Antimicrobial Potential of Single Metabolites of Curcuma longa Assessed in the Total Extract by Thin-Layer Chromatography-Based Bioautography and Image Analysis
Abstract
:1. Introduction
2. Results and Discussion
2.1. LC-ESI-Q-TOF-MS Analysis of the Extracts’ Qualitative and Quantitative Composition
2.2. TLC Bioautography Tests and Identification of Active Metabolites by TLC-LC-ESI-TOF-MS
2.3. Purification of Single Curcuminoids by CPC Chromatography
2.4. Antibacterial Potential of Turmeric Metabolites in Relation to the Commonly-Used Antibiotic Vancomycin
3. Materials and Methods
3.1. General
3.2. Chemicals and Reagents
3.3. Plant Extract Preparation
3.4. Thin-Layer Chromatography (TLC) Conditions
3.5. Organisms and Media
3.6. Bioautographic Methods
Determination of the MIQ Values
3.7. Image Processing
3.8. Quantitative and Qualitative Analysis of the Samples by Mass Spectrometry
3.9. Application of CPC Chromatography in the Isolation of Active Components of Turmeric Extract
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Gupta, P.D.; Birdi, T.J. Development of botanicals to combat antibiotic resistance. J. Ayurveda Integr. Med. 2017, 8, 266–275. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, I.; Biswas, K.; Bandyopadhyay, U.; Banerjee, R.K. Turmeric and curcumin: Biological actions and medicinal applications. Curr. Sci. 2004, 87, 44–53. [Google Scholar]
- Anand, P.; Kunnumakkara, A.B.; Newman, R.A.; Aggarwal, B.B. Bioavailability of curcumin: Problems and promises. Mol. Pharm. 2007, 4, 807–818. [Google Scholar] [CrossRef] [PubMed]
- Amalraja, A.; Pius, A.; Gopi, S.; Gopi, S. Biological activities of curcuminoids, other biomolecules from turmeric and their derivatives—A review. J. Tradit. Complement Med. 2017, 7, 205–233. [Google Scholar] [CrossRef] [PubMed]
- Ruby, A.J.; Kuttan, G.; Babu, K.G.; Rajasekharan, K.N.; Kuttan, R. Anti-tumour and antioxidant activity of natural curcuminoids. Cancer Lett. 1995, 94, 79–83. [Google Scholar] [CrossRef]
- Suvarna, R.; Bhat, S.S.; Hegde, K.H. Antibacterial Activity of Turmeric against Enterococcus faecalis An In vitro Study. Int. J. Curr. Microbiol. App. Sci. 2014, 3, 498–504. [Google Scholar]
- Mohammed, N.A.; Habil, N.Y. Evaluation of antimicrobial activity of Curcumin against two oral bacteria. Autom. Control Intell. Syst. 2015, 3, 18–21. [Google Scholar]
- Moghadamtousi, S.Z.; Kadir, H.A.; Hassandarvish, P.; Tajik, H.; Abubakar, S.; Zandi, K. A Review on Antibacterial, Antiviral, and Antifungal Activity of Curcumin. BioMed Res. Int. 2014, 2014, 186864. [Google Scholar] [CrossRef]
- Guptaa, A.; Mahajana, S.; Sharmab, R. Evaluation of antimicrobial activity of Curcuma longa rhizome extract against Staphylococcus aureus. Biotechnol. Rep. 2015, 6, 51–55. [Google Scholar] [CrossRef]
