Metabolomic Profiling of the Host Response of Tomato (Solanum lycopersicum) Following Infection by Ralstonia solanacearum
Abstract
:1. Introduction
2. Results
2.1. Cultivar Information
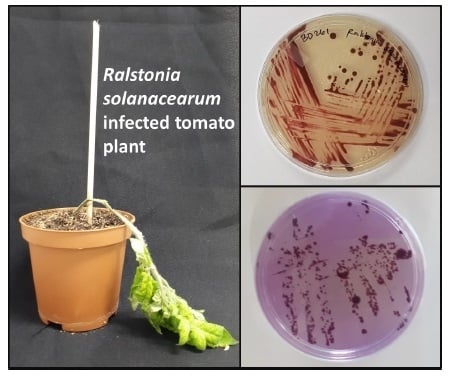
2.2. Symptoms and Characterization
2.3. Ultrahigh-performance Liquid Chromatography Coupled to Mass Spectrometry
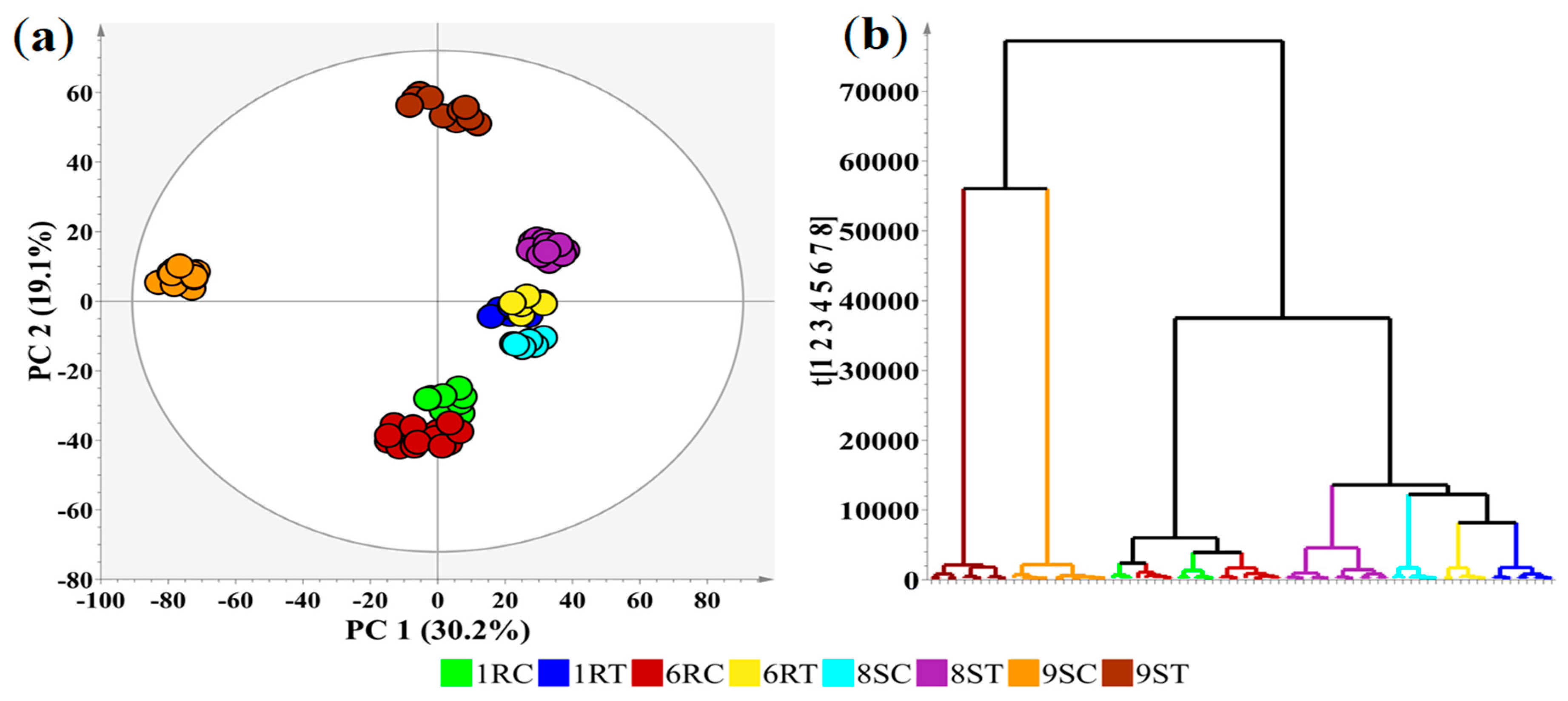
2.4. Multivariate Data Analyses
2.5. Heatmap Visualisation
3. Discussion
3.1. Hydroxybenzoic Acids
3.2. Hydroxycinnamic Acids
3.3. Phenylamides
4. Materials and Methods
4.1. Plant Cultivation
4.2. Pathogen Inoculation
4.3. Bacterial Re-Isolation and Pathogenicity Trials
4.4. Symptom Index
4.5. Metabolite Extraction and Sample Preparation
4.6. Ultra-High Performance Liquid Chromatography-Mass Spectrometry Analyses
4.7. Quadrupole Time-Of-Flight Mass Spectrometry (q-TOF-MS) Analyses
4.8. Data Analyses
4.9. Metabolite Annotation and Qualitative Comparison
4.10. Relative Quantification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Grandillo, S.; Zamir, D.; Tanksley, S.D. Genetic improvement of processing tomatoes: A 20 years perspective. Euphytica 1999, 110, 85–97. [Google Scholar] [CrossRef]
- Li, Y.; Wang, H.; Zhang, Y.; Martin, C. Can the world’s favorite fruit, tomato, provide an effective biosynthetic chassis for high-value metabolites? Plant Cell Rep. 2018, 37, 1443–1450. [Google Scholar] [CrossRef] [PubMed]
- Doughari, J. An overview of plant immunity. J. Plant Pathol. Microbiol. 2015, 6. [Google Scholar] [CrossRef]
- Arie, T.; Takahashi, H.; Kodama, M.; Teraoka, T. Tomato as a model plant for plant-pathogen interactions. Plant Biotechnol. 2007, 24, 135–147. [Google Scholar] [CrossRef]
- Kim, S.G.; Hur, O.-S.; Ro, N.-Y.; Ko, H.-C.; Rhee, J.-H.; Sung, J.S.; Ryu, K.-Y.; Lee, S.-Y.; Baek, H.J. Evaluation of resistance to Ralstonia solanacearum in tomato genetic resources at seedling stage. Plant Pathol. J. 2016, 32, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Barman, A.; Phukan, T.; Kabyashree, K.; Singh, N.; Jha, G.; Sonti, R.V.; Genin, S.; Ray, S.K. Ralstonia solanacearum virulence in tomato seedlings inoculated by leaf clipping. Plant Pathol. 2017, 66, 835–841. [Google Scholar] [CrossRef]
- Mansfield, J.; Genin, S.; Magori, S.; Citovsky, V.; Sriariyanum, M.; Ronald, P.; Dow, M.; Verdier, V.; Beer, S.V.; Machado, M.A.; et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 614–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuluaga, A.P.; Puigvert, M.; Valls, M. Novel plant inputs influencing Ralstonia solanacearum during infection. Front. Microbiol. 2013, 4, 349. [Google Scholar] [CrossRef]
- Caldwell, D.; Kim, B.S.; Iyer-Pascuzzi, A.S. Ralstonia solanacearum differentially colonizes roots of resistant and susceptible tomato plants. Phytopathology 2017, 107, 528–536. [Google Scholar] [CrossRef]
