Prognostic Relevance of RIP140 and ERβ Expression in Unifocal Versus Multifocal Breast Cancers: A Preliminary Report
Abstract
:1. Introduction
2. Results
2.1. RIP140 and LCoR Expression in Unifocal vs. Multifocal Tumors
2.2. Correlation with Clinical and Biological Parameters
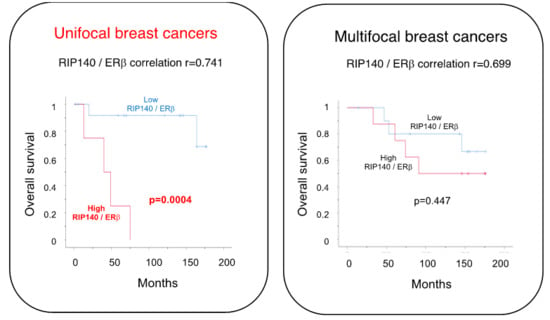
2.3. Correlation with Patient Survival
3. Discussion
4. Materials and Methods
4.1. Collective
4.2. Immunohistochemistry
4.3. Statistical Analyses
4.4. Survival Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BC | breast cancer |
| DFS | disease free survival |
| DCIS | ductal carcinoma in situ |
| ER | estrogen receptor |
| HER2 | human epidermal growth factor receptor 2 |
| IRS | immunoreactive score |
| LCoR | ligand dependent corepressor |
| MUC-1 | epithelial mucin-1 |
| NST | non-special type |
| pN | lymph node involvement |
| LMU | Ludwig Maximilians University |
| M | Metastasis |
| NMRI | nuclear magnetic resonance imaging |
| NR | nuclear receptor |
| OS | overall survival |
| PBS | phosphate buffered saline |
| pT | primary tumor size |
| PR | progesterone receptor |
| RFS | recurrence free survival |
| ROC-curve | receiver operating characteristic curve |
| RIP140 | receptor interacting protein of 140 kDa |
| TNM status | tumor node metastasis status |
References
- Cserni, G.; Bori, R.; Sejben, I.; Vörös, A.; Kaiser, L.; Hamar, S.; Csörgő, E.; Kulka, J. Unifocal, multifocal and diffuse carcinomas: A reproducibility study of breast cancer distribution. Breast 2013, 22, 34–38. [Google Scholar] [CrossRef] [PubMed]
- Tot, T.; Pekár, G. Multifocality in “basal-like” breast carcinomas and its influence on lymph node status. Ann. Surg. Oncol. 2011, 18, 1671–1677. [Google Scholar] [CrossRef] [PubMed]
- Lynch, S.P.; Lei, X.; Hsu, L.; Meric-Bernstam, F.; Buchholz, T.A.; Zhang, H.; Hortobágyi, G.N.; Gonzalez-Angulo, A.M.; Valero, V. Breast cancer multifocality and multicentricity and locoregional recurrence. Oncologist 2013, 18, 1167–1173. [Google Scholar] [CrossRef] [PubMed]
- Salgado, R.; Aftimos, P.; Sotiriou, C.; Desmedt, C. Evolving paradigms in multifocal breast cancer. Semin. Cancer Biol. 2015, 31, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Manavathi, B.; Dey, O.; Gajulapalli, V.N.R.; Bhatia, R.S.; Bugide, S.; Kumar, R. Derailed estrogen signaling and breast cancer: An authentic couple. Endocr. Rev. 2013, 34, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Lang, Z.; Wu, Y.; Li, C.; Li, X.; Wang, X.; Qu, G. Multifocal and Multicentric Breast Carcinoma: A Significantly More Aggressive Tumor than Unifocal Breast Cancer. Anticancer Res. 2017, 37, 4593–4598. [Google Scholar] [PubMed]
- Vera-Badillo, F.E.; Napoleone, M.; Ocana, A.; Templeton, A.J.; Seruga, B.; Al-Mubarak, M.; AlHashem, H.; Tannock, I.F.; Amir, E. Effect of multifocality and multicentricity on outcome in early stage breast cancer: A systematic review and meta-analysis. Breast Cancer Res. Treat. 2014, 146, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Boros, M.; Voidazan, S.; Moldovan, C.; Georgescu, R.; Toganel, C.; Moncea, D.; Molnar, C.V.; Podoleanu, C.; Eniu, A.; Stolnicu, S. Clinical implications of multifocality as a prognostic factor in breast carcinoma—A multivariate analysis study comprising 460 cases. Int. J. Clin. Exp. Med. 2015, 8, 9839–9846. [Google Scholar] [PubMed]
