Expression Analysis of PIN Genes in Root Tips and Nodules of Lotus japonicus
Abstract
:1. Introduction
2. Results
2.1. Bioinformatic Analysis of L. japonicus PIN Sequences Reveals Significant Similarity to Their M. truncatula and A. thaliana Orthologs and a Typical Transmembrane Topology
2.2. Real-Time Quantitative Polymerase Chain Reaction (qPCR) Expression Analysis of LjPINs Reveals Differences among Root Tips and Root Nodules at Both Tested Developmental Stages
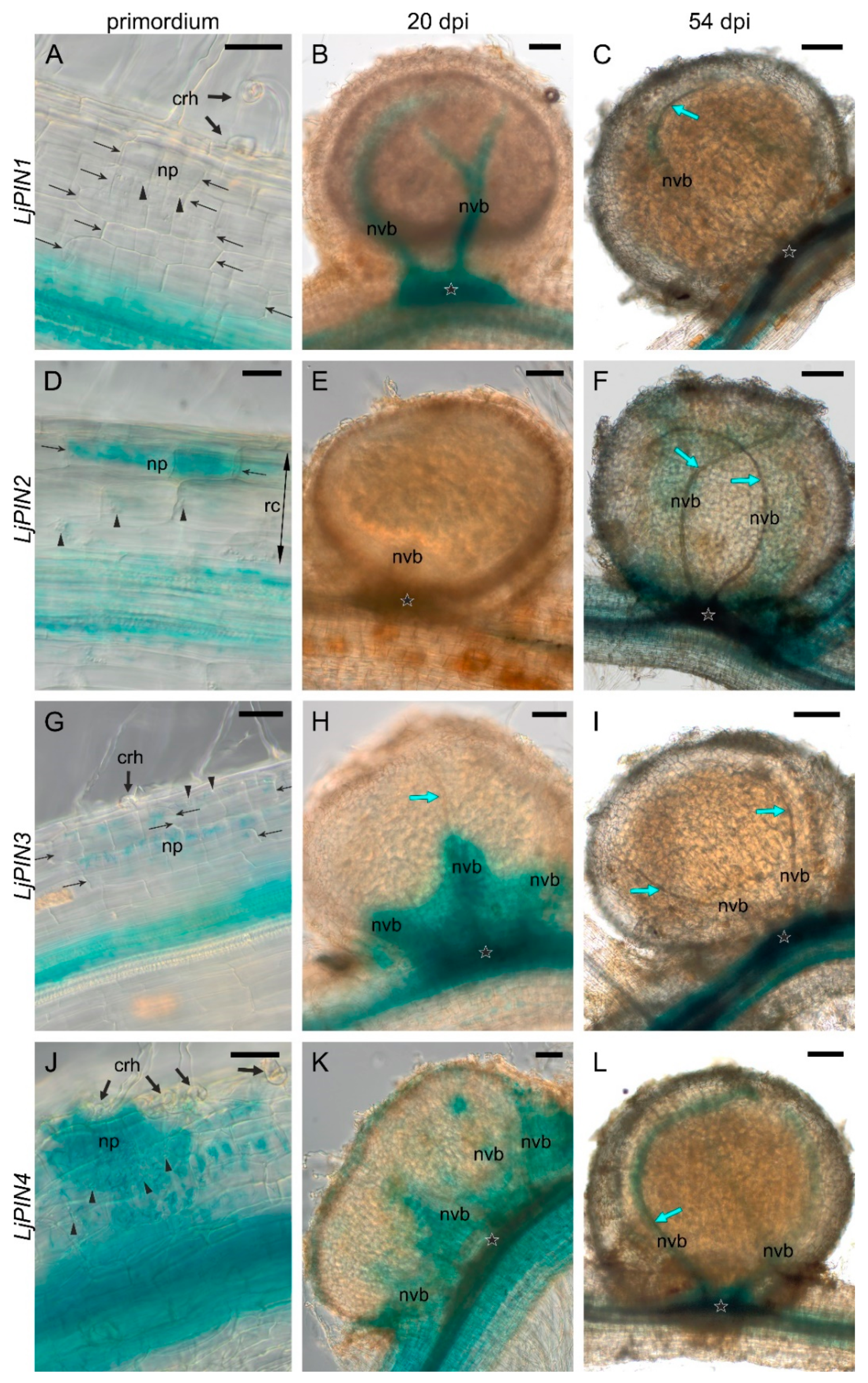
2.3. Spatial Expression Patterns of PIN Genes Strongly Indicate Their Essential Role in the Development of L. Japonicus Root Nodules and in the Formation of the Polar Auxin Transport Pattern in the Root Tips
2.4. Twenty Days Post Inoculation L. japonicus Nodules Still Demonstrate Meristematic Activity
3. Discussion
4. Materials and Methods
4.1. Plant Material, Inoculation with Rhizobia and Growth Conditions
4.2. Phylogenetic Analysis of LjPINs and Transmembrane Domain Prediction
4.3. RNA Isolation and cDNA Synthesis
4.4. Reverse Transcriptase PCR (RT-PCR) and Real-Time qPCR
4.5. Preparation of Genetic Constructs
4.6. Transient Transformation of L. japonicus Roots and Histochemical Localization of GUS Activity
4.7. Microscopic Analysis of Mitotic Divisions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ABC | ATP-binding cassette |
| BLAST | Basic Local Alignment Search Tool |
| bp | base pairs |
| CDK | cyclin-dependent kinases |
| CDKB | class B of cyclin-dependent kinases |
| cDNA | complementary deoxyribonucleic acid |
| CDS | coding sequence |
| cll | columella |
| crh | curled root hair |
| cv. | cultivar |
| CYC1 | cyclin 1 |
| DNA | deoxyribonucleic acid |
| DNase | deoxyribonuclease |
| dpi | day/days postrhizobial inoculation |
| ER | endoplasmic reticulum |
| GUS | β-glucuronidase |
| IAA | indole-3-acetic acid |
| lc | lateral root cap |
| LECA | lightweight expanded clay aggregate |
| NCBI | National Center for Biotechnology Information |
| nc | nodule cortex |
| np | nodule primordium |
| nvb | nodule’s vascular bundle |
| OD | optical density |
| p | probability value |
| PAT | polar auxin transport |
| pc | procambium |
| PCR | polymerase chain reaction |
| PEO-IAA | α-(phenyl ethyl-2-one)-indole-3-acetic acid |
| PIN | PIN-formed protein |
| PPFD | photosynthetic photon flux density |
| prc | meristematic primary root cortex |
| qPCR | quantitive polymerase chain reaction |
| RAM | root apical meristem |
| rc | root primary cortex |
| RNA | ribonucleic acid |
| RT-PCR | reverse transcriptase polymerase chain reaction |
| SE | standard error |
| v. | version |
| vs. | versus |
| w/v | weight per volume |
References