- Jiang, G.; Wei, Z.; Xu, J.; Chen, H.; Zhang, Y.; She, X.; Macho, A.P.; Ding, W.; Liao, B. Bacterial wilt in China: History, current status, and future perspectives. Front. Plant Sci. 2017, 8, 1549. [Google Scholar] [CrossRef]
- Vanitha, S.C.; Niranjana, S.R.; Mortensen, C.N.; Umesha, S. Bacterial wilt of tomato in Karnataka and its management by Pseudomonas fluorescens. BioControl 2009, 54, 685–695. [Google Scholar] [CrossRef]
- Wei, Y.; Caceres-Moreno, C.; Jimenez-Gongora, T.; Wang, K.; Sang, Y.; Lozano-Duran, R.; Macho, A.P. The Ralstonia solanacearum csp22 peptide, but not flagellin-derived peptides, is perceived by plants from the Solanaceae family. Plant Biotechnol. J. 2018, 16, 1349–1362. [Google Scholar] [CrossRef]
- Plener, L.; Boistard, P.; González, A.; Boucher, C.; Genin, S. Metabolic adaptation of Ralstonia solanacearum during plant infection: A methionine biosynthesis case study. PLoS ONE 2012, 7, e36877. [Google Scholar] [CrossRef]
- Yahiaoui, N.; Chéron, J.-J.; Ravelomanantsoa, S.; Hamza, A.A.; Petrousse, B.; Jeetah, R.; Jaufeerally-Fakim, Y.; Félicité, J.; Fillatre, J.; Hostachy, B.; et al. Genetic diversity of the Ralstonia solanacearum species complex in the Southwest Indian ocean islands. Front. Plant Sci. 2017, 8, 2139. [Google Scholar] [CrossRef]
- Khokhani, D.; Lowe-Power, T.M.; Tran, T.M.; Allen, C. A single regulator mediates strategic switching between attachment / spread and growth / virulence in the plant. mBio 2017, 8, e00895-17. [Google Scholar] [CrossRef]
- Jacobs, J.M.; Milling, A.; Mitra, R.M.; Hogan, C.S.; Ailloud, F.; Prior, P.; Allen, C. Ralstonia solanacearum requires PopS, an ancient AvrE-family effector, for virulence and to overcome salicylic acid-mediated defenses during tomato pathogenesis. mBio 2013, 4, e00875-13. [Google Scholar] [CrossRef]
- Milling, A.; Babujee, L.; Allen, C. Ralstonia solanacearum extracellular polysaccharide is a specific elicitor of defense responses in wilt-resistant tomato plants. PLoS ONE 2011, 6, e15853. [Google Scholar] [CrossRef]
- Yao, J.; Allen, C. Chemotaxis is required for virulence and competitive fitness of the bacterial wilt pathogen Ralstonia solanacearum. J. Bacteriol. 2006, 188, 3697–3708. [Google Scholar] [CrossRef]
- Turner, M.; Jauneau, A.; Genin, S.; Tavella, M.-J.; Vailleau, F.; Gentzbittel, L.; Jardinaud, M.-F. Dissection of Bacterial Wilt on Medicago truncatula revealed two type III secretion system effectors acting on root infection process and disease development. Plant Physiol. 2009, 150, 1713–1722. [Google Scholar] [CrossRef]
- Álvarez, B.; Biosca, E.G.; López, M.M. On the life of Ralstonia solanacearum, a destructive bacterial plant pathogen. Technol. Educ. Top. Appl. Microbiol. Microb. Biotechnol. 2010, 1, 267–279. [Google Scholar] [CrossRef]
