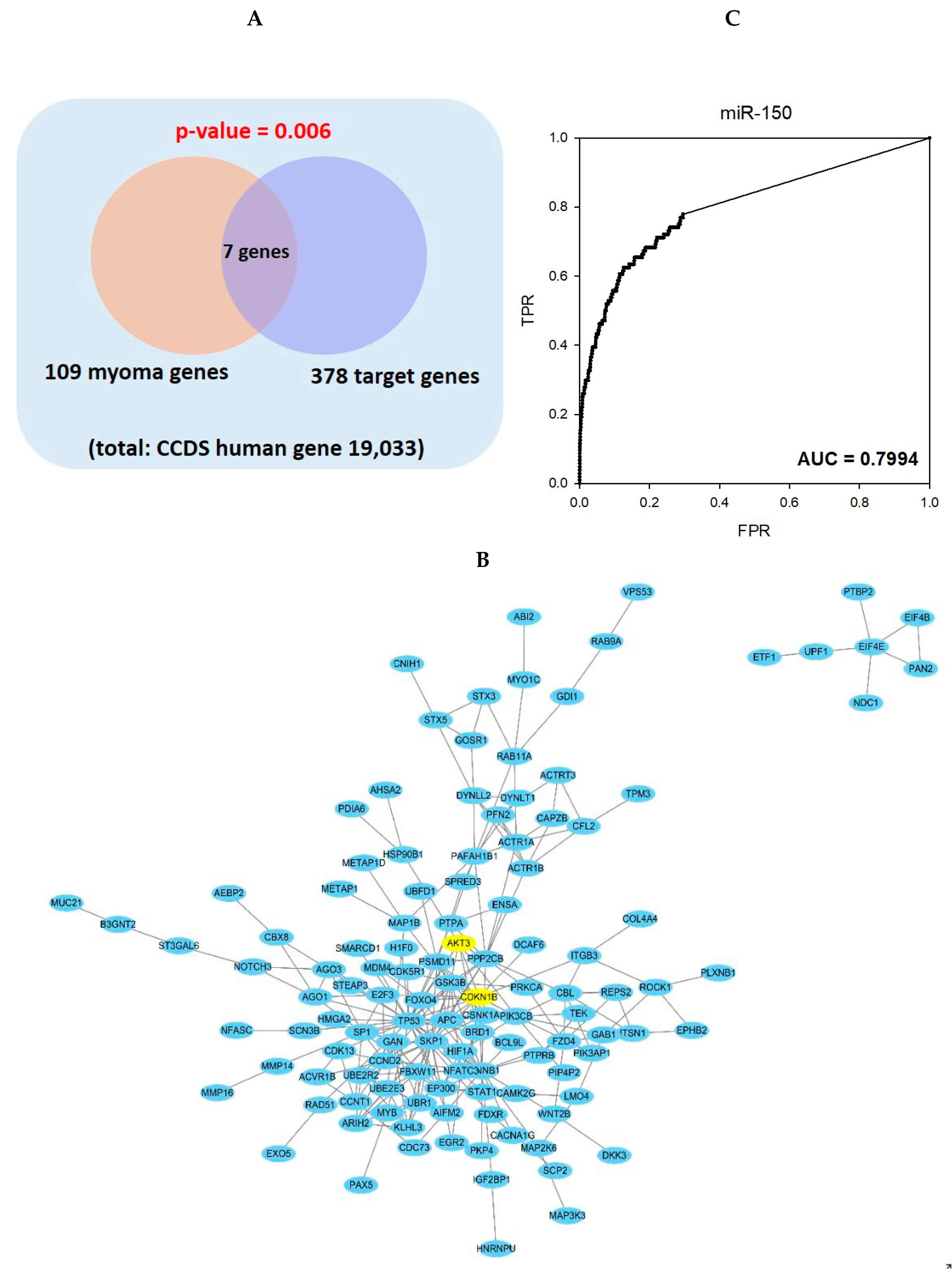
2.3. Gene Set Analysis and Network Analysis of miR-150 Predicted Target Genes and Leiomyoma Related Genes
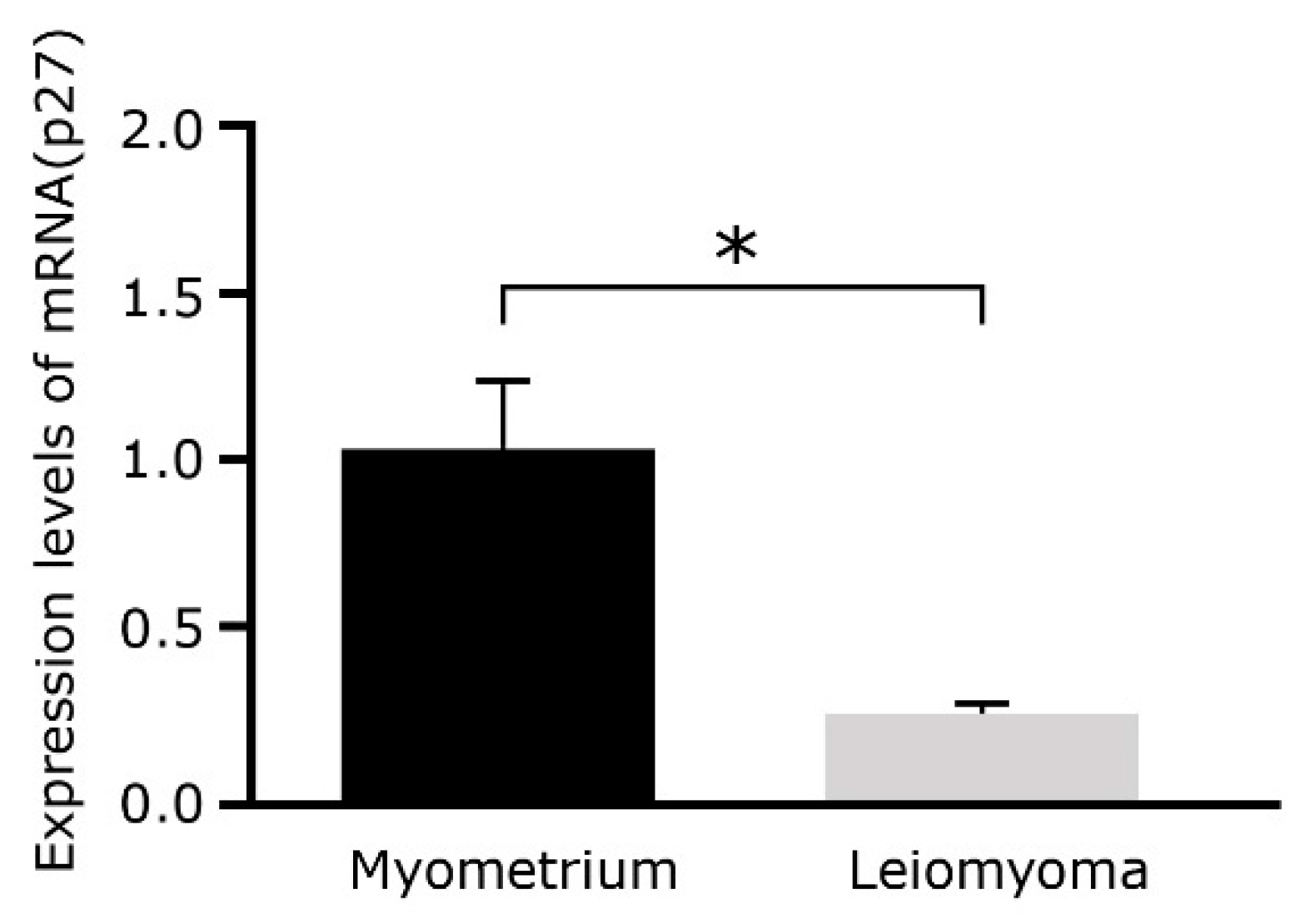
2.4. p27Kip1 mRNA Expression Levels Decrease in Leiomyoma
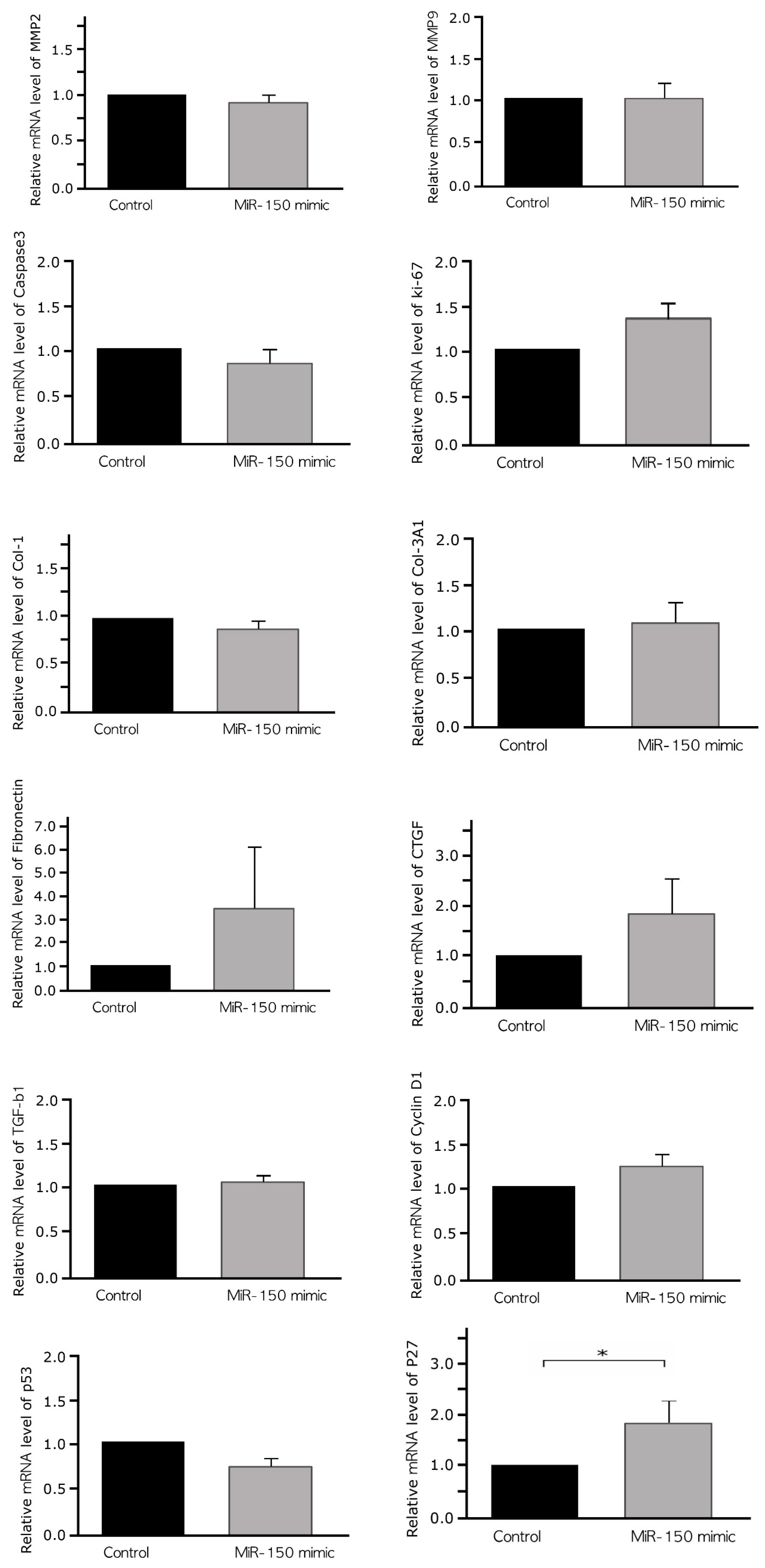
2.5. Effects of a miR-150 Mimic on the Expression of Markers of Cell Cycle, Invasion, Apoptosis, and Fibrosis in Cultured Leiomyoma Cells
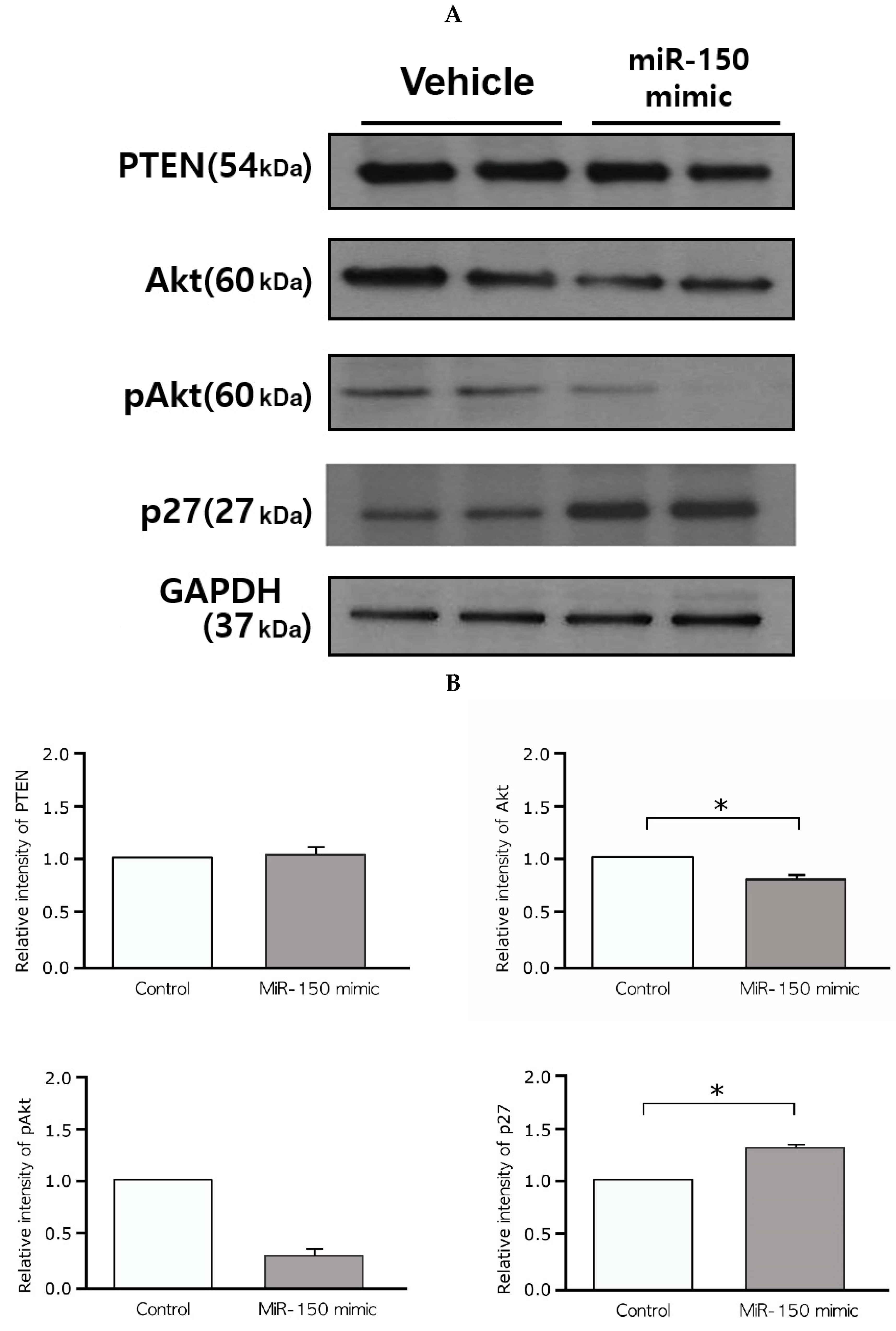
2.6. Effects of the miR-150 Mimic on Akt and p27Kip1 Expression in Leiomyoma
3. Discussion
4. Materials and Methods
4.1. Study Subjects and Tissue Specimens
4.2. Cell Culture of Leiomyoma and Myometrial Smooth Muscle Cells
4.3. miR Microarray Analysis
4.4. RNA Isolation and Quantitative Real-Time PCR
4.5. Target Gene Prediction for miR-150
4.6. Protein Isolation and Western Blotting Analysis
4.7. MiR Transfection
4.8. Migration Assay
4.9. Collagen Gel Contraction Assay
4.10. Wound Healing Assay
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| FC | Fold change |
| miR | microRNA |
| RT-PCR | Real-time polymerase chain reaction |
| UTR | Untranslated region |
References
- Hurskainen, R.; Teperi, J.; Rissanen, P.; Aalto, A.-M.; Grénman, S.; Kivelä, A.; Kujansuu, E.; Vuorma, S.; Yliskoski, M.; Paavonen, J. Quality of life and cost-effectiveness of levonorgestrel-releasing intrauterine system versus hysterectomy for treatment of menorrhagia: A randomised trial. Lancet 2001, 357, 273–277. [Google Scholar] [CrossRef]
- Wallach, E.E.; Vlahos, N.F. Uterine Myomas: An Overview of Development, Clinical Features, and Management. Obstet. Gynecol. 2004, 104, 393–406. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.S.; Protic, O.; Stortoni, P.; Grechi, G.; Lamanna, P.; Petraglia, F.; Castellucci, M.; Ciarmela, P. Complex networks of multiple factors in the pathogenesis of uterine leiomyoma. Fertil. Steril. 2013, 100, 178–193. [Google Scholar] [CrossRef] [PubMed]
- Baird, D.D.; Dunson, D.B.; Hill, M.C.; Cousins, D.; Schectman, J.M. High cumulative incidence of uterine leiomyoma in black and white women: Ultrasound evidence. Am. J. Obstet. Gynecol. 2003, 188, 100–107. [Google Scholar] [CrossRef]