- Ferguson, B.J.; Indrasumunar, A.; Hayashi, S.; Lin, M.-H.; Lin, Y.-H.; Reid, D.E.; Gresshoff, P.M. Molecular analysis of legume nodule development and autoregulation. J. Integr. Plant Biol. 2010, 52, 61–76. [Google Scholar] [CrossRef] [PubMed]
- Sprent, J.I.; Sprent, P. Nitrogen Fixing Organisms: Pure and Applied Aspects; Chapman and Hall Ltd.: London, UK, 1990. [Google Scholar]
- Szczyglowski, K.; Shaw, R.S.; Wopereis, J.; Copeland, S.; Hamburger, D.; Kasiborski, B.; Dazzo, F.B.; de Bruijn, F.J. Nodule Organogenesis and Symbiotic Mutants of the Model Legume Lotus japonicus. Mol. Plant Microbe Interact. 1998, 11, 684–697. [Google Scholar] [CrossRef]
- Oke, V.; Long, S.R. Bacteroid formation in the Rhizobium-legume symbiosis. Curr. Opin. Microbiol. 1999, 2, 641–646. [Google Scholar] [CrossRef]
- Rolfe, B.G.; Gresshoff, P.M. Genetic analysis of legume nodule initiation. Annu. Rev. Plant Physiol. Plant Mol. Biol. (USA) 1988, 39, 297–319. [Google Scholar] [CrossRef]
- Vieten, A.; Sauer, M.; Brewer, P.B.; Friml, J. Molecular and cellular aspects of auxin-transport-mediated development. Trends Plant Sci. 2007, 12, 160–168. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Wang, B.; Moreno, I.; Dupláková, N.; Simon, S.; Carraro, N.; Reemmer, J.; Pěnčík, A.; Chen, X.; Tejos, R.; et al. ER-localized auxin transporter PIN8 regulates auxin homeostasis and male gametophyte development in Arabidopsis. Nat. Commun. 2012, 3, 941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mravec, J.; Skůpa, P.; Bailly, A.; Hoyerová, K.; Křeček, P.; Bielach, A.; Petrášek, J.; Zhang, J.; Gaykova, V.; Stierhof, Y.-D.; et al. Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature 2009, 459. [Google Scholar] [CrossRef] [PubMed]
- Sawchuk, M.G.; Edgar, A.; Scarpella, E. Patterning of leaf vein networks by convergent auxin transport pathways. PLoS Genet. 2013, 9, e1003294. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, A.M.; Bhuvaneswari, T.V.; Torrey, J.G.; Bisseling, T. Early nodulin genes are induced in alfalfa root outgrowths elicited by auxin transport inhibitors. Proc. Natl. Acad. Sci. USA 1989, 86, 1244–1248. [Google Scholar] [CrossRef] [Green Version]
- Huo, X.; Schnabel, E.; Hughes, K.; Frugoli, J. RNAi Phenotypes and the Localization of a Protein::GUS Fusion Imply a Role for Medicago truncatula PIN Genes in Nodulation. J. Plant Growth Regul. 2006, 25, 156–165. [Google Scholar] [CrossRef]
- Rightmyer, A.P.; Long, S.R. Pseudonodule formation by wild-type and symbiotic mutant Medicago truncatula in response to auxin transport inhibitors. Mol. Plant Microbe Interact. 2011, 24, 1372–1384. [Google Scholar] [CrossRef] [PubMed]
- Sańko-Sawczenko, I.; Łotocka, B.; Czarnocka, W. Expression Analysis of PIN Genes in Root Tips and Nodules of Medicago truncatula. Int. J. Mol. Sci. 2016, 17, 1197. [Google Scholar] [CrossRef]