- Yuliar, Y.A.N.; Toyota, K. Recent Trends in control methods for bacterial wilt diseases caused by Ralstonia solanacearum. Microbes Environ. 2015, 30, 1–11. [Google Scholar] [CrossRef]
- Courant, F.; Antignac, J.-P.; Dervilly-Pinel, G.; Le Bizec, B.; Dervilly-Pinel, G. Basics of mass spectrometry based metabolomics. Proteomics 2014, 14, 2369–2388. [Google Scholar] [CrossRef]
- Zampieri, M.; Sekar, K.; Zamboni, N.; Sauer, U. Frontiers of high-throughput metabolomics. Curr. Opin. Chem. Biol. 2017, 36, 15–23. [Google Scholar] [CrossRef]
- Zhou, B.; Xiao, J.F.; Tuli, L.; Ressom, H.W. LC-MS-based metabolomics. Mol. Biosyst. 2012, 8, 470–481. [Google Scholar] [CrossRef]
- Liu, X.; Locasale, J.W. Metabolomics—A primer. Trends Biochem. Sci. 2017, 42, 274–284. [Google Scholar] [CrossRef]
- Tugizimana, F.; Piater, L.; Dubery, I. Plant metabolomics: A new frontier in phytochemical analysis. South Afr. J. Sci. 2013, 109, 1–11. [Google Scholar] [CrossRef]
- Peng, B.; Li, H.; Peng, X.-X. Functional metabolomics: From biomarker discovery to metabolome reprogramming. Protein Cell 2015, 6, 628–637. [Google Scholar] [CrossRef]
- Saito, K.; Matsuda, F. Metabolomics for functional genomics, systems biology, and biotechnology. Annu. Rev. Plant Biol. 2010, 61, 463–489. [Google Scholar] [CrossRef]
- Allwood, J.W.; Ellis, D.I.; Goodacre, R. Metabolomic technologies and their application to the study of plants and plant-host interactions. Physiol. Plant. 2008, 132, 117–135. [Google Scholar] [CrossRef]
- Zeiss, D.R.; Mhlongo, M.I.; Tugizimana, F.; Steenkamp, P.A.; Dubery, I.A. Comparative metabolic phenotyping of tomato (Solanum lycopersicum) for the identification of metabolic signatures in cultivars differing in resistance to Ralstonia solanacearum. Int. J. Mol. Sci. 2018, 19, 2558. [Google Scholar] [CrossRef]
- Ahmed, N.N.; Islam, M.R.; Hossain, M.A.; Meah, M.B.; Hossain, M.M. Determination of races and biovars of Ralstonia solanacearum causing bacterial wilt disease of potato. J. Agric. Sci. 2013, 5, 86–93. [Google Scholar] [CrossRef]
- Mori, T.; Fujiyoshi, T.; Inada, T.; Matsusaki, H.; Ogawa, K.; Matsuzoe, N. Phenotypic conversion of resistant and susceptible solanum plants. Environ. Control Biol. 2011, 49, 165–176. [Google Scholar] [CrossRef]
- Shutt, V.; Shin, G.; Van Der Waals, J.; Goszczynska, T.; Coutinho, T. Characterization of Ralstonia strains infecting tomato plants in South Africa. Crop. Prot. 2018, 112, 56–62. [Google Scholar] [CrossRef]
- Ren, S.; Hinzman, A.A.; Kang, E.L.; Szczesniak, R.D.; Lu, L.J. Computational and statistical analysis of metabolomics data. Metabolomics 2015, 11, 1492–1513. [Google Scholar] [CrossRef]
- Saccenti, E.; Hoefsloot, H.C.J.; Smilde, A.K.; Westerhuis, J.A.; Hendriks, M.M.W.B. Reflections on univariate and multivariate analysis of metabolomics data. Metabolomics 2014, 10, 361–374. [Google Scholar] [CrossRef]
