Computer-Aided Strategies for Determining the Amino Acid Composition of Medium for Chinese Hamster Ovary Cell-Based Biomanufacturing Platforms
Abstract
:1. Introduction
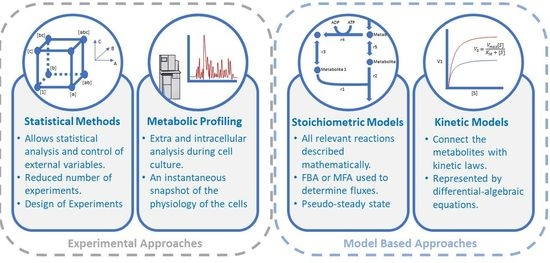
2. Brief Overview of the Established Practice in Determining the Amino Acid Composition of the CHO Cell Medium
2.1. Statistical Methods: Utilizing Data-Driven Models in Experimental Design
2.2. Metabolic Profiling
2.3. Modification of the Amino Acid Transporters
3. Mechanistic Insight by Model-Assisted CHO Cell Medium Development and Optimization
3.1. Kinetic Models to Assist CHO Cell Upstream Bioprocess Development and Control
3.2. Stoichiometry-Based Modelling to Explore Amino Acid Requirements for Medium Development and Bioprocess Design
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Gils, A.; Bertolotto, A.; Mulleman, D.; Bejan-Angoulvant, T.; Declerck, P.J. Biopharmaceuticals. Ther. Drug Monit. 2017, 39, 308–315. [Google Scholar] [CrossRef] [PubMed]
- Walsh, G. Biopharmaceutical benchmarks 2018. Nat. Biotechnol. 2018, 36, 1136–1145. [Google Scholar] [CrossRef] [PubMed]
- Carrillo-Cocom, L.M.; Genel-Rey, T.; Araíz-Hernández, D.; López-Pacheco, F.; López-Meza, J.; Rocha-Pizaña, M.R.; Ramírez-Medrano, A.; Alvarez, M.M. Amino acid consumption in naïve and recombinant CHO cell cultures: Producers of a monoclonal antibody. Cytotechnology 2015, 67, 809–820. [Google Scholar] [CrossRef] [PubMed]
- Selvarasu, S.; Ho, Y.S.; Chong, W.P.K.; Wong, N.S.C.; Yusufi, F.N.K.; Lee, Y.Y.; Yap, M.G.S.; Lee, D.-Y. Combined in silico modeling and metabolomics analysis to characterize fed-batch CHO cell culture. Biotechnol. Bioeng. 2012, 109, 1415–1429. [Google Scholar] [CrossRef] [PubMed]
- Mulukutla, B.C.; Mitchell, J.; Geoffroy, P.; Harrington, C.; Krishnan, M.; Kalomeris, T.; Morris, C.; Zhang, L.; Pegman, P.; Hiller, G.W. Metabolic engineering of Chinese hamster ovary cells towards reduced biosynthesis and accumulation of novel growth inhibitors in fed-batch cultures. Metab. Eng. 2019, 54, 54–68. [Google Scholar] [CrossRef] [PubMed]
- Pereira, S.; Kildegaard, H.F.; Andersen, M.R. Impact of CHO Metabolism on Cell Growth and Protein Production: An Overview of Toxic and Inhibiting Metabolites and Nutrients. Biotechnol. J. 2018, 13, 1700499. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Jimenez Del Val, I.; Müller, C.; Wagtberg Sen, J.; Rasmussen, S.K.; Kontoravdi, C.; Weilguny, D.; Andersen, M.R. Amino acid and glucose metabolism in fed-batch CHO cell culture affects antibody production and glycosylation. Biotechnol. Bioeng. 2015, 112, 521–535. [Google Scholar] [CrossRef]
- Salazar, A.; Keusgen, M.; von Hagen, J. Amino acids in the cultivation of mammalian cells. Amino Acids 2016, 48, 1161–1171. [Google Scholar] [CrossRef] [Green Version]
- Ritacco, F.V.; Wu, Y.; Khetan, A. Cell culture media for recombinant protein expression in Chinese hamster ovary (CHO) cells: History, key components, and optimization strategies. Biotechnol. Prog. 2018, 34, 1407–1426. [Google Scholar] [CrossRef]
- Fox, S.R.; Patel, U.A.; Yap, M.G.S.; Wang, D.I.C. Maximizing Interferon-γ Production by Chinese Hamster Ovary Cells through Temperature Shift Optimization: Experimental and Modeling. Biotechnol. Bioeng. 2004, 85, 177–184. [Google Scholar] [CrossRef]