- Leather, A.T.; Studd, J.W.; Watson, N.R.; Holland, E.F. The prevention of bone loss in young women treated with GnRH analogues with “add-back” estrogen therapy. Obstet. Gynecol. 1993, 81, 104–107. [Google Scholar]
- Donnez, J.; Arriagada, P.; Marciniak, M.; Larrey, D. Liver safety parameters of ulipristal acetate for the treatment of uterine fibroids: A comprehensive review of the clinical development program. Expert Opin. Drug Saf. 2018, 17, 1225–1232. [Google Scholar] [CrossRef]
- Luo, X.; Chegini, N. The expression and potential regulatory function of microRNAs in the pathogenesis of leiomyoma. Semin. Reprod. Med. 2008, 26, 500–514. [Google Scholar] [CrossRef]
- Georgieva, B.; Milev, I.; Minkov, I.; Dimitrova, I.; Bradford, A.P.; Baev, V. Characterization of the uterine leiomyoma microRNAome by deep sequencing. Genomics 2012, 99, 275–281. [Google Scholar] [CrossRef] [Green Version]
- Chuang, T.D.; Khorram, O. miR-200c Regulates IL8 Expression by Targeting IKBKB: A Potential Mediator of Inflammation in Leiomyoma Pathogenesis. PLoS ONE 2014, 9, e95370. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.-D.; Khorram, O.; Information, P.E.K.F.C. Mechanisms underlying aberrant expression of miR-29c in uterine leiomyoma. Fertil. Steril. 2016, 105, 236–245.e1. [Google Scholar] [CrossRef]
- Chuang, T.-D.; Panda, H.; Luo, X.; Chegini, N. miR-200c is aberrantly expressed in leiomyomas in an ethnic-dependent manner and targets ZEBs, VEGFA, TIMP2, and FBLN5. Endocr. Relat. Cancer 2012, 19, 541–556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.J.; Kim, Y.Y.; Shin, J.H.; Kim, H.; Ku, S.-Y.; Suh, C.S. Variation in MicroRNA Expression Profile of Uterine Leiomyoma with Endometrial Cavity Distortion and Endometrial Cavity Non-Distortion. Int. J. Mol. Sci. 2018, 19, 2524. [Google Scholar] [CrossRef] [PubMed]
- Janssen, H.L.; Reesink, H.W.; Lawitz, E.J.; Zeuzem, S.; Rodriguez-Torres, M.; Patel, K.; Van Der Meer, A.J.; Patick, A.K.; Chen, A.; Zhou, Y.; et al. Treatment of HCV Infection by Targeting MicroRNA. N. Engl. J. Med. 2013, 368, 1685–1694. [Google Scholar] [CrossRef] [PubMed]