- Trygg, J.; Holmes, E.; Lundstedt, T. Chemometrics in Metabonomics. J. Proteome Res. 2007, 6, 469–479. [Google Scholar] [CrossRef] [PubMed]
- Triba, M.; Le Moyec, L.; Amathieu, R.; Goossens, C.; Bouchemal, N.; Nahon, P.; Rutledge, D.; Savarin, P. PLS/OPLS models in metabolomics: Impact of permutation of dataset rows on the K-fold cross-validation quality parameters. Mol. Biosyst. 2014, 11, 13–19. [Google Scholar] [CrossRef]
- Goodacre, R.; Broadhurst, D.; Smilde, A.K.; Kristal, B.S.; Baker, J.D.; Beger, R.; Bessant, C.; Connor, S.; Capuani, G.; Craig, A.; et al. Proposed minimum reporting standards for data analysis in metabolomics. Metabolomics 2007, 3, 231–241. [Google Scholar] [CrossRef]
- Tugizimana, F.; Steenkamp, P.A.; Piater, L.A.; Dubery, I.A. A conversation on data mining strategies in LC-MS untargeted metabolomics: pre-processing and pre-treatment steps. Metabolites 2016, 6, 40. [Google Scholar] [CrossRef]
- Mhlongo, M.I.; Steenkamp, P.A.; Piater, L.A.; Madala, N.E.; Dubery, I.A. Profiling of altered metabolomic states in Nicotiana tabacum cells induced by priming agents. Front. Plant Sci. 2016, 7, 1527. [Google Scholar] [CrossRef]
- Westerhuis, J.A.; Hoefsloot, H.C.J.; Smit, S.; Vis, D.J.; Smilde, A.K.; Van Velzen, E.J.J.; Van Duijnhoven, J.P.M.; Van Dorsten, F.A.; Velzen, E.J.J.; Duijnhoven, J.P.M.; et al. Assessment of PLSDA cross validation. Metabolomics 2008, 4, 81–89. [Google Scholar] [CrossRef] [Green Version]
- Trivedi, D.K.; Iles, R.K. The application of SIMCA P+ in shotgun metabolomics analysis of ZICⓇHILIC-MS spectra of human urine—Experience with the Shimadzu IT-TOF and profiling solutions data extraction software. J. Chromatogr. Sep. Tech. 2012, 3, 145. [Google Scholar] [CrossRef]
- Tugizimana, F.; Ncube, E.N.; Steenkamp, P.A.; Dubery, I.A. Metabolomics-derived insights into the manipulation of terpenoid synthesis in Centella asiatica cells by methyl jasmonate. Plant Biotechnol. Rep. 2015, 9, 125–136. [Google Scholar] [CrossRef]
- Widhalm, J.R.; Dudareva, N. A Familiar Ring to It: Biosynthesis of plant benzoic acids. Mol. Plant 2015, 8, 83–97. [Google Scholar] [CrossRef]
- Gómez-Romero, M.; Carretero, A.S.; Fernández-Gutiérrez, A. Metabolite profiling and quantification of phenolic compounds in methanol extracts of tomato fruit. Phytochemistry 2010, 71, 1848–1864. [Google Scholar] [CrossRef] [PubMed]
- Mhlongo, M.I.; Piater, L.A.; Madala, N.E.; Steenkamp, P.A.; Dubery, I.A. Phenylpropanoid defences in Nicotiana tabacum cells: overlapping metabolomes indicate common aspects to priming responses induced by lipopolysaccharides, chitosan and flagellin-22. PLoS ONE 2016, 11, e0151350. [Google Scholar] [CrossRef] [PubMed]