- Badsha, M.B.; Kurata, H.; Onitsuka, M.; Oga, T.; Omasa, T. Metabolic analysis of antibody producing Chinese hamster ovary cell culture under different stresses conditions. J. Biosci. Bioeng. 2016, 122, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Stamatis, C.; Goldrick, S.; Gruber, D.; Turner, R.; Titchener-Hooker, N.J.; Farid, S.S. High throughput process development workflow with advanced decision-support for antibody purification. J. Chromatogr. A 2019, 1596, 104–116. [Google Scholar] [CrossRef] [PubMed]
- Pollock, J.; Bolton, G.; Coffman, J.; Ho, S.V.; Bracewell, D.G.; Farid, S.S. Optimising the design and operation of semi-continuous affinity chromatography for clinical and commercial manufacture. J. Chromatogr. A 2013, 1284, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Shirahata, H.; Diab, S.; Sugiyama, H.; Gerogiorgis, D.I. Dynamic modelling, simulation and economic evaluation of two CHO cell-based production modes towards developing biopharmaceutical manufacturing processes. Chem. Eng. Res. Des. 2019, 150, 218–233. [Google Scholar] [CrossRef]
- Bunnak, P.; Allmendinger, R.; Ramasamy, S.V.; Lettieri, P.; Titchener-Hooker, N.J. Life-cycle and cost of goods assessment of fed-batch and perfusion-based manufacturing processes for mAbs. Biotechnol. Prog. 2016, 32, 1324–1335. [Google Scholar] [CrossRef]
- Bielser, J.M.; Wolf, M.; Souquet, J.; Broly, H.; Morbidelli, M. Perfusion mammalian cell culture for recombinant protein manufacturing—A critical review. Biotechnol. Adv. 2018, 36, 1328–1340. [Google Scholar] [CrossRef]
- Gronemeyer, P.; Ditz, R.; Strube, J. Trends in Upstream and Downstream Process Development for Antibody Manufacturing. Bioengineering 2014, 1, 188–212. [Google Scholar] [CrossRef] [Green Version]
- Dikicioglu, D.; Kırdar, B.; Oliver, S.G. Biomass composition: The “elephant in the room” of metabolic modelling. Metabolomics 2015, 11, 1690–1701. [Google Scholar] [CrossRef]
- Cankorur-Cetinkaya, A.; Dikicioglu, D.; Oliver, S.G. Metabolic modeling to identify engineering targets for Komagataella phaffii: The effect of biomass composition on gene target identification. Biotechnol. Bioeng. 2017, 114, 2605–2615. [Google Scholar] [CrossRef]
- Savizi, I.S.P.; Soudi, T.; Shojaosadati, S.A. Systems biology approach in the formulation of chemically defined media for recombinant protein overproduction. Appl. Microbiol. Biotechnol. 2019, 103, 8315–8326. [Google Scholar] [CrossRef]
- Altamirano, C.; Paredes, C.; Cairo, J.J.; Godia, F. Improvement of CHO Cell Culture Medium Formulation: Simultaneous Substitution of Glucose and Glutamine. Biotechnol. Prog. 2000, 16, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Sun, X.; Zhang, Y. Insight into metabolism of CHO cells at low glucose concentration on the basis of the determination of intracellular metabolites. Process Biochem. 2005, 40, 1917–1921. [Google Scholar] [CrossRef]
- Tsao, Y.-S.; Cardoso, A.G.; Condon, R.G.G.; Voloch, M.; Lio, P.; Lagos, J.C.; Kearns, B.G.; Liu, Z. Monitoring Chinese hamster ovary cell culture by the analysis of glucose and lactate metabolism. J. Biotechnol. 2005, 118, 316–327. [Google Scholar] [CrossRef] [PubMed]
