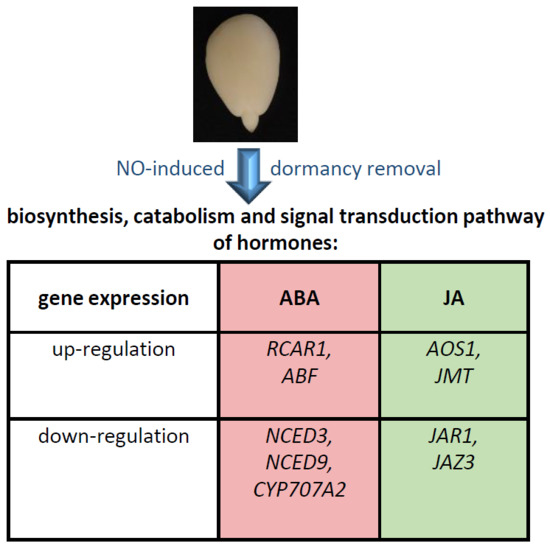
Nitric Oxide-Induced Dormancy Removal of Apple Embryos Is Linked to Alterations in Expression of Genes Encoding ABA and JA Biosynthetic or Transduction Pathways and RNA Nitration
Abstract
:1. Introduction
2. Results
2.1. Dormancy Removal of Apple Embryos by Short-Term NO Treatment Was Associated with Alterations in the Expression of Genes Encoding Enzymes of ABA Biosynthetic Pathway and ABA Degradation
2.1.1. Short-Term Fumigation with NO Decreased NCED3 and NCED9 Transcripts Level in Embryonic Axes of Apple Seeds
2.1.2. Short-term Fumigation with NO Had no Effect on CYP707A1 and Decreased CYP707A2 Transcripts Level in Embryonic Axes of Apple Seeds
2.2. Dormancy Removal of Apple Embryos by Short-Term NO Treatment Was Associated with Alterations in the Expression of Genes Encoding Elements of ABA Signal Transduction Pathway
2.2.1. Short-Term Fumigation of Apple Embryos with NO Increased the Expression of RCAR1 but Did Not Influence the Expression of Other Genes Encoding ABA Co-Receptors (PYL1, PYL2, RCAR3) in Embryonic Axes
2.2.2. Short-term Fumigation of Apple Embryos with NO Did Not Influence the Expression of Genes (PP2CA, ABI1, ABI2) Encoding Protein Phosphatases PP2Cs and SnRK2 Encoding Protein Kinase in Embryonic Axes
2.2.3. Short-term Fumigation of Apple Embryos with NO Prevented Increase of the Expression Level of Gene Encoding ABF While Had No Effect on the Expression of Genes Encoding Other ABA Transcription Factors (ABI5 and AREB3) in Embryonic Axes
2.3. Dormancy Removal of Apple Embryos by NO Fumigation Was Linked to Alterations in Expression of Genes Encoding Enzymes of JA Biosynthesis and JA Derivatives Formation
Dormancy Removal of Apple Embryos by NO Fumigation was Linked to Up-Regulation of Genes Encoding Enzymes of JA Biosynthesis (AOS1) and MeJA Formation (JMT)
2.4. Dormancy Removal of Apple Embryos by NO Fumigation Was Linked to Alterations in Expression of Genes Encoding Elements of JA Signal Transduction Pathway
Dormancy Removal of Apple Embryos by NO Fumigation Was Linked to Alterations in the Expression of JAZ3 Encoding Negative Regulator of JA Signal Transduction Pathway
2.5. NO Fumigation of Apple Embryos Increased RNA Nitration Level in Embryonic Axes
3. Discussion
4. Materials and Methods
4.1. Experimental Material
Fumigation of Dormant Apple Embryos with NO
4.2. Analysis of Gene Expression
4.2.1. RNA Isolation from Axes of Apple Embryos
4.2.2. RT-PCR Conditions
4.3. Measurement of Nitrated RNA Content
4.4. Statistical Analysis
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ABF | ABA-Responsive Elements Binding Factor |
| ABI | Abscisic Acid Insensitive |
| ABI5 | ABA Insensitive 5 |
| AOS | Allene Oxide Synthase |
| AREB | ABA Response Element Binding Factor |
| COI | Coronatine Insensitive 1 |
| CYP707 | ABA 8’-hydroxylase |
| GA | Gibberellins |
| GDR | The Genome Database for Rosaceae |
| JA | Jasmonic Acid |
| JAR1 | Jasmonate-Resistant 1 |
| JAZ | Jasmonate ZIM-domain |
| JMT | Jasmonate O-methyltransferase |
| NO | Nitric Oxide |
| NCED | 9-cis-Epoxycarotenoid Dioxygenase |
| PP2C | Protein Phosphatase 2C |
| PYL | Pyrabactin resistance 1-like |
| PYR1 | Pyrabactin resistance 1 |
| RCAR | Regulatory Component ABA Receptor 1 |
| RNS | Reactive Nitrogen Species |
| ROS | Reactive Oxygen Species |
| SIN-1 | 3-Morpholinosydnonimine |
| SNP | Sodium Nitroprusside |
| SNAP | S-nitroso-N-acetylpenicillamine |
| SnRK2 | SNF1-Related Protein Kinase 2 |
References
- Nonogaki, H. Seed dormancy and germination emerging mechanisms and new hypotheses. Front. Plant Sci. 2014, 5, 233. [Google Scholar] [CrossRef] [PubMed]
- Nonogaki, H. Seed biology updates–highlights and new discoveries in seed dormancy and germination research. Front. Plant Sci. 2017, 8, 524. [Google Scholar] [CrossRef] [PubMed]
- Kucera, B.; Cohn, M.A.; Leubner-Metzger, G. Plant hormone interactions during seed dormancy release and germination. Seed Sci. Res. 2005, 15, 281–307. [Google Scholar] [CrossRef]
- Krasuska, U.; Ciacka, K.; Gniazdowska, A. Nitric oxide-polyamines cross-talk during dormancy release and germination of apple embryos. Nitric Oxide 2017, 68, 38–50. [Google Scholar] [CrossRef] [PubMed]