- Segars, J.H.; Parrott, E.C.; Nagel, J.D.; Guo, X.C.; Gao, X.; Birnbaum, L.S.; Pinn, V.W.; Dixon, D. Proceedings from the Third National Institutes of Health International Congress on Advances in Uterine Leiomyoma Research: Comprehensive review, conference summary and future recommendations. Hum. Reprod. Updat. 2014, 20, 309–333. [Google Scholar] [CrossRef] [PubMed]
- McWilliams, M.M.; Chennathukuzhi, V.M. Recent Advances in Uterine Fibroid Etiology. Semin. Reprod. Med. 2017, 35, 181–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Commandeur, A.E.; Styer, A.K.; Teixeira, J.M. Epidemiological and genetic clues for molecular mechanisms involved in uterine leiomyoma development and growth. Hum. Reprod. Update 2015, 21, 593–615. [Google Scholar] [CrossRef] [Green Version]
- Borahay, M.A.; Al-Hendy, A.; Kilic, G.S.; Boehning, D. Signaling Pathways in Leiomyoma: Understanding Pathobiology and Implications for Therapy. Mol. Med. 2015, 21, 242–256. [Google Scholar] [CrossRef]
- Laganà, A.S.; Vergara, D.; Favilli, A.; La Rosa, V.L.; Tinelli, A.; Gerli, S.; Noventa, M.; Vitagliano, A.; Triolo, O.; Rapisarda, A.M.C.; et al. Epigenetic and genetic landscape of uterine leiomyomas: A current view over a common gynecological disease. Arch. Gynecol. Obstet. 2017, 296, 855–867. [Google Scholar] [CrossRef]
- Zavadil, J.; Ye, H.; Liu, Z.; Wu, J.; Lee, P.; Hernando, E.; Soteropoulos, P.; Toruner, G.A.; Wei, J.-J. Profiling and Functional Analyses of MicroRNAs and Their Target Gene Products in Human Uterine Leiomyomas. PLoS ONE 2010, 5, e12362. [Google Scholar] [CrossRef]
- Chuang, T.-D.; Khorram, O. Regulation of Cell Cycle Regulatory Proteins by MicroRNAs in Uterine Leiomyoma. Reprod. Sci. 2018, 26, 250–258. [Google Scholar] [CrossRef]
- Qiang, W.; Liu, Z.; Serna, V.A.; Druschitz, S.A.; Liu, Y.; Espona-Fiedler, M.; Wei, J.-J.; Kurita, T. Down-Regulation of miR-29b Is Essential for Pathogenesis of Uterine Leiomyoma. J. Clin. Endocrinol. Metab. 2014, 155, 663–669. [Google Scholar] [CrossRef]
- Peng, Y.; Laser, J.; Shi, G.; Mittal, K.; Melamed, J.; Lee, P.; Wei, J.-J. Antiproliferative Effects by Let-7 Repression of High-Mobility Group A2 in Uterine Leiomyoma. Mol. Cancer Res. 2008, 6, 663–673. [Google Scholar] [CrossRef] [PubMed]