- Wilkens, C.A.; Altamirano, C.; Gerdtzen, Z.P. Comparative metabolic analysis of lactate for CHO cells in glucose and galactose. Biotechnol. Bioprocess Eng. 2011, 16, 714–724. [Google Scholar] [CrossRef]
- Cankorur-Cetinkaya, A.; Dias, J.M.L.; Kludas, J.; Slater, N.K.H.; Rousu, J.; Oliver, S.G.; Dikicioglu, D. CamOptimus: A tool for exploiting complex adaptive evolution to optimize experiments and processes in biotechnology. Microbiology 2017, 163, 829–839. [Google Scholar] [CrossRef] [PubMed]
- Parampalli, A.; Eskridge, K.; Smith, L.; Meagher, M.M.; Mowry, M.C.; Subramanian, A. Developement of serum-free media in CHO-DG44 cells using a central composite statistical design. Cytotechnology 2007, 54, 57–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González-Leal, I.J.; Carrillo-Cocom, L.M.; Ramírez-Medrano, A.; López-Pacheco, F.; Bulnes-Abundis, D.; Webb-Vargas, Y.; Alvarez, M.M. Use of a Plackett-Burman statistical design to determine the effect of selected amino acids on monoclonal antibody production in CHO cells. Biotechnol. Prog. 2011, 27, 1709–1717. [Google Scholar] [CrossRef]
- Torkashvand, F.; Vaziri, B.; Maleknia, S.; Heydari, A.; Vossoughi, M.; Davami, F.; Mahboudi, F. Designed Amino Acid Feed in Improvement of Production and Quality Targets of a Therapeutic Monoclonal Antibody. PLoS ONE 2015, 10, e0140597. [Google Scholar] [CrossRef]
- Yu, M.; Hu, Z.; Pacis, E.; Vijayasankaran, N.; Shen, A.; Li, F. Understanding the intracellular effect of enhanced nutrient feeding toward high titer antibody production process. Biotechnol. Bioeng. 2011, 108, 1078–1088. [Google Scholar] [CrossRef]
- Read, E.K.; Bradley, S.A.; Smitka, T.A.; Agarabi, C.D.; Lute, S.C.; Brorson, K.A. Fermentanomics informed amino acid supplementation of an antibody producing mammalian cell culture. Biotechnol. Prog. 2013, 29, 745–753. [Google Scholar] [CrossRef]
- Sun, Y.-T.; Zhao, L.; Liu, X.-P.; Hu, D.; Fan, L.; Tan, W.-S. Application of improved top-down approach in maximizing CHO cell mass and productivity in fed-batch culture. J. Chem. Technol. Biotechnol. 2013, 88, 1237–1247. [Google Scholar] [CrossRef]
- Kandasamy, P.; Gyimesi, G.; Kanai, Y.; Hediger, M.A. Amino acid transporters revisited: New views in health and disease. Trends Biochem. Sci. 2018, 43, 752–789. [Google Scholar] [CrossRef] [PubMed]
- Geoghegan, D.; Arnall, C.; Hatton, D.; Noble-Longster, J.; Sellick, C.; Senussi, T.; James, D.C. Control of amino acid transport into Chinese hamster ovary cells. Biotechnol. Bioeng. 2018, 115, 2908–2929. [Google Scholar] [CrossRef] [PubMed]
- Kyriakopoulos, S.; Polizzi, K.M.; Kontoravdi, C. Comparative analysis of amino acid metabolism and transport in CHO variants with different levels of productivity. J. Biotechnol. 2013, 168, 543–551. [Google Scholar] [CrossRef] [PubMed]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selişteanu, D.; Șendrescu, D.; Georgeanu, V.; Roman, M. Mammalian Cell Culture Process for Monoclonal Antibody Production: Nonlinear Modelling and Parameter Estimation. Biomed. Res. Int. 2015, 2015, 598721. [Google Scholar] [CrossRef]
- Hunter, M.; Yuan, P.; Vavilala, D.; Fox, M. Optimization of Protein Expression in Mammalian Cells. Curr. Protoc. Protein Sci. 2019, 95, e77. [Google Scholar] [CrossRef]