- Naz, S.; Jabeen, S.; Ilyas, S.; Manzoor, F.; Pak, J. Antibacterial activity of Curcuma longa varieties against different strains of bacteria. Pakistan J. of Bot. 2010, 42, 455–462. [Google Scholar]
- Gul, P.; Bakht, J. Antimicrobial activity of turmeric extract and its potential use in food industry. J. Food Sci. Technol. 2015, 52, 2272–2279. [Google Scholar] [CrossRef]
- Negi, P.S.; Jayaprakasha, G.K.; Rao, L.J.M.; Sakariah, K.K. Antibacterial Activity of Turmeric Oil: A Byproduct from Curcumin Manufacture. J. Agric. Food Chem. 1999, 47, 4297–4300. [Google Scholar] [CrossRef] [PubMed]
- Apisariyakul, A.; Vanittanakom, N.; Buddhasukh, D. Antifungal activity of turmeric oil extracted from Curcuma longa (Zingiberaceae). J. Ethnopharmacol. 1995, 49, 163–169. [Google Scholar] [CrossRef]
- Singh, R.; Chandra, R.; Bose, M.; Luthra, P.M. Antibacterial activity of Curcuma longa rhizome extract on pathogenic bacteria. Curr Sci. 2002, 83, 737–740. [Google Scholar]
- Dosoky, N.S.; Setzer, W.N. Chemical Composition and Biological Activities of Essential Oils of Curcuma Species. Nutrients 2018, 10, 1196. [Google Scholar] [CrossRef] [PubMed]
- Vaughn, A.R.; Haas, K.N.; Burney, W.; Andersen, E.; Clark, A.K.; Crawford, R.; Sivamani, R.K. Potential Role of Curcumin Against Biofilm-Producing Organisms on the Skin: A Review. Phytother. Res. 2017, 31, 1807–1816. [Google Scholar] [CrossRef] [PubMed]
- Neelakantan, P.; Subbarao, C.; Sharma, S.; Subbarao, C.V.; Garcia-Godoy, F.; Gutmann, J.L. Effectiveness of curcumin against Enterococcus faecalis biofilm. Acta Odontol. Scand. 2013, 71, 1453–1457. [Google Scholar] [CrossRef]
- Seo, M.J.; Park, J.E.; Jang, M.S. Optimization of sponge cake added with turmeric (Curcuma longa L.) powder using mixture design. Food Sci. Biotechnol 2010, 19, 617–625. [Google Scholar] [CrossRef]
- Péret-Almeida, L.; Naghetini, C.C.; de Aguiar Nunan, E.; Junqueira, R.G.; Glória, M.B.A. In vitro antimicrobial activity of the ground rhizome, curcuminoid pigments and essential oil of Curcuma longa L. Ciêne. Agrotec 2008, 32, 875–881. [Google Scholar]
- Jayaprakasha, G.K.; Jagan Mohan Rao, L.; Sakariah, K.K. Improved HPLC Method for the Determination of Curcumin, Demethoxycurcumin, and Bisdemethoxycurcumin. J. Agric. Food Chem. 2002, 50, 3668–3672. [Google Scholar] [CrossRef]
- Jiang, H.; Timmermann, B.N.; Gang, D.R. Use of liquid chromatography-electrospray ionization tandem mass spectrometry to identify diarylheptanoids in turmeric (Curcuma longa L.) rhizome. J. Chromatogr. A 2006, 7, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Kukuła-Koch, W.; Grabarska, A.; Łuszczki, J.; Czernicka, L.; Nowosadzka, E.; Gumbarewicz, E.; Jarząb, A.; Audo, G.; Upadhyay, S.; Głowniak, K.; et al. Superior anticancer activity is demonstrated by total extract of Curcuma longa L. as opposed to individual curcuminoids separated by centrifugal partition chromatography. Phytother. Res. 2018, 32, 933–942. [Google Scholar] [CrossRef] [PubMed]