- Shi, G.; Perle, M.A.; Mittal, K.; Chen, H.; Zou, X.; Narita, M.; Hernando, E.; Lee, P.; Wei, J.J. Let-7 repression leads to HMGA2 overexpression in uterine leiomyosarcoma. J. Cell. Mol. Med. 2009, 13, 3898–3905. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, J.B.; Chennathukuzhi, V.; Koohestani, F.; Nowak, R.A.; Christenson, L.K. Role of microRNA-21 and programmed cell death 4 in the pathogenesis of human uterine leiomyomas. Fertil. Steril. 2012, 98, 726–734.e2. [Google Scholar] [CrossRef] [Green Version]
- Cardozo, E.R.; Foster, R.; Karmon, A.E.; Lee, A.E.; Gatune, L.W.; Rueda, B.R.; Styer, A.K. MicroRNA 21a-5p overexpression impacts mediators of extracellular matrix formation in uterine leiomyoma. Reprod. Boil. Endocrinol. 2018, 16, 46. [Google Scholar] [CrossRef]
- Gao, Y.; Dai, M.; Liu, H.; He, W.; Lin, S.; Yuan, T.; Chen, H.; Dai, S. Diagnostic value of circulating miR-21: An update meta-analysis in various cancers and validation in endometrial cancer. Oncotarget 2016, 7, 68894–68908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maccani, M.A.; Padbury, J.F.; Marsit, C.J. miR-16 and miR-21 Expression in the Placenta Is Associated with Fetal Growth. PLoS ONE 2011, 6, e21210. [Google Scholar] [CrossRef] [PubMed]
- Lasabová, Z.; Vazan, M.; Zibolenova, J.; Svecova, I. Overexpression of miR-21 and miR-122 in preeclamptic placentas. Neuro Endocrinol. Lett. 2015, 36, 695–699. [Google Scholar]
- Marsh, E.E.; Lin, Z.; Yin, P.; Milad, M.; Chakravarti, D.; Bulun, S.E. Differential expression of microRNA species in human uterine leiomyoma versus normal myometrium. Fertil. Steril. 2008, 89, 1771–1776. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, A.; Tagawa, H.; Yamashita, J.; Teshima, K.; Nara, M.; Iwamoto, K.; Kume, M.; Kameoka, Y.; Takahashi, N.; Nakagawa, T.; et al. The role of microRNA-150 as a tumor suppressor in malignant lymphoma. Leukemia 2011, 25, 1324–1334. [Google Scholar] [CrossRef] [Green Version]