- Sha, S.; Huang, Z.; Wang, Z.; Yoon, S. Mechanistic modeling and applications for CHO cell culture development and production. Curr. Opin. Chem. Eng. 2018, 22, 54–61. [Google Scholar] [CrossRef]
- Galleguillos, S.N.; Ruckerbauer, D.; Gerstl, M.P.; Borth, N.; Hanscho, M.; Zanghellini, J. What can mathematical modelling say about CHO metabolism and protein glycosylation? Comput. Struct. Biotechnol. J. 2017, 15, 212–221. [Google Scholar] [CrossRef]
- Smallbone, K.; Messiha, H.L.; Carroll, K.M.; Winder, C.L.; Malys, N.; Dunn, W.B.; Murabito, E.; Swainston, N.; Dada, J.O.; Khan, F.; et al. A model of yeast glycolysis based on a consistent kinetic characterisation of all its enzymes. FEBS Lett. 2013, 587, 2832–2841. [Google Scholar] [CrossRef]
- López-Meza, J.; Araíz-Hernández, D.; Carrillo-Cocom, L.M.; López-Pacheco, F.; del Refugio Rocha-Pizaña, M.; Alvarez, M.M. Using simple models to describe the kinetics of growth, glucose consumption, and monoclonal antibody formation in naive and infliximab producer CHO cells. Cytotechnology 2016, 68, 1287–1300. [Google Scholar] [CrossRef] [PubMed]
- Kontoravdi, C.; Wong, D.; Lam, C.; Lee, Y.Y.; Yap, M.G.S.; Pistikopoulos, E.N.; Mantalaris, A. Modeling Amino Acid Metabolism in Mammalian Cells-Toward the Development of a Model Library. Biotechnol. Prog. 2007, 23, 1261–1269. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-H.; Bi, J.-X.; Zeng, A.-P.; Yuan, J.-Q. A simple kinetic model for myeloma cell culture with consideration of lysine limitation. Bioprocess Biosyst. Eng. 2008, 31, 569–577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Craven, S.; Shirsat, N.; Whelan, J.; Glennon, B. Process model comparison and transferability across bioreactor scales and modes of operation for a mammalian cell bioprocess. Biotechnol. Prog. 2013, 29, 186–196. [Google Scholar] [CrossRef] [PubMed]
- Poli, R.; Kennedy, J.; Blackwell, T. Particle swarm optimization. Swarm Intell. 2007, 1, 33–57. [Google Scholar] [CrossRef]
- Robitaille, J.; Chen, J.; Jolicoeur, M. A Single Dynamic Metabolic Model Can Describe mAb Producing CHO Cell Batch and Fed-Batch Cultures on Different Culture Media. PLoS ONE 2015, 10, e0136815. [Google Scholar] [CrossRef] [PubMed]
- Simon, L.; Karim, M.N. Modeling and control of amino acid starvation-induced apoptosis in CHO cell cultures. In Proceedings of the 2002 American Control Conference (IEEE Cat. No.CH37301), Anchorage, AK, USA, 8–10 May 2002; Volume 2, pp. 1579–1584. [Google Scholar]
- Almquist, J.; Cvijovic, M.; Hatzimanikatis, V.; Nielsen, J.; Jirstrand, M. Kinetic models in industrial biotechnology—Improving cell factory performance. Metab. Eng. 2014, 24, 38–60. [Google Scholar] [CrossRef]
- Orth, J.D.; Thiele, I.; Palsson, B.Ø. What is flux balance analysis? Nat. Biotechnol. 2010, 28, 245–248. [Google Scholar] [CrossRef]
- Basler, G.; Fernie, A.R.; Nikoloski, Z. Advances in metabolic flux analysis toward genome-scale profiling of higher organisms. Biosci. Rep. 2018, 38. [Google Scholar] [CrossRef]
- Quek, L.-E.; Dietmair, S.; Krömer, J.O.; Nielsen, L.K. Metabolic flux analysis in mammalian cell culture. Metab. Eng. 2010, 12, 161–171. [Google Scholar] [CrossRef]