- Weissenbacher, T.M.; Zschage, M.; Janni, W.; Jeschke, U.; Dimpfl, T.; Mayr, D.; Rack, B.; Schindlbeck, C.; Friese, K.; Dian, D. Multicentric and multifocal versus unifocal breast cancer: Is the tumor-node-metastasis classification justified? Breast Cancer Res. Treat. 2010, 122, 27–34. [Google Scholar] [CrossRef]
- Weissenbacher, T.; Hirte, E.; Kuhn, C.; Janni, W.; Mayr, D.; Karsten, U.; Rack, B.; Friese, K.; Jeschke, U.; Heublein, S.; et al. Multicentric and multifocal versus unifocal breast cancer: Differences in the expression of E-cadherin suggest differences in tumor biology. BMC Cancer 2013, 13, 361. [Google Scholar] [CrossRef]
- Park, B.-W.; Kim, K.-S.; Heo, M.-K.; Ko, S.-S.; Hong, S.W.; Yang, W.-I.; Kim, J.-H.; Kim, G.E.; Lee, K.S. Expression of estrogen receptor-beta in normal mammary and tumor tissues: Is it protective in breast carcinogenesis? Breast Cancer Res. Treat. 2003, 80, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Park, B.-W.; Kim, K.-S.; Heo, M.-K.; Yang, W.-I.; Kim, S.I.; Kim, J.-H.; Kim, G.E.; Lee, K.S. The changes of estrogen receptor-beta variants expression in breast carcinogenesis: Decrease of estrogen receptor-beta2 expression is the key event in breast cancer development. J. Surg. Oncol. 2006, 93, 504–510. [Google Scholar] [CrossRef] [PubMed]
- Doan, T.B.; Eriksson, N.A.; Graham, D.; Funder, J.W.; Simpson, E.R.; Kuczek, E.S.; Clyne, C.; Leedman, P.J.; Tilley, W.D.; Fuller, P.J.; et al. Breast cancer prognosis predicted by nuclear receptor-coregulator networks. Mol. Oncol. 2014, 8, 998–1013. [Google Scholar] [CrossRef] [Green Version]
- Cavaillès, V.; Dauvois, S.; L’Horset, F.; Lopez, G.; Hoare, S.; Kushner, P.J.; Parker, M.G. Nuclear factor RIP140 modulates transcriptional activation by the estrogen receptor. EMBO J. 1995, 14, 3741–3751. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, I.; Flores, A.M.; Aneskievich, B.J. Corepressors of agonist-bound nuclear receptors. Toxicol. Appl. Pharmacol. 2007, 223, 288–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Docquier, A.; Harmand, P.-O.; Fritsch, S.; Chanrion, M.; Darbon, J.-M.; Cavaillès, V. The transcriptional coregulator RIP140 represses E2F1 activity and discriminates breast cancer subtypes. Clin. Cancer Res. 2010, 16, 2959–2970. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.-J.; Peng, R.-J.; Kuang, B.-H.; Yuan, Z.-Y.; Qin, T.; Liu, W.-S.; Guo, Y.-M.; Han, H.-Q.; Lian, Y.-F.; Deng, C.-C.; et al. NOP14 suppresses breast cancer progression by inhibiting NRIP1/Wnt/β-catenin pathway. Oncotarget 2015, 6, 25701–25714. [Google Scholar] [CrossRef] [Green Version]
- Lapierre, M.; Docquier, A.; Castet-Nicolas, A.; Gitenay, D.; Jalaguier, S.; Teyssier, C.; Cavaillès, V. The emerging role of the transcriptional coregulator RIP140 in solid tumors. Biochim. Biophys. Acta 2015, 1856, 144–150. [Google Scholar] [CrossRef]