- Karra, L.; Shushan, A.; Ben-Meir, A.; Rojansky, N.; Klein, B.Y.; Shveiky, D.; Levitzki, R.; Ben-Bassat, H. Changes related to phosphatidylinositol 3-kinase/Akt signaling in leiomyomas: Possible involvement of glycogen synthase kinase 3α and cyclin D2 in the pathophysiology. Fertil. Steril. 2010, 93, 2646–2651. [Google Scholar] [CrossRef]
- Peng, L.; Wen, Y.; Han, Y.; Wei, A.; Shi, G.; Mizuguchi, M.; Lee, P.; Hernando, E.; Mittal, K.; Wei, J.-J. Expression of insulin-like growth factors (IGFs) and IGF signaling: Molecular complexity in uterine leiomyomas. Fertil. Steril. 2009, 91, 2664–2675. [Google Scholar] [CrossRef] [PubMed]
- Makker, A.; Goel, M.M.; Das, V.; Agarwal, A. PI3K-Akt-mTOR and MAPK signaling pathways in polycystic ovarian syndrome, uterine leiomyomas and endometriosis: An update. Gynecol. Endocrinol. 2012, 28, 175–181. [Google Scholar] [CrossRef]
- Sefton, E.C.; Qiang, W.; Serna, V.; Kurita, T.; Wei, J.-J.; Chakravarti, D.; Kim, J.J. MK-2206, an AKT Inhibitor, Promotes Caspase-Independent Cell Death and Inhibits Leiomyoma Growth. Endocrinology 2013, 154, 4046–4057. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Lu, Z.; Qiang, W.; Vidimar, V.; Kong, B.; Kim, J.J.; Wei, J.-J. Inactivation of AKT Induces Cellular Senescence in Uterine Leiomyoma. Endocrinology 2014, 155, 1510–1519. [Google Scholar] [CrossRef]
- Denicourt, C.; Dowdy, S.F. Cip/Kip proteins: More than just CDKs inhibitors. Genes Dev. 2004, 18, 851–855. [Google Scholar] [CrossRef]
- Viglietto, G.; Motti, M.L.; Bruni, P.; Melillo, R.M.; D’Alessio, A.; Califano, D.; Vinci, F.; Chiappetta, G.; Tsichlis, P.; Bellacosa, A.; et al. Cytoplasmic relocalization and inhibition of the cyclin-dependent kinase inhibitor p27Kip1 by PKB/Akt-mediated phosphorylation in breast cancer. Nat. Med. 2002, 8, 1136–1144. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Yu, L.; Castro, L.; Tucker, C.J.; Moore, A.B.; Xiao, H.; Dixon, D. An essential role of p27 downregulation in fenvalerate-induced cell growth in human uterine leiomyoma and smooth muscle cells. Am. J. Physiol. Metab. 2012, 303, E1025–E1035. [Google Scholar] [CrossRef]