- Sengupta, N.; Rose, S.T.; Morgan, J.A. Metabolic flux analysis of CHO cell metabolism in the late non-growth phase. Biotechnol. Bioeng. 2011, 108, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Niu, H.; Amribt, Z.; Fickers, P.; Tan, W.; Bogaerts, P. Metabolic pathway analysis and reduction for mammalian cell cultures—Towards macroscopic modeling. Chem. Eng. Sci. 2013, 102, 461–473. [Google Scholar] [CrossRef]
- Ahn, W.S.; Antoniewicz, M.R. Metabolic flux analysis of CHO cells at growth and non-growth phases using isotopic tracers and mass spectrometry. Metab. Eng. 2011, 13, 598–609. [Google Scholar] [CrossRef] [PubMed]
- Sheikholeslami, Z.; Jolicoeur, M.; Henry, O. Probing the metabolism of an inducible mammalian expression system using extracellular isotopomer analysis. J. Biotechnol. 2013, 164, 469–478. [Google Scholar] [CrossRef] [PubMed]
- Sheikholeslami, Z.; Jolicoeur, M.; Henry, O. Elucidating the effects of postinduction glutamine feeding on the growth and productivity of CHO cells. Biotechnol. Prog. 2014, 30, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Naderi, S.; Meshram, M.; Wei, C.; McConkey, B.; Ingalls, B.; Budman, H.; Scharer, J. Development of a mathematical model for evaluating the dynamics of normal and apoptotic Chinese hamster ovary cells. Biotechnol. Prog. 2011, 27, 1197–1205. [Google Scholar] [CrossRef]
- Wahrheit, J.; Nicolae, A.; Heinzle, E. Dynamics of growth and metabolism controlled by glutamine availability in Chinese hamster ovary cells. Appl. Microbiol. Biotechnol. 2014, 98, 1771–1783. [Google Scholar] [CrossRef]
- Xing, Z.; Kenty, B.; Koyrakh, I.; Borys, M.; Pan, S.-H.; Li, Z.J. Optimizing amino acid composition of CHO cell culture media for a fusion protein production. Process Biochem. 2011, 46, 1423–1429. [Google Scholar] [CrossRef]
- Hagrot, E.; Oddsdóttir, H.Æ.; Mäkinen, M.; Forsgren, A.; Chotteau, V. Novel column generation-based optimization approach for poly-pathway kinetic model applied to CHO cell culture. Metab. Eng. Commun. 2019, 8, e00083. [Google Scholar] [CrossRef]
- Hefzi, H.; Ang, K.S.; Hanscho, M.; Bordbar, A.; Ruckerbauer, D.; Lakshmanan, M.; Orellana, C.A.; Baycin-Hizal, D.; Huang, Y.; Ley, D.; et al. A Consensus Genome-scale Reconstruction of Chinese Hamster Ovary Cell Metabolism. Cell Syst. 2016, 3, 434–443. [Google Scholar] [CrossRef]
- Calmels, C.; McCann, A.; Malphettes, L.; Andersen, M.R. Application of a curated genome-scale metabolic model of CHO DG44 to an industrial fed-batch process. Metab. Eng. 2019, 51, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Fouladiha, H.; Marashi, S.-A.; Torkashvand, F.; Mahboudi, F.; Lewis, N.E.; Vazir, B. A metabolic network-based approach for developing feeding strategies for CHO cells to increase monoclonal antibody production. bioRxiv 2019. [Google Scholar] [CrossRef]
- Traustason, B. Amino Acid Requirements of the Chinese Hamster Ovary Cell Metabolism during Recombinant Protein Production. bioRxiv 2019, 796490. [Google Scholar] [CrossRef]