- Schnabel, E.L.; Frugoli, J. The PIN and LAX families of auxin transport genes in Medicago truncatula. Mol. Genet. Genom. 2004, 272, 420–432. [Google Scholar] [CrossRef] [PubMed]
- Mathesius, U.; Schlaman, H.R.; Spaink, H.P.; Of Sautter, C.; Rolfe, B.G.; Djordjevic, M.A. Auxin transport inhibition precedes root nodule formation in white clover roots and is regulated by flavonoids and derivatives of chitin oligosaccharides. Plant J. 1998, 14, 23–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ng, J.L.P.; Hassan, S.; Truong, T.T.; Hocart, C.H.; Laffont, C.; Frugier, F.; Mathesius, U. Flavonoids and Auxin Transport Inhibitors Rescue Symbiotic Nodulation in the Medicago truncatula Cytokinin Perception Mutant cre1. Plant Cell 2015, 27, 2210–2226. [Google Scholar] [CrossRef] [PubMed]
- Pii, Y.; Crimi, M.; Cremonese, G.; Spena, A.; Pandolfini, T. Auxin and nitric oxide control indeterminate nodule formation. BMC Plant Biol. 2007, 7, 21. [Google Scholar] [CrossRef] [PubMed]
- Bustos-Sanmamed, P.; Mao, G.; Deng, Y.; Elouet, M.; Khan, G.A.; Bazin, J.; Turner, M.; Subramanian, S.; Yu, O.; Crespi, M.; Lelandais-Brière, C. Overexpression of miR160 affects root growth and nitrogen-fixing nodule number in Medicago truncatula. Funct. Plant Biol. 2013, 40, 1208–1220. [Google Scholar] [CrossRef]
- Mao, G.; Turner, M.; Yu, O.; Subramanian, S. miR393 and miR164 influence indeterminate but not determinate nodule development. Plant Signal. Behav. 2013, 8. [Google Scholar] [CrossRef] [Green Version]
- Turner, M.; Nizampatnam, N.R.; Baron, M.; Coppin, S.; Damodaran, S.; Adhikari, S.; Arunachalam, S.P.; Yu, O.; Subramanian, S. Ectopic expression of miR160 results in auxin hypersensitivity, cytokinin hyposensitivity, and inhibition of symbiotic nodule development in soybean. Plant Physiol. 2013, 162, 2042–2055. [Google Scholar] [CrossRef]
- Ng, J.L.P.; Mathesius, U. Acropetal Auxin Transport Inhibition Is Involved in Indeterminate but Not Determinate Nodule Formation. Front. Plant Sci. 2018, 9, 169. [Google Scholar] [CrossRef]
- Pacios-Bras, C.; Schlaman, H.R.M.; Boot, K.; Admiraal, P.; Langerak, J.M.; Stougaard, J.; Spaink, H.P. Auxin distribution in Lotus japonicus during root nodule development. Plant Mol. Biol. 2003, 52, 1169–1180. [Google Scholar] [CrossRef] [PubMed]
- Takanashi, K.; Sugiyama, A.; Sato, S.; Tabata, S.; Yazaki, K. LjABCB1, an ATP-binding cassette protein specifically induced in uninfected cells of Lotus japonicus nodules. J. Plant Physiol. 2012, 169, 322–326. [Google Scholar] [CrossRef]
- Takanashi, K.; Sugiyama, A.; Yazaki, K. Involvement of auxin distribution in root nodule development of Lotus japonicus. Planta 2011, 234, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Takanashi, K.; Sugiyama, A.; Yazaki, K. Auxin distribution and lenticel formation in determinate nodule of Lotus japonicus. Plant Signal. Behav. 2011, 6, 1405–1407. [Google Scholar] [CrossRef] [PubMed]