- Prasad, S.B.; Yadav, S.S.; Das, M.; Modi, A.; Kumari, S.; Pandey, L.K.; Singh, S.; Pradhan, S.; Narayan, G. PI3K/AKT pathway-mediated regulation of p27Kip1 is associated with cell cycle arrest and apoptosis in cervical cancer. Cell. Oncol. 2015, 38, 215–225. [Google Scholar] [CrossRef]
- Lee, H.-G.; Baek, J.-W.; Shin, S.-J.; Kwon, S.-H.; Cha, S.-D.; Park, W.-J.; Chung, R.; Choi, E.-S.; Lee, G.-H.; Cho, C.-H. Antitumor Effects of Flavopiridol on Human Uterine Leiomyoma In Vitro and in a Xenograft Model. Reprod. Sci. 2014, 21, 1153–1160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chalhoub, N.; Baker, S.J. PTEN and the PI3-Kinase Pathway in Cancer. Annu. Pathol. Mech. 2009, 4, 127–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, D.Z.; Zhang, H.Y.; Long, X.L.; Zou, S.L.; Zhang, X.Y.; Han, G.Y.; Cui, Z.G. MIR-150 promotes prostate cancer stem cell development via suppressing p27Kip1. Eur. Rev. Med. Pharmacol. Sci. 2015, 19, 4344–4352. [Google Scholar] [PubMed]
- Wu, Q.; Jin, H.; Yang, Z.; Luo, G.; Lu, Y.; Li, K.; Ren, G.; Su, T.; Pan, Y.; Feng, B.; et al. MiR-150 promotes gastric cancer proliferation by negatively regulating the pro-apoptotic gene EGR2. Biochem. Biophys. Commun. 2010, 392, 340–345. [Google Scholar] [CrossRef]
- Huang, S.; Chen, Y.; Wu, W.; Ouyang, N.; Chen, J.; Li, H.; Liu, X.; Su, F.; Lin, L.; Yao, Y. miR-150 Promotes Human Breast Cancer Growth and Malignant Behavior by Targeting the Pro-Apoptotic Purinergic P2X7 Receptor. PLoS ONE 2013, 8, e80707. [Google Scholar] [CrossRef]
- Zhang, N.; Wei, X.; Xu, L. miR-150 promotes the proliferation of lung cancer cells by targeting P53. FEBS Lett. 2013, 587, 2346–2351. [Google Scholar] [CrossRef] [PubMed]
- Yokobori, T.; Suzuki, S.; Tanaka, N.; Inose, T.; Sohda, M.; Sano, A.; Sakai, M.; Nakajima, M.; Miyazaki, T.; Kato, H. MiR-150 is associated with poor prognosis in esophageal squamous cell carcinoma via targeting the EMT inducer ZEB 1. Cancer Sci. 2013, 104, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.J.; Lin, J.; Lwin, T.; Yang, H.; Guo, J.; Kong, W.; Dessureault, S.; Moscinski, L.C.; Rezania, D.; Dalton, W.S. microRNA expression profile and identification of miR-29 as a prognostic marker and pathogenetic factor by targeting CDK6 in mantle cell lymphoma. Blood 2010, 115, 2630–2639. [Google Scholar] [CrossRef]