- Mhlongo, M.I.; Piater, L.A.; Steenkamp, P.A.; Madala, N.E.; Dubery, I.A. Metabolomic fingerprinting of primed tobacco cells provide the first evidence for the biological origin of cis-chlorogenic acid. Biotechnol. Lett. 2015, 37, 205–209. [Google Scholar] [CrossRef] [PubMed]
- Sequera-Mutiozabal, M.; Antoniou, C.; Tiburcio, A.F.; Alcázar, R.; Fotopoulos, V. Polyamines: emerging hubs promoting drought and salt stress tolerance in plants. Curr. Mol. Biol. Rep. 2017, 3, 28–36. [Google Scholar] [CrossRef]
- Torras-Claveria, L.; Jáuregui, O.; Codina, C.; Tiburcio, A.F.; Bastida, J.; Viladomat, F. Analysis of phenolic compounds by high-performance liquid chromatography coupled to electrospray ionization tandem mass spectrometry in senescent and water-stressed tobacco. Plant Sci. 2012, 182, 71–78. [Google Scholar] [CrossRef]
- Rider, J.E.; Hacker, A.; Mackintosh, C.A.; Pegg, A.E.; Woster, P.M.; Casero, R.A. Spermine and spermidine mediate protection against oxidative damage caused by hydrogen peroxide. Amino Acids 2007, 33, 231–240. [Google Scholar] [CrossRef]
- Campos, L.; Lisón, P.; López-Gresa, M.P.; Rodrigo, I.; Zacarés, L.; Conejero, V.; Bellés, J.M. Transgenic tomato plants overexpressing tyramine N-hydroxycinnamoyltransferase exhibit elevated hydroxycinnamic acid amide levels and enhanced resistance to Pseudomonas syringae. Mol. Plant-Microbe Interact. 2014, 27, 1159–1169. [Google Scholar] [CrossRef] [PubMed]
- Ly, D.; Kang, K.; Choi, J.-Y.; Ishihara, A.; Back, K.; Lee, S.-G. HPLC Analysis of serotonin, tryptamine, tyramine, and the hydroxycinnamic acid amides of serotonin and tyramine in food vegetables. J. Med. Food 2008, 11, 385–389. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.; Ishihara, A.; Back, K. Production of coumaroylserotonin and feruloylserotonin in transgenic rice expressing pepper hydroxycinnamoyl-coenzyme A: serotonin N-(hydroxycinnamoyl)transferase. Plant Physiol. 2004, 135, 346–356. [Google Scholar] [CrossRef] [PubMed]
- Von Roepenack-Lahaye, E.; Schornack, S.; Hammond-Kosack, K.E.; Lahaye, T.; Jones, J.D.G.; Daniels, M.J.; Newman, M.-A.; Dow, J.M. p-Coumaroylnoradrenaline, a novel plant metabolite implicated in tomato defense against pathogens. J. Biol. Chem. 2003, 278, 43373–43383. [Google Scholar] [CrossRef] [PubMed]
- Zacarés, L.; López-Gresa, M.P.; Fayos, J.; Primo, J.; Bellés, J.M.; Conejero, V. Induction of p-Coumaroyldopamine and feruloyldopamine, two novel metabolites, in tomato by the bacterial pathogen Pseudomonas syringae. Mol. Plant-Microbe Interact. 2007, 20, 1439–1448. [Google Scholar] [CrossRef]
- Miedes, E.; Vanholme, R.; Boerjan, W.; Molina, A. The role of the secondary cell wall in plant resistance to pathogens. Front. Plant Sci. 2014, 5, 358. [Google Scholar] [CrossRef] [Green Version]
- Sanabria, N.M.; Huang, J.C.; Dubery, I.A. Self/non-self perception in plants in innate immunity and defense. Self Nonself. 2012, 1, 40–54. [Google Scholar] [CrossRef]