- Suzaki, T.; Yano, K.; Ito, M.; Umehara, Y.; Suganuma, N.; Kawaguchi, M. Positive and negative regulation of cortical cell division during root nodule development in Lotus japonicus is accompanied by auxin response. Development 2012, 139, 3997–4006. [Google Scholar] [CrossRef] [PubMed]
- Kohlen, W.; Ng, J.L.P.; Deinum, E.E.; Mathesius, U. Auxin transport, metabolism, and signalling during nodule initiation: Indeterminate and determinate nodules. J. Exp. Bot. 2017. [Google Scholar] [CrossRef] [PubMed]
- Shirasawa, K.; Isobe, S.; Tabata, S.; Hirakawa, H. Kazusa Marker DataBase: A database for genomics, genetics, and molecular breeding in plants. Breed. Sci. 2014, 64, 264–271. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Nakamura, Y.; Kaneko, T.; Asamizu, E.; Kato, T.; Nakao, M.; Sasamoto, S.; Watanabe, A.; Ono, A.; Kawashima, K.; et al. Genome structure of the legume, Lotus japonicus. DNA Res. 2008, 15, 227–239. [Google Scholar] [CrossRef] [PubMed]
- Paponov, I.A.; Teale, W.D.; Trebar, M.; Blilou, I.; Palme, K. The PIN auxin efflux facilitators: Evolutionary and functional perspectives. Trends Plant Sci. 2005, 10, 170–177. [Google Scholar] [CrossRef]
- Krecek, P.; Skupa, P.; Libus, J.; Naramoto, S.; Tejos, R.; Friml, J.; Zazímalová, E. The PIN-FORMED (PIN) protein family of auxin transporters. Genome Biol. 2009, 10, 249. [Google Scholar] [CrossRef] [Green Version]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Hirt, H.; Mink, M.; Pfosser, M.; Bögre, L.; Györgyey, J.; Jonak, C.; Gartner, A.; Dudits, D.; Heberle-Bors, E. Alfalfa cyclins: Differential expression during the cell cycle and in plant organs. Plant Cell 1992, 4, 1531–1538. [Google Scholar] [CrossRef]
- Magyar, Z.; Mészáros, T.; Miskolczi, P.; Deák, M.; Fehér, A.; Brown, S.; Kondorosi, E.; Athanasiadis, A.; Pongor, S.; Bilgin, M.; et al. Cell cycle phase specificity of putative cyclin-dependent kinase variants in synchronized alfalfa cells. Plant Cell 1997, 9, 223–235. [Google Scholar] [CrossRef] [PubMed]
- Petrásek, J.; Friml, J. Auxin transport routes in plant development. Development 2009, 136, 2675–2688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Chen, J.; Weng, L.; Li, X.; Cao, X.; Hu, X.; Luo, D.; Yang, J. Multiple Components are Integrated to Determine Leaf Complexity in Lotus japonicus. J. Integr. Plant Biol. 2013, 55, 419–433. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chai, C.; Valliyodan, B.; Maupin, C.; Annen, B.; Nguyen, H.T. Genome-wide analysis and expression profiling of the PIN auxin transporter gene family in soybean (Glycine max). BMC Genom. 2015, 16, 951. [Google Scholar] [CrossRef] [PubMed]