- Ito, M.; Teshima, K.; Ikeda, S.; Kitadate, A.; Watanabe, A.; Nara, M.; Yamashita, J.; Ohshima, K.; Sawada, K.; Tagawa, H. MicroRNA-150 inhibits tumor invasion and metastasis by targeting the chemokine receptor CCR6 in advanced cutaneous T-cell lymphoma. Blood 2014, 123, 1499–1511. [Google Scholar] [CrossRef]
- Wang, M.; Yang, W.; Li, M.; Li, Y. Low expression of miR-150 in pediatric intestinal Burkitt lymphoma. Exp. Mol. Pathol. 2014, 96, 261–266. [Google Scholar] [CrossRef]
- Ramachandran, S.; Kwon, K.-Y.; Shin, S.-J.; Kwon, S.-H.; Cha, S.-D.; Bae, I.; Cho, C.-H. Cyclin-Dependent Kinase Inhibitor p27Kip1Controls Growth and Cell Cycle Progression in Human Uterine Leiomyoma. J. Korean Med. Sci. 2008, 23, 667–673. [Google Scholar] [CrossRef]
- Cui, L.; Ren, Y.; Yin, H.; Wang, Y.; Li, D.; Liu, M.; Zhu, Y.; Lin, W.; Tang, X.D.; Gui, Y.; et al. Increased expression of tuberin in human uterine leiomyoma. Fertil. Steril. 2011, 95, 1805–1808. [Google Scholar] [CrossRef]
- Moon, J.; Lee, S.-T.; Kong, I.G.; Byun, J.-I.; Sunwoo, J.-S.; Shin, J.-W.; Shim, J.-Y.; Park, J.-H.; Jeon, D.; Jung, K.-H.; et al. Early diagnosis of Alzheimer’s disease from elevated olfactory mucosal miR-206 level. Sci. Rep. 2016, 6, 20364. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.; Loudet, J.C.; Hanusse, P.; Poulin, P. A Probabilistic Functional Network of Yeast Genes. Science 2004, 306, 1555–1558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, S.; Mutlu, L.; Zhou, Y.; Taylor, H.S. Aromatase inhibitor regulates let-7 expression and let-7f induced cell migration in endometrial cells from women with endometriosis. Fertil. Steril. 2016, 106, 673–680. [Google Scholar] [CrossRef] [PubMed]
- Mochitate, K.; Pawelek, P.; Grinnell, F. Stress relaxation of contracted collagen gels: Disruption of actin filament bundles, release of cell surface fibronectin, and down-regulation of DNA and protein synthesis. Exp. Cell 1991, 193, 198–207. [Google Scholar] [CrossRef]