- Al Sinani, S.; Eltayeb, E. The steroidal glycoalkaloids solamargine and solasonine in Solanum plants. South Afr. J. Bot. 2017, 112, 253–269. [Google Scholar] [CrossRef]
- Klessig, D.F.; Choi, H.W.; Dempsey, D.A.; Dempsey, D.A. Systemic acquired resistance and salicylic acid: past, present, and future. Mol. Plant-Microbe Interact. 2018, 31, 871–888. [Google Scholar] [CrossRef]
- Bari, R.; Jones, J.D.G. Role of plant hormones in plant defence responses. Plant Mol. Biol. 2009, 69, 473–488. [Google Scholar] [CrossRef]
- Bellés, J.M.; Garro, R.; Fayos, J.; Navarro, P.; Primo, J.; Conejero, V. Gentisic acid as a pathogen-inducible signal, additional to salicylic acid for activation of plant defenses in tomato. Mol. Plant-Microbe Interact. 1999, 12, 227–235. [Google Scholar] [CrossRef]
- Fayos, J.; Bellés, J.M.; López-Gresa, M.P.; Primo, J.; Conejero, V. Induction of gentisic acid 5-O-β-d-xylopyranoside in tomato and cucumber plants infected by different pathogens. Phytochemistry 2006, 67, 142–148. [Google Scholar] [CrossRef]
- Zhu, G.-Q.; Liu, Q.; Chen, L.; Li, Y.-J.; Hou, B.-K.; Huang, X. Modulation of plant salicylic acid-associated immune responses via glycosylation of dihydroxybenzoic acids. Plant Physiol. 2018, 176, 3103–3119. [Google Scholar]
- Mhlongo, M.I.; Piater, L.A.; Steenkamp, P.A.; Madala, N.E.; Dubery, I.A. Priming agents of plant defence stimulate the accumulation of mono- and di-acylated quinic acids in cultured tobacco cells. Physiol. Mol. Plant Pathol. 2014, 88, 61–66. [Google Scholar] [CrossRef]
- Harris, V.; Jiranek, V.; Ford, C.M.; Grbin, P.R. Inhibitory effect of hydroxycinnamic acids on Dekkera spp. Appl. Microbiol. Biotechnol. 2010, 86, 721–729. [Google Scholar] [CrossRef]
- Setti, L.; Faulds, C.; Giuliani, S. Hydroxycinnamic acids as natural antioxidants. Sci. Technol. 2001, 83, 1–5. [Google Scholar]
- Lowe, T.M.; Ailloud, F.; Allen, C. Hydroxycinnamic acid degradation, a broadly conserved trait, protects Ralstonia solanacearum from chemical plant defenses and contributes to root colonization and virulence. Mol. Plant-Microbe Interact. 2015, 28, 286–297. [Google Scholar] [CrossRef]
- Fry, S.C.; Willis, S.C.; Paterson, A.E.J. Intraprotoplasmic and wall-localised formation of arabinoxylan-bound diferulates and larger ferulate coupling-products in maize cell-suspension cultures. Planta 2000, 211, 679–692. [Google Scholar] [CrossRef]
- Yang, L.; Wu, L.; Yao, X.; Zhao, S.; Wang, J.; Li, S.; Ding, W. Hydroxycoumarins: new, effective plant-derived compounds reduce Ralstonia pseudosolanacearum populations and control tobacco bacterial wilt. Microbiol. Res. 2018, 215, 15–21. [Google Scholar] [CrossRef]