- Müller, A.; Guan, C.; Gälweiler, L.; Tänzler, P.; Huijser, P.; Marchant, A.; Parry, G.; Bennett, M.; Wisman, E.; Palme, K. AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J. 1998, 17, 6903–6911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahman, A.; Takahashi, M.; Shibasaki, K.; Wu, S.; Inaba, T.; Tsurumi, S.; Baskin, T.I. Gravitropism of Arabidopsis thaliana roots requires the polarization of PIN2 toward the root tip in meristematic cortical cells. Plant Cell 2010, 22, 1762–1776. [Google Scholar] [CrossRef] [PubMed]
- Friml, J.; Vieten, A.; Sauer, M.; Weijers, D.; Schwarz, H.; Hamann, T.; Offringa, R.; Jürgens, G. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 2003, 426, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Blilou, I.; Xu, J.; Wildwater, M.; Willemsen, V.; Paponov, I.; Friml, J.; Heidstra, R.; Aida, M.; Palme, K.; Scheres, B. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 2005, 433, 39–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grieneisen, V.A.; Xu, J.; Marée, A.F.M.; Hogeweg, P.; Scheres, B. Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 2007, 449, 1008–1013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gälweiler, L.; Guan, C.; Müller, A.; Wisman, E.; Mendgen, K.; Yephremov, A.; Palme, K. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 1998, 282, 2226–2230. [Google Scholar] [CrossRef]
- Friml, J.; Wiśniewska, J.; Benková, E.; Mendgen, K.; Palme, K. Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 2002, 415, 806–809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, K.-X.; Xu, H.-H.; Yuan, T.-T.; Zhang, L.; Lu, Y.-T. Blue-light-induced PIN3 polarization for root negative phototropic response in Arabidopsis. Plant J. 2013, 76, 308–321. [Google Scholar] [CrossRef]
- Zádníková, P.; Petrásek, J.; Marhavy, P.; Raz, V.; Vandenbussche, F.; Ding, Z.; Schwarzerová, K.; Morita, M.T.; Tasaka, M.; Hejátko, J.; et al. Role of PIN-mediated auxin efflux in apical hook development of Arabidopsis thaliana. Development 2010, 137, 607–617. [Google Scholar] [CrossRef] [PubMed]
- Nadzieja, M.; Kelly, S.; Stougaard, J.; Reid, D. Epidermal auxin biosynthesis facilitates rhizobial infection in Lotus japonicus. Plant J. 2018, 95, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Simon, S.; Skůpa, P.; Viaene, T.; Zwiewka, M.; Tejos, R.; Klíma, P.; Čarná, M.; Rolčík, J.; Rycke, R.D.; Moreno, I.; et al. PIN6 auxin transporter at endoplasmic reticulum and plasma membrane mediates auxin homeostasis and organogenesis in Arabidopsis. New Phytol. 2016, 211, 65–74. [Google Scholar] [CrossRef] [Green Version]
- Hagen, G.; Martin, G.; Li, Y.; Guilfoyle, T.J. Auxin-induced expression of the soybean GH3 promoter in transgenic tobacco plants. Plant Mol. Biol. 1991, 17, 567–579. [Google Scholar] [CrossRef]