- Zampieri, G.; Coggins, M.; Valle, G.; Angione, C.; Zampieri, G.; Coggins, M.; Valle, G.; Angione, C. A poly-omics machine-learning method to predict metabolite production in CHO cells. In Proceedings of the 2nd International Electronic Conference on Metabolomics, 20–27 November 2017; MDPI: Basel, Switzerland; Volume 2, p. 4993. [Google Scholar]
- Lachance, J.C.; Lloyd, C.J.; Monk, J.M.; Yang, L.; Sastry, A.V.; Seif, Y.; Palsson, B.O.; Rodrigue, S.; Feist, A.M.; King, Z.A.; et al. BOFdat: Generating biomass objective functions for genome-scale metabolic models from experimental data. PLoS Comput. Biol. 2019, 15, e1006971. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Ebrahim, A.; Lloyd, C.J.; Saunders, M.A.; Palsson, B.O. DynamicME: Dynamic simulation and refinement of integrated models of metabolism and protein expression. BMC Syst. Biol. 2019, 13, 2. [Google Scholar] [CrossRef]
- Yang, L.; Yurkovich, J.T.; King, Z.A.; Palsson, B.O. Modeling the multi-scale mechanisms of macromolecular resource allocation. Curr. Opin. Microbiol. 2018, 45, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Kiparissides, A.; Hatzimanikatis, V. Thermodynamics-based Metabolite Sensitivity Analysis in metabolic networks. Metab. Eng. 2017, 39, 117–127. [Google Scholar] [CrossRef] [Green Version]
- Hatzimanikatis, V.; Saez-Rodriguez, J. Integrative approaches for signalling and metabolic networks. Integr. Biol. 2015, 7, 844–845. [Google Scholar] [CrossRef]
- Miskovic, L.; Tokic, M.; Savoglidis, G.; Hatzimanikatis, V. Control Theory Concepts for Modeling Uncertainty in Enzyme Kinetics of Biochemical Networks. Ind. Eng. Chem. Res. 2019. [Google Scholar] [CrossRef]
- Liu, S.; Farid, S.S.; Papageorgiou, L.G. Integrated Optimization of Upstream and Downstream Processing in Biopharmaceutical Manufacturing under Uncertainty: A Chance Constrained Programming Approach. Ind. Eng. Chem. Res. 2016, 55, 4599–4612. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Simaria, A.S.; Farid, S.S.; Papageorgiou, L.G. Mathematical programming approaches for downstream processing optimisation of biopharmaceuticals. Chem. Eng. Res. Des. 2015, 94, 18–31. [Google Scholar] [CrossRef]
- Hassan, S.; Huang, H.; Warren, K.; Mahdavi, B.; Smith, D.; Jong, S.; Farid, S.S. Process change evaluation framework for allogeneic cell therapies: Impact on drug development and commercialization. Regen. Med. 2016, 11, 287–305. [Google Scholar] [CrossRef] [PubMed]
- Popova, D.; Stonier, A.; Pain, D.; Titchener-Hooker, N.J.; Farid, S.S. Integrated economic and experimental framework for screening of primary recovery technologies for high cell density CHO cultures. Biotechnol. J. 2016, 11, 899–909. [Google Scholar] [CrossRef] [PubMed]
- Jankauskas, K.; Papageorgiou, L.G.; Farid, S.S. Continuous-Time Heuristic Model for Medium-Term Capacity Planning of a Multi-Suite, Multi-Product Biopharmaceutical Facility. In Computer Aided Chemical Engineering; Elsevier B.V.: Amsterdam, The Netherlands, 2017; Volume 40, pp. 1303–1308. [Google Scholar]
- Zampieri, G.; Vijayakumar, S.; Yaneske, E.; Angione, C. Machine and deep learning meet genome-scale metabolic modeling. PLoS Comput. Biol. 2019, 15, e1007084. [Google Scholar] [CrossRef]

| Details on Kinetic Model | Metabolites/Pathways Involved | Reference |
|---|---|---|