- Beckman, C.H. Phenolic-storing cells: Keys to programmed cell death and periderm formation in wilt disease resistance and in general defence responses in plants? Physiol. Mol. Plant Pathol. 2000, 57, 101–110. [Google Scholar] [CrossRef]
- Bassard, J.-E.; Ullmann, P.; Bernier, F.; Werck-Reichhart, D. Phenolamides: Bridging polyamines to the phenolic metabolism. Phytochemistry 2010, 71, 1808–1824. [Google Scholar] [CrossRef]
- Edreva, A.M.; Velikova, V.B.; Tsonev, T.D.; Velikova, V.; Tsonev, T. Phenylamides in plants. Russ. J. Plant Physiol. 2007, 54, 287–301. [Google Scholar] [CrossRef]
- Gupta, K.; Dey, A.; Gupta, B. Plant polyamines in abiotic stress responses. Acta Physiol. Plant. 2013, 35, 2015–2036. [Google Scholar] [CrossRef]
- Kusano, T.; Berberich, T.; Tateda, C.; Takahashi, Y. Polyamines: Essential factors for growth and survival. Planta 2008, 228, 367–381. [Google Scholar] [CrossRef]
- Hussain, S.S.; Ali, M.; Ahmad, M.; Siddique, K.H. Polyamines: Natural and engineered abiotic and biotic stress tolerance in plants. Biotechnol. Adv. 2011, 29, 300–311. [Google Scholar] [CrossRef]
- Walters, D.R. Polyamines and plant disease. Phytochemistry 2003, 64, 97–107. [Google Scholar] [CrossRef]
- Marinova, E.; Georgiev, L.; Totseva, I.; Seizova, K.; Milkova, T. Antioxidant activity and mechanism of action of some synthesised phenolic acid amides of aromatic amines. Czech J. Food Sci. 2013, 31, 5–13. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Hyuk, J.; Hung, W.; Zheng, X.; Wang, Y.; Ho, C. Use of UHPLC-TripleQ with synthetic standards to profile anti-inflammatory hydroxycinnamic acid amides in root barks and leaves of Lycium barbarum. J. Food Drug Anal. 2017, 26, 572–582. [Google Scholar] [CrossRef]
- Almoulah, N.F.; Voynikov, Y.; Gevrenova, R.; Schohn, H.; Tzanova, T.; Yagi, S.; Thomas, J.; Mignard, B.; Ahmed, A.; El Siddig, M.; et al. Antibacterial, antiproliferative and antioxidant activity of leaf extracts of selected Solanaceae species. S. Afr. J. Bot. 2017, 112, 368–374. [Google Scholar] [CrossRef]
- Chochkova, M.; Stoykova, B.; Petrova, P.; Gyoshkova, N.; Ivanova, G.; Štícha, M.; Milkova, T. Synthesis and radical scavenging activity of cinnamic acid esters. Bulg. Chem. Commun. 2017, 1, 68–73. [Google Scholar]
- Ghuge, S.A.; Tisi, A.; Carucci, A.; Rodrigues-Pousada, R.A.; Franchi, S.; Tavladoraki, P.; Angelini, R.; Cona, A. Cell wall amine oxidases: new players in root xylem differentiation under stress conditions. Plants 2015, 4, 489–504. [Google Scholar] [CrossRef]
- Yoda, H.; Fujimura, K.; Takahashi, H.; Munemura, I.; Uchimiya, H.; Sano, H. Polyamines as a common source of hydrogen peroxide in host- and nonhost hypersensitive response during pathogen infection. Plant Mol. Biol. 2009, 70, 103–112. [Google Scholar] [CrossRef]