- Singh, V.K.; Jain, M.; Garg, R. Genome-wide analysis and expression profiling suggest diverse roles of GH3 genes during development and abiotic stress responses in legumes. Front. Plant Sci. 2015, 5. [Google Scholar] [CrossRef] [Green Version]
- Tank, J.G.; Thaker, V.S. Cyclin dependent kinases and their role in regulation of plant cell cycle. Biol. Plant. 2011, 55, 201. [Google Scholar] [CrossRef]
- Boudolf, V.; Vlieghe, K.; Beemster, G.T.S.; Magyar, Z.; Torres Acosta, J.A.; Maes, S.; Van Der Schueren, E.; Inzé, D.; De Veylder, L. The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell 2004, 16, 2683–2692. [Google Scholar] [CrossRef] [PubMed]
- Vincent, J.M. A Manual for the Practical Study of Root-Nodule Bacteria; International Biological Programme; Blackwell Scientific: Hoboken, NJ, USA, 1970. [Google Scholar]
- Magrane, M.; Consortium, U. UniProt Knowledgebase: A hub of integrated protein data. Database (Oxford) 2011, 2011. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.-F.; Guindon, S.; Lefort, V.; Lescot, M.; et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008, 36, W465–W469. [Google Scholar] [CrossRef] [PubMed]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef] [Green Version]
- Untergasser, A.; Nijveen, H.; Rao, X.; Bisseling, T.; Geurts, R.; Leunissen, J.A.M. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007, 35, W71–W74. [Google Scholar] [CrossRef]
- Ramakers, C.; Ruijter, J.M.; Deprez, R.H.L.; Moorman, A.F.M. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003, 339, 62–66. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef]
- Karimi, M.; Inzé, D.; Depicker, A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002, 7, 193–195. [Google Scholar] [CrossRef]
- Hartley, J.L.; Temple, G.F.; Brasch, M.A. DNA Cloning Using In Vitro Site-Specific Recombination. Genome Res. 2000, 10, 1788–1795. [Google Scholar] [CrossRef] [Green Version]
- Díaz, C.L.; Grønlund, M.; Schlaman, H.R.M.; Spaink, H.P. Induction of hairy roots for symbiotic gene expression studies. In Lotus japonicus Handbook; Márquez, A.J., Ed.; Springer Netherlands: Dordrecht, The Netherlands, 2005; pp. 261–277. ISBN 978-1-4020-3735-1. [Google Scholar]
- Broda, B. Metody Histochemii Roślinnej [Methods in Plant Histochemistry]; PZWiL: Warsaw, Poland, 1971. [Google Scholar]






| L. japonicus Protein/Gene | A. thaliana Orthologous Sequences Identified Using BLAST Search | M. truncatula Orthologous Sequences Identified Using BLAST Search |
|---|---|---|
| LjPIN1 (Lj4g3v3114900.1, Lj2g3v0661480.1) | AtPIN1 (At1g73590) | MtPIN4 (MTR_6g069510) |
| LjPIN2 (Lj4g3v2139970.1) | AtPIN2 (At5g57090) | MtPIN2 (MTR_4g127100) |
| LjPIN3 (Lj0g3v0320849.2) | AtPIN3 (At1g70940) | MtPIN3 (MTR_1g030890) |