- Augereau, P.; Badia, E.; Carascossa, S.; Castet, A.; Fritsch, S.; Harmand, P.-O.; Jalaguier, S.; Cavaillès, V. The nuclear receptor transcriptional coregulator RIP140. Nucl. Recept. Signal. 2006, 4, e024. [Google Scholar] [CrossRef] [PubMed]
- Docquier, A.; Garcia, A.; Savatier, J.; Boulahtouf, A.; Bonnet, S.; Bellet, V.; Busson, M.; Margeat, E.; Jalaguier, S.; Royer, C.; et al. Negative regulation of estrogen signaling by ERβ and RIP140 in ovarian cancer cells. Mol. Endocrinol. 2013, 27, 1429–1441. [Google Scholar] [CrossRef] [PubMed]
- Ruddy, S.C.; Lau, R.; Cabrita, M.A.; McGregor, C.; McKay, B.C.; Murphy, L.C.; Wright, J.S.; Durst, T.; Pratt, M.A.C. Preferential estrogen receptor β ligands reduce Bcl-2 expression in hormone-resistant breast cancer cells to increase autophagy. Mol. Cancer Ther. 2014, 13, 1882–1893. [Google Scholar] [CrossRef] [PubMed]
- Madak-Erdogan, Z.; Charn, T.-H.; Jiang, Y.; Liu, E.T.; Katzenellenbogen, J.A.; Katzenellenbogen, B.S. Integrative genomics of gene and metabolic regulation by estrogen receptors α and β, and their coregulators. Mol. Syst. Biol. 2013, 9, 676. [Google Scholar] [CrossRef] [PubMed]
- Nautiyal, J.; Steel, J.H.; Mane, M.R.; Oduwole, O.; Poliandri, A.; Alexi, X.; Wood, N.; Poutanen, M.; Zwart, W.; Stingl, J.; et al. The transcriptional co-factor RIP140 regulates mammary gland development by promoting the generation of key mitogenic signals. Development 2013, 140, 1079–1089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandes, I.; Bastien, Y.; Wai, T.; Nygard, K.; Lin, R.; Cormier, O.; Lee, H.S.; Eng, F.; Bertos, N.R.; Pelletier, N.; et al. Ligand-dependent nuclear receptor corepressor LCoR functions by histone deacetylase-dependent and -independent mechanisms. Mol. Cell 2003, 11, 139–150. [Google Scholar] [CrossRef]
- Song, Y.; Shan, S.; Zhang, Y.; Liu, W.; Ding, W.; Ren, W.; Xia, H.; Li, X.; Zhang, Q.; Zhao, L.; et al. Ligand-dependent corepressor acts as a novel corepressor of thyroid hormone receptor and represses hepatic lipogenesis in mice. J. Hepatol. 2012, 56, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Asim, M.; Hafeez, B.B.; Siddiqui, I.A.; Gerlach, C.; Patz, M.; Mukhtar, H.; Baniahmad, A. Ligand-dependent corepressor acts as a novel androgen receptor corepressor, inhibits prostate cancer growth, and is functionally inactivated by the Src protein kinase. J. Biol. Chem. 2011, 286, 37108–37117. [Google Scholar] [CrossRef] [PubMed]
- Jalaguier, S.; Teyssier, C.; Nait Achour, T.; Lucas, A.; Bonnet, S.; Rodriguez, C.; Elarouci, N.; Lapierre, M.; Cavaillès, V. Complex regulation of LCoR signaling in breast cancer cells. Oncogene 2017, 36, 4790–4801. [Google Scholar] [CrossRef]
- Sixou, S.; Müller, K.; Jalaguier, S.; Kuhn, C.; Harbeck, N.; Mayr, D.; Engel, J.; Jeschke, U.; Ditsch, N.; Cavaillès, V. Importance of RIP140 and LCoR Sub-Cellular Localization for Their Association With Breast Cancer Aggressiveness and Patient Survival. Transl. Oncol. 2018, 11, 1090–1096. [Google Scholar] [CrossRef]