- Paneth, A.; Frączek, T.; Grzegorczyk, A.; Janowska, D.; Malm, A.; Paneth, P. A search for dual action HIV-1 reverse transcriptase, bacterial RNA polymerase inhibitors. Molecules 2017, 22, 1808. [Google Scholar] [CrossRef] [PubMed]
- Isnard, C.; Dhalluin, A.; Malandain, D.; Bruey, Q.; Auzou, M.; Michon, J.; Giard, J.C.; Guérin, F.; Cattoir, V. In vitro activity of novel anti-MRSA cephalosporins and comparator antimicrobial agents against staphylococci involved in prosthetic joint infections. J. Glob. Antimicrob. Resist. 2018, 13, 221–225. [Google Scholar] [CrossRef] [PubMed]
- Sá, S.; Chaul, L.T.; Alves, V.F.; Fiuza, T.S.; Tresvenzol, L.M.F.; Vaz, B.G.; Ferri, P.H.; Borges, L.L.; Paula, J.R. Phytochemistry and antimicrobial activity of Campomanesia adamantium. Braz. J. Pharmacogn. 2018, 28, 303–311. [Google Scholar] [CrossRef]
- Andrews, J.M. Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 2001, 48, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Skalicka-Woźniak, K.; Grzegorczyk, A.; Świątek, Ł.; Walasek, M.; Widelski, J.; Rajtar, B.; Polz-Dacewicz, M.; Malm, A.; Elansary, H.O. Biological activity and safety profile of the essential oil from fruits of Heracleum mantegazzianum Sommier & Levier (Apiaceae). Food Chem. Toxicol. 2017, 109, 820–826. [Google Scholar]
- Olech, M.; Komsta, Ł.; Nowak, R.; Cieśla, Ł.; Waksmundzka-Hajnos, M. Investigation of antiradical activity of plant material by thin-layer chromatography with image processing. Food Chem. 2012, 132, 549–553. [Google Scholar] [CrossRef]
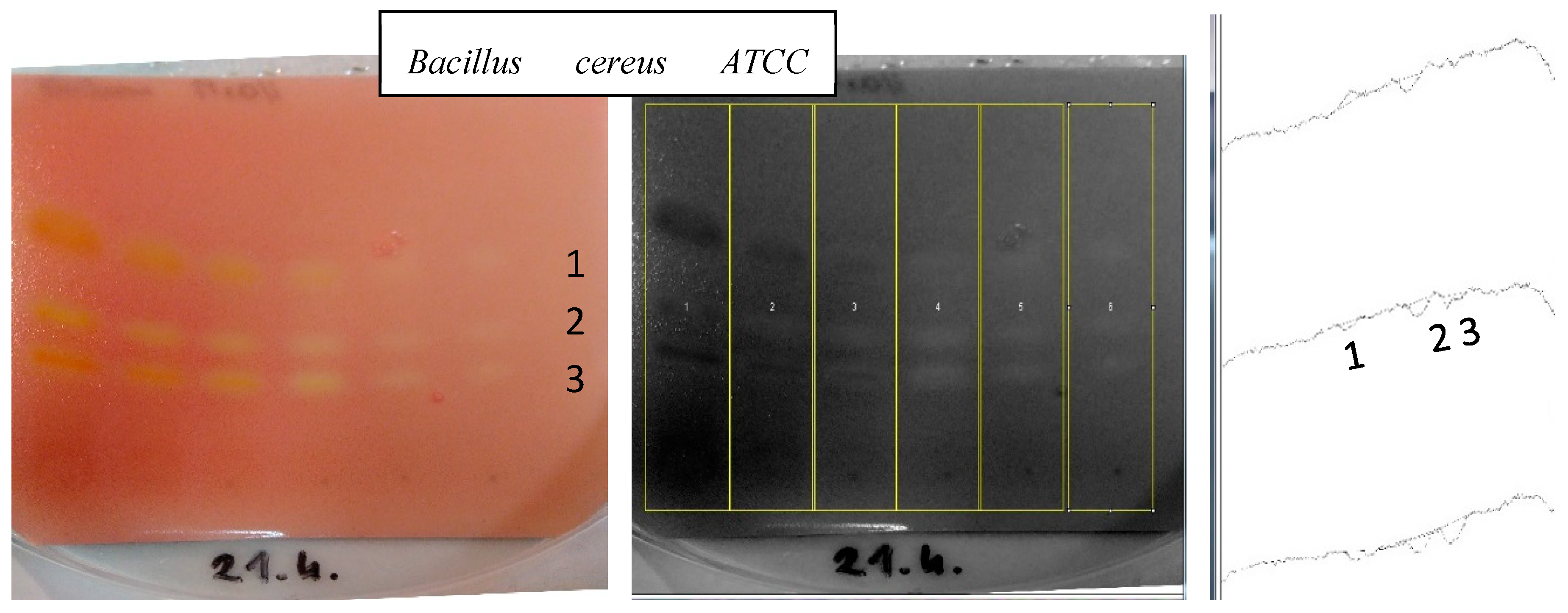
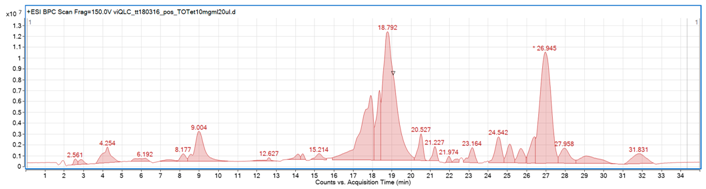 | |||||||
| The total ion chromatogram of diethyl ether extract from turmeric tuber | |||||||
| No. | Compound | Molecular Formula | Experimental Mass [M+H]+ | Calculated Mass [M+H]+ | Mass Error (ppm) | DBE | MS/MS Fragments |
| 1 | CU | C21H20O6 | 369.1336 | 369.1333 | −0.91 | 12 | 177, 145 |
| 2 | DMCU | C20H18O5 | 339.1233 | 339.1227 | −1.77 | 12 | 177, 147 |
| 3 | BDMCU | C19H16O4 | 309.1122 | 309.1121 | −0.21 | 12 | 147, 119 |
| 4 | TUR | C15H20O | 217.1586 | 217.1587 | 0.42 | 12 | 119, 91 |
| 5 | Dihydro-BDMCU | C19H18O4 | 311.1286 | 311.1278 | −2.63 | 11 | 225, 147 |
| 6 | Dihydro-CU | C21H22O6 | 371.1468 | 371.1489 | 5.71 | 11 | 245, 177, 147 |
| 7 | Dihydro-DMCU | C20H20O5 | 341.1377 | 341.1384 | 1.91 | 11 | 177, 147 |
| 8 | Tetrahydro-BDMCU | C19H20O4 | 313.1286 | 313.1434 | 3.0 | 10 | - |