| LjPIN4 (Lj4g3v0633470.1) | AtPIN3 (At1g70940) | MtPIN1 (MTR_4g084870) |
| LjPIN5 (Lj1g3v2809230.1) | AtPIN5 (At5g16530) | MtPIN9 (MTR_7g079720) |
| LjPIN6: LjPIN6a (Lj0g3v0178829.1), LjPIN6b (Lj1g3v0264160.1) | AtPIN6 (At1g77110) | MtPIN6 (MTR_1g029190) |
| LjPIN7 (Lj1g3v4106960.1) | AtPIN1 (At1g73590) | MtPIN10 (MTR_7g089360) |
| LjPIN8 (Lj2g3v1034600.1) | AtPIN8 (At5g15100) | MtPIN11 (MTR_6g011400) |
| Nodule Developmental Stage | Expression Pattern of LjPIN Genes | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| LjPIN1 | LjPIN2 | LjPIN3 | LjPIN4 | LjPIN5 | LjPIN6 | LjPIN7 | LjPIN8 | ||
| Active initial primordia | − | + | + | + | w | + | + | + | |
| “Hidden” primordia (tissues not differentiated) | + | + | + | + | − | + | + | w | |
| “Emerged” primordia (tissues discernible, nondifferentiated) | Vascular bundle | + | + | − | + | + | + | + | w |
| Cortex | − | + | s | + | − | + | − | w | |
| Vascular bundle–stele connection | + | + | + | + | + | + | + | + | |
| Nodules 20 dpi (young, tissues differentiated) | Vascular bundle | + | − | − | s | + | + | s | − |
| Cortex | − | − | − | − | − | − | − | − | |
| Vascular bundle–stele connection | + | − | + | + | + | + | + | + | |
| Nodules 54 dpi (mature) | Vascular bundle | w | ws | − | w | − | s | − | − |
| Cortex | − | − | − | − | − | − | − | − | |
| Vascular bundle–stele connection | w | w | − | w | + | + | − | + | |
| Species | Parts of the Root Tip | ||||
|---|---|---|---|---|---|
| Procambium | Columella Initials-Procambial Initials Boundary | Periblem | Columella | Lateral Root Cap/Protoderm | |
| A. thaliana | AtPIN1, (AtPIN3, AtPIN4, AtPIN7) | AtPIN1, (AtPIN3, AtPIN4, AtPIN7) | AtPIN1, AtPIN2 | AtPIN1, (AtPIN3, AtPIN4, AtPIN7) | AtPIN1, AtPIN2 |
| J. japonicus | LjPIN1, LjPIN2, LjPIN3, LjPIN6 | LjPIN2, LjPIN3, LjPIN6 | LjPIN2, LjPIN3, LjPIN6 | LjPIN1, LjPIN2, LjPIN6 | LjPIN2, (LjPIN3, LjPIN4), LjPIN5, LjPIN6 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sańko-Sawczenko, I.; Dmitruk, D.; Łotocka, B.; Różańska, E.; Czarnocka, W. Expression Analysis of PIN Genes in Root Tips and Nodules of Lotus japonicus. Int. J. Mol. Sci. 2019, 20, 235. https://doi.org/10.3390/ijms20020235
Sańko-Sawczenko I, Dmitruk D, Łotocka B, Różańska E, Czarnocka W. Expression Analysis of PIN Genes in Root Tips and Nodules of Lotus japonicus. International Journal of Molecular Sciences. 2019; 20(2):235. https://doi.org/10.3390/ijms20020235
Chicago/Turabian StyleSańko-Sawczenko, Izabela, Dominika Dmitruk, Barbara Łotocka, Elżbieta Różańska, and Weronika Czarnocka. 2019. "Expression Analysis of PIN Genes in Root Tips and Nodules of Lotus japonicus" International Journal of Molecular Sciences 20, no. 2: 235. https://doi.org/10.3390/ijms20020235