- Alcázar, R.; Altabella, T.; Marco, F.; Bortolotti, C.; Reymond, M.; Koncz, C.; Carrasco, P.; Tiburcio, A.F. Polyamines: Molecules with regulatory functions in plant abiotic stress tolerance. Planta 2010, 231, 1237–1249. [Google Scholar] [CrossRef]
- Cao, F.Y.; Yoshioka, K.; Desveaux, D. The roles of ABA in plant–pathogen interactions. J. Plant Res. 2011, 124, 489–499. [Google Scholar] [CrossRef]
- Fariduddin, Q.; Varshney, P.; Yusuf, M.; Ahmad, A. Polyamines: Potent modulators of plant responses to stress. J. Plant Interact. 2013, 8, 1–16. [Google Scholar] [CrossRef]
- Williamson, L.; Nakaho, K.; Hudelson, B.; Allen, C. Ralstonia solanacearum race 3, biovar 2 strains isolated from geranium are pathogenic on potato. Plant Dis. 2002, 86, 987–991. [Google Scholar] [CrossRef]
- Pontes, N.C.; Fujinawa, M.F.; Oliveira, J.R. Selective media for detection and quantification of Brazilian Ralstonia solanacearum isolates in soil. Hortic. Bras. 2017, 35, 41–47. [Google Scholar] [CrossRef]
- Chen, H.; Song, Q.-Q.; Tang, G.; Feng, Q.; Lin, L. The combined optimization of Savitzky-Golay smoothing and multiplicative scatter correction for FT-NIR PLS Models. Int. Sch. Res. Not. 2013, 2013, 642190. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Furumichi, M.; Morishima, K.; Tanabe, M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019, 47, D590–D595. [Google Scholar] [CrossRef]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef]
- Spicer, R.A.; Salek, R.; Steinbeck, C. Compliance with minimum information guidelines in public metabolomics repositories. Sci. Data 2017, 4, 170137. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef]





© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeiss, D.R.; Mhlongo, M.I.; Tugizimana, F.; Steenkamp, P.A.; Dubery, I.A. Metabolomic Profiling of the Host Response of Tomato (Solanum lycopersicum) Following Infection by Ralstonia solanacearum. Int. J. Mol. Sci. 2019, 20, 3945. https://doi.org/10.3390/ijms20163945
Zeiss DR, Mhlongo MI, Tugizimana F, Steenkamp PA, Dubery IA. Metabolomic Profiling of the Host Response of Tomato (Solanum lycopersicum) Following Infection by Ralstonia solanacearum. International Journal of Molecular Sciences. 2019; 20(16):3945. https://doi.org/10.3390/ijms20163945
Chicago/Turabian StyleZeiss, Dylan R., Msizi I. Mhlongo, Fidele Tugizimana, Paul A. Steenkamp, and Ian A. Dubery. 2019. "Metabolomic Profiling of the Host Response of Tomato (Solanum lycopersicum) Following Infection by Ralstonia solanacearum" International Journal of Molecular Sciences 20, no. 16: 3945. https://doi.org/10.3390/ijms20163945
APA StyleZeiss, D. R., Mhlongo, M. I., Tugizimana, F., Steenkamp, P. A., & Dubery, I. A. (2019). Metabolomic Profiling of the Host Response of Tomato (Solanum lycopersicum) Following Infection by Ralstonia solanacearum. International Journal of Molecular Sciences, 20(16), 3945. https://doi.org/10.3390/ijms20163945