| Strains of Bacteria | MIQ (µg) | |||
|---|---|---|---|---|
| CU | DMCU | BDMCU | TUR | |
| Staphylococcus aureus ATCC 25923 | 4.42 | 0.91 | 0.57 | - |
| Staphylococcus aureus ATCC 6538 | 0.55 | 0.23 | 0.28 | - |
| Staphylococcus aureus ATCC 43300 | 8.84 | 1.83 | 0.57 | 0.23 |
| Staphylococcus epidermidis ATCC 12228 | 0.55 | 0.23 | 0.14 | 1.80 |
| Bacillus subtilis ATCC 6633 | 4.42 | 0.46 | 0.28 | 1.83 |
| Bacillus cereus ATCC 10876 | <0.28 | <0.11 | <0.14 | - |
| MIQ (µg) | ||
|---|---|---|
| Strains of Bacteria | CU | Vancomycin |
| Staphylococcus aureus ATCC 25923 | 25 | 0.098 |
| Staphylococcus aureus ATCC 6538 | 6.25 | 0.049 |
| Staphylococcus aureus ATCC 43300 | 50 | 0.049 |
| Staphylococcus epidermidis ATCC 12228 | 6.25 | 0.098 |
| Bacillus subtilis ATCC 6633 | 25 | 0.024 |
| Bacillus cereus ATCC 10876 | 25 | 0.098 |
| Strains of Bacteria | Calibration Curve Equation (Vancomycin) | Vancomycin Equivalents (TLC Bioutography) | Vancomycin Equivalents (Microdilution Assay) | |
|---|---|---|---|---|
| CU (µg ± SD) | RSD (%) | CU [µg] | ||
| Staphylococcus aureus ATCC 25923 | y = 9731ln(x) − 12434 | 206 ± 12 | 5.8 | 255 |
| Staphylococcus aureus ATCC 6538 | y = 17492ln(x) − 7990.8 | 142 ± 10 | 6.9 | 127 |
| Staphylococcus aureus ATCC 43300 | y = 1381.9ln(x) + 5925.8 | 1310 ± 69 | 5.3 | 1020 |
| Staphylococcus epidermidis ATCC 12228 | y = 1340.3ln(x) + 128.79 | 49 ± 4 | 8.7 | 64 |
| Bacillus subtilis ATCC 6633 | y = 13889ln(x) + 22510 | 1328 ± 56 | 4.2 | 1041 |
| Bacillus cereus ATCC 10876 | y = 629.12ln(x) + 10915 | 202 ± 13 | 6.5 | 255 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Czernicka, L.; Grzegorczyk, A.; Marzec, Z.; Antosiewicz, B.; Malm, A.; Kukula-Koch, W. Antimicrobial Potential of Single Metabolites of Curcuma longa Assessed in the Total Extract by Thin-Layer Chromatography-Based Bioautography and Image Analysis. Int. J. Mol. Sci. 2019, 20, 898. https://doi.org/10.3390/ijms20040898
Czernicka L, Grzegorczyk A, Marzec Z, Antosiewicz B, Malm A, Kukula-Koch W. Antimicrobial Potential of Single Metabolites of Curcuma longa Assessed in the Total Extract by Thin-Layer Chromatography-Based Bioautography and Image Analysis. International Journal of Molecular Sciences. 2019; 20(4):898. https://doi.org/10.3390/ijms20040898
Chicago/Turabian StyleCzernicka, Lidia, Agnieszka Grzegorczyk, Zbigniew Marzec, Beata Antosiewicz, Anna Malm, and Wirginia Kukula-Koch. 2019. "Antimicrobial Potential of Single Metabolites of Curcuma longa Assessed in the Total Extract by Thin-Layer Chromatography-Based Bioautography and Image Analysis" International Journal of Molecular Sciences 20, no. 4: 898. https://doi.org/10.3390/ijms20040898