- Shaaban, A.M.; Green, A.R.; Karthik, S.; Alizadeh, Y.; Hughes, T.A.; Harkins, L.; Ellis, I.O.; Robertson, J.F.; Paish, E.C.; Saunders, P.T.K.; et al. Nuclear and cytoplasmic expression of ERbeta1, ERbeta2, and ERbeta5 identifies distinct prognostic outcome for breast cancer patients. Clin. Cancer Res. 2008, 14, 5228–5235. [Google Scholar] [CrossRef]
- Yan, M.; Rayoo, M.; Takano, E.A.; kConFab Investigators; Fox, S.B. Nuclear and cytoplasmic expressions of ERβ1 and ERβ2 are predictive of response to therapy and alters prognosis in familial breast cancers. Breast Cancer Res. Treat. 2011, 126, 395–405. [Google Scholar] [CrossRef]
- Shanle, E.K.; Onitilo, A.A.; Huang, W.; Kim, K.; Zang, C.; Engel, J.M.; Xu, W.; Wisinski, K.B. Prognostic significance of full-length estrogen receptor beta expression in stage I-III triple negative breast cancer. Am J. Transl. Res. 2015, 7, 1246–1259. [Google Scholar] [PubMed]
- Dall, G.V.; Hawthorne, S.; Seyed-Razavi, Y.; Vieusseux, J.; Wu, W.; Gustafsson, J.-A.; Byrne, D.; Murphy, L.; Risbridger, G.P.; Britt, K.L. Estrogen receptor subtypes dictate the proliferative nature of the mammary gland. J. Endocrinol. 2018, 237, 323–336. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.-J.; Lee, A.; Choi, Y.-J.; Song, B.J.; Yim, H.W.; Kang, C.S. Prognostic Significance of High Expression of ER-beta in Surgically Treated ER-Positive Breast Cancer Following Endocrine Therapy. J Breast Cancer 2012, 15, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Zhu, Q.; Yilamu, D.; Jakulin, A.; Liu, S.; Liang, T. Expression and prognostic value of estrogen receptor beta in breast cancer patients. Int. J. Clin. Exp. Med. 2014, 7, 3730–3736. [Google Scholar] [PubMed]
- Tan, W.; Li, Q.; Chen, K.; Su, F.; Song, E.; Gong, C. Estrogen receptor beta as a prognostic factor in breast cancer patients: A systematic review and meta-analysis. Oncotarget 2016, 7, 10373–10385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elston, E.W.; Ellis, I.O. Method for grading breast cancer. J. Clin. Pathol. 1993, 46, 189–190. [Google Scholar] [CrossRef] [PubMed]
- Mylonas, I.; Makovitzky, J.; Jeschke, U.; Briese, V.; Friese, K.; Gerber, B. Expression of Her2/neu, steroid receptors (ER and PR), Ki67 and p53 in invasive mammary ductal carcinoma associated with ductal carcinoma In Situ (DCIS) Versus invasive breast cancer alone. Anticancer Res. 2005, 25, 1719–1723. [Google Scholar]
- Bock, C.; Kuhn, C.; Ditsch, N.; Krebold, R.; Heublein, S.; Mayr, D.; Doisneau-Sixou, S.; Jeschke, U. Strong correlation between N-cadherin and CD133 in breast cancer: Role of both markers in metastatic events. J. Cancer Res. Clin. Oncol. 2014, 140, 1873–1881. [Google Scholar] [CrossRef]



| Parameters | Unifocal (n = 21) | Multifocal (n = 21) |
|---|---|---|
| Age | ||
| Mean (years) | 58.5 | 63.7 |
| Histological type | ||
| Ductal | 13 (61.9%) | 16 (71.4%) |
| Lobular | 3 (14.3%) | 4 (19%) |
| Ductal-Lobular | 1 (4.8%) | 2 (9.5%) |
| Medullary | 1 (4.8%) | 0 (0%) |