- Tian, B.; Lessan, K.; Kahm, J.; Kleidon, J.; Henke, C. b1 Integrin regulates fibroblast viability during collagen matrix contraction through a phosphatidylinositol 3-kinase AKT/protein kinase B signaling pathway. J. Biol. Chem. 2002, 277, 24667–24675. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.-H.; Xie, D.-D.; Xu, S.; Xia, M.-Z.; Zhang, Z.-Q.; Geng, H.; Chen, L.; Wang, D.-M.; Wei, W.; Yu, D.-X.; et al. Total glucosides of paeony inhibits lipopolysaccharide-induced proliferation, migration and invasion in androgen insensitive prostate cancer cells. PLoS ONE 2017, 12, e0182584. [Google Scholar] [CrossRef] [PubMed]


| Variables | N = 13 |
|---|---|
| Age | 44 (37–48) |
| Name of operation | Hysterectomy |
| Site of leiomyoma | Intramural |
| Size of leiomyoma | 7.4 cm (3.7–12.2) |
| Follicular phase | 10 |
| Proliferative phase | 3 |
| Upregulated | Downregulated | ||||
|---|---|---|---|---|---|
| miRNA | FC (SD) | p Value | miRNA | FC (SD) | p Value |
| hsa-miR-196b-3p | 2.16 (0.39) | 0.04 | hsa-miR-139-5p | 5.06 (2.31) | 0.04 |
| hsa-miR-483-5p | 3.1 (0.63) | 0.04 | hsa-miR-140-3p | 2.08 (0.94) | 0.02 |
| hsa-miR-378d | 1.82 (0.52) | 0.04 | hsa-miR-150-5p | 4.79 (2.29) | 0.03 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J.H.; Choi, Y.S.; Park, J.H.; Kim, H.; Lee, I.; Won, Y.B.; Yun, B.H.; Park, J.H.; Seo, S.K.; Lee, B.S.; et al. MiR-150-5p May Contribute to Pathogenesis of Human Leiomyoma via Regulation of the Akt/p27Kip1 Pathway In Vitro. Int. J. Mol. Sci. 2019, 20, 2684. https://doi.org/10.3390/ijms20112684
Lee JH, Choi YS, Park JH, Kim H, Lee I, Won YB, Yun BH, Park JH, Seo SK, Lee BS, et al. MiR-150-5p May Contribute to Pathogenesis of Human Leiomyoma via Regulation of the Akt/p27Kip1 Pathway In Vitro. International Journal of Molecular Sciences. 2019; 20(11):2684. https://doi.org/10.3390/ijms20112684
Chicago/Turabian StyleLee, Jae Hoon, Young Sik Choi, Ji Hyun Park, Heeyon Kim, Inha Lee, Young Bin Won, Bo Hyon Yun, Joo Hyun Park, Seok Kyo Seo, Byung Seok Lee, and et al. 2019. "MiR-150-5p May Contribute to Pathogenesis of Human Leiomyoma via Regulation of the Akt/p27Kip1 Pathway In Vitro" International Journal of Molecular Sciences 20, no. 11: 2684. https://doi.org/10.3390/ijms20112684