| Monod-kinetics, Luedeking-Piret model for associating rates of growth and product formation | Growth, glucose uptake, lactate secretion, product formation | [41] |
| Dynamic model, particle-swarm optimization for parameter estimation | Alanine, glutamine, glutamate, aspartate, asparagine, and proline utilization, product formation | [36,45] |
| 34-reaction model, multiplicative Michaelis-Menten kinetics | Role of amino acids in central carbon metabolism, tricarboxylic acid (TCA) cycle, recombinant monoclonal antibody (mAb) production, and cellular growth | [46] |
| Kalman filters for approximating the state variables represented by dynamic mathematical models, model predictive control on the apoptotic cell density, neural networks to express apoptotic cells as a function of state variables | Viable cell density, glutamine and asparagine concentration to predict apoptotic cell population | [47] |
| Method | Scale | Reference |
|---|---|---|
| Metabolic flux analysis + 13C analysis | Small scale: 272 reactions and 228 metabolites | [51] |
| Metabolic flux analysis + 13C analysis | Small scale: 58 reactions and 50 metabolites | [52] |
| Elementary flux mode analysis/extreme pathways | Small scale: 24 extracellular, 13 intracellular species, 35 reactions | [53] |
| Metabolic flux analysis + 13C analysis | Small scale: 73 reactions and 77 metabolites | [54] |
| Metabolic flux analysis + 13/14/15C-labelling | Small scale: 68 reactions and 21 metabolites (19 amino acids) | [55] |
| Metabolic flux analysis + 13C-labelled glucose and glutamine | Small scale: 37 reactions | [56] |
| Metabolic flux analysis | Small scale: 34 reactions and 30 metabolites | [57] |
| Metabolic flux analysis in response to varying levels of glutamine supplementation | Small scale: 40 reactions and 37 intracellular, 23 extracellular metabolites | [58] |
| Metabolic flux analysis to modify medium amino acid composition | Small scale: 23 reactions and 23 metabolites | [59] |
| Elementary flux mode analysis + kinetic modelling | Small scale: 166 reactions and 29 extracellular, 89 intracellular metabolites | [60] |
| Flux balance analysis | Genome scale: 1540 reactions and 1302 metabolites | [4] |
| Flux balance analysis | Genome scale: 1766 genes, 6663 reactions, and 4456 metabolites (in different subcellular compartments) | [61,62,63,64,65] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Traustason, B.; Cheeks, M.; Dikicioglu, D. Computer-Aided Strategies for Determining the Amino Acid Composition of Medium for Chinese Hamster Ovary Cell-Based Biomanufacturing Platforms. Int. J. Mol. Sci. 2019, 20, 5464. https://doi.org/10.3390/ijms20215464
Traustason B, Cheeks M, Dikicioglu D. Computer-Aided Strategies for Determining the Amino Acid Composition of Medium for Chinese Hamster Ovary Cell-Based Biomanufacturing Platforms. International Journal of Molecular Sciences. 2019; 20(21):5464. https://doi.org/10.3390/ijms20215464
Chicago/Turabian StyleTraustason, Bergthor, Matthew Cheeks, and Duygu Dikicioglu. 2019. "Computer-Aided Strategies for Determining the Amino Acid Composition of Medium for Chinese Hamster Ovary Cell-Based Biomanufacturing Platforms" International Journal of Molecular Sciences 20, no. 21: 5464. https://doi.org/10.3390/ijms20215464