| Micropapillary | 2 (9.5%) | 0 (0%) |
| Unknown | 1 (4.8%) | 0 (0%) |
| Tumor size | ||
| pT1 a, b, c | 18 (85.7%) | 18 (85.7%) |
| pT2 | 2 (9.5%) | 2 (9.5%) |
| pT3 | 0 (0%) | 0 (0%) |
| pT4a, b, c, d | 1 (4.8%) | 1 (4.8%) |
| Grade | ||
| I | 0 (0%) | 0 (0%) |
| II | 17 (81.0%) | 17 (81.0%) |
| III | 4 (19.0%) | 4 (19.0%) |
| Lymph node metastasis | ||
| No | 16 (76.2%) | 16 (76.2%) |
| Yes | 4 (19%) | 4 (19%) |
| Unknown | 1 (4.8%) | 1 (4.8%) |
| Local recurrence | ||
| No | 13 (61.9%) | 17 (81%) |
| Yes | 8 (38.1%) | 4 (19%) |
| Overall survival | ||
| Mean time (months) | 130.79 | 133.41 |
| ERα status | ||
| Negative | 5 (23.8%) | 5 (23.8%) |
| Positive | 16 (76.2%) | 16 (76.2%) |
| ERβ status | ||
| Negative | 10 (47.6%) | 5 (23.8%) |
| Positive | 11 (52.4%) | 16 (76.2%) |
| HER2 status | ||
| Negative | 18 (85.7%) | 16 (76.2%) |
| Positive | 3 (14.3%) | 5 (23.8%) |
| Parameters | Unifocal (n = 21) | Multifocal (n = 21) |
|---|---|---|
| RIP140 expression | ||
| Mean IRS ± SE | 2.61 ± 1.74 | 2.98 +/− 2.12 |
| Low expressing tumors n (%) | 16 (76.2%) | 12 (57.1%) |
| High expressing tumors n (%) | 5 (23.8%) | 9 (42.9%) |
| LCoR expression | ||
| Mean IRS ± SE | 2.38 * ± 1.49 | 3.38 ± 1.93 |
| Low expressing tumors n (%) | 17 * (80.1%) | 9 (42.9%) |
| High expressing tumors n (%) | 4 * (19%) | 12 (57.1%) |
| Correlation between RIP140 and LCoR | ||
| Spearman’s Rho correlation coefficient | 0.714 ** | 0.686 ** |
| RIP140 | LCoR | |||
|---|---|---|---|---|
| Unifocal | Multifocal | Unifocal | Multifocal | |
| Recurrence Status | 0.368 | −0.01 | 0.353 | 0.091 |
| pT | −0.157 | 0.083 | 0.017 | 0.091 |
| pN | −0.204 | −0.054 | 0.018 | −0.096 |
| pM | −0.257 | 0.159 | −0.34 | 0.011 |
| Grade | −0.152 | −0.01 | 0.071 | 0.081 |
| Histology | 0.034 | 0.124 | −0.12 | 0.004 |
| ERα | 0.012 | 0.126 | 0.181 | −0.212 |
| ERβ | 0.741 ** | 0.699 ** | 0.783 ** | 0.612 ** |
| HER2 | 0.397 | 0.196 | 0.239 | 0.309 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Müller, K.; Sixou, S.; Kuhn, C.; Jalaguier, S.; Mayr, D.; Ditsch, N.; Weissenbacher, T.; Harbeck, N.; Mahner, S.; Cavaillès, V.; et al. Prognostic Relevance of RIP140 and ERβ Expression in Unifocal Versus Multifocal Breast Cancers: A Preliminary Report. Int. J. Mol. Sci. 2019, 20, 418. https://doi.org/10.3390/ijms20020418
Müller K, Sixou S, Kuhn C, Jalaguier S, Mayr D, Ditsch N, Weissenbacher T, Harbeck N, Mahner S, Cavaillès V, et al. Prognostic Relevance of RIP140 and ERβ Expression in Unifocal Versus Multifocal Breast Cancers: A Preliminary Report. International Journal of Molecular Sciences. 2019; 20(2):418. https://doi.org/10.3390/ijms20020418
Chicago/Turabian StyleMüller, Katharina, Sophie Sixou, Christina Kuhn, Stephan Jalaguier, Doris Mayr, Nina Ditsch, Tobias Weissenbacher, Nadia Harbeck, Sven Mahner, Vincent Cavaillès, and et al. 2019. "Prognostic Relevance of RIP140 and ERβ Expression in Unifocal Versus Multifocal Breast Cancers: A Preliminary Report" International Journal of Molecular Sciences 20, no. 2: 418. https://doi.org/10.3390/ijms20020418