- Krasuska, U.; Ciacka, K.; Andryka-Dudek, P.; Bogatek, R.; Gniazdowska, A. “Nitrosative Door” in seed dormancy alleviation and germination. In Reactive Oxygen and Nitrogen Species Signaling and Communication in Plants, Signaling and Communication in Plants, Vol. 23; Gupta, K.J., Igamberdiev, A.U., Eds.; Springer: Cham, Switzerland, 2015; pp. 215–237. [Google Scholar]
- Šírová, J.; Sedlářová, M.; Piterková, J.; Luhová, L.; Petřivalský, M. The role of nitric oxide in the germination of plant seeds and pollen. Plant Sci. 2011, 181, 560–572. [Google Scholar] [CrossRef] [PubMed]
- Arc, E.; Sechet, J.; Corbineau, F.; Rajjou, L.; Marion-Poll, A. ABA crosstalk with ethylene and nitric oxide in seed dormancy and germination. Front. Plant Sci. 2013, 4, 63. [Google Scholar] [CrossRef] [PubMed]
- Arc, E.; Galland, M.; Godin, B.; Cueff, G.; Rajjou, L. Nitric oxide implication in the control of seed dormancy and germination. Front. Plant Sci. 2013, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanz, L.; Albertos, P.; Mateos, I.; Sánchez-Vicente, I.; Lechón, T.; Fernández-Marcos, M.; Lorenzo, O. Nitric oxide (NO) and phytohormones crosstalk during early plant development. J. Exp. Bot. 2015, 66, 2857–2868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dębska, K.; Krasuska, U.; Budnicka, K.; Bogatek, R.; Gniazdowska, A. Dormancy removal of apple seeds by cold stratification is associated with fluctuation in H2O2, NO production and protein carbonylation level. J. Plant Physiol. 2013, 170, 480–488. [Google Scholar] [CrossRef] [PubMed]
- Lewak, S. Metabolic control of embryonic dormancy in apple seed: Seven decades of research. Acta Physiol. Plant. 2011, 33, 1–24. [Google Scholar] [CrossRef]
- Gniazdowska, A.; Krasuska, U.; Bogatek, R. Dormancy removal in apple embryos by nitric oxide or cyanide involves modifications in ethylene biosynthetic pathway. Planta 2010, 232, 1397–1407. [Google Scholar] [CrossRef] [PubMed]
- Krasuska, U.; Ciacka, K.; Orzechowski, S.; Fettke, J.; Bogatek, R.; Gniazdowska, A. Modification of the endogenous NO level influences apple embryos dormancy by alterations of nitrated and biotinylated protein patterns. Planta 2016, 244, 877–891. [Google Scholar] [CrossRef] [PubMed]
- Bethke, P.C.; Libourel, I.G.L.; Aoyama, N.; Chung, Y.-Y.; Still, D.W.; Jones, R.L. The Arabidopsis aleurone layer responds to nitric oxide, gibberellin, and abscisic acid and is sufficient and necessary for seed dormancy. Plant Physiol. 2007, 143, 1173–1188. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Shi, L.; Ye, N.; Liu, R.; Jia, W.; Zhang, J. Nitric oxide-induced rapid decrease of abscisic acid concentration is required in breaking seed dormancy in Arabidopsis. New Phytol. 2009, 183, 1030–1042. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, J.V.; Barrero, J.M.; Hughes, T.; Julkowska, M.; Taylor, J.M.; Xu, Q.; Gubler, F. Roles for blue light, jasmonate and nitric oxide in the regulation of dormancy and germination in wheat grain (Triticum aestivum L.). Planta 2013, 238, 121–138. [Google Scholar] [CrossRef] [PubMed]
- Gniazdowska, A.; Krasuska, U.; Czajkowska, K.; Bogatek, R. Nitric oxide, hydrogen cyanide and ethylene are required in the control of germination and undisturbed development of young apple seedlings. Plant Growth Regul. 2010, 61, 75–84. [Google Scholar] [CrossRef]
- Jones, L.H. Chemistry and biology of biomolecule nitration. Chem. Biol. 2012, 19, 1086–1092. [Google Scholar] [CrossRef] [PubMed]
- Izbiańska, K.; Floryszak-Wieczorek, J.; Gajewska, J.; Meller, B.; Kuźnicki, D.; Arasimowicz-Jelonek, M. RNA and mRNA nitration as a novel metabolic link in potato immune response to Phytophthora infestans. Front. Plant Sci. 2018, 9, 672. [Google Scholar] [CrossRef] [PubMed]
- Wurtmann, E.J.; Wolin, S.L. RNA under attack: Cellular handling of RNA damage. Crit. Rev. Biochem. Mol. Biol. 2009, 44, 34–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Maarouf-Bouteau, H.; Meimoun, P.; Job, C.; Job, D.; Bailly, C. Role of protein and mRNA oxidation in seed dormancy and germination. Front. Plant Sci. 2013, 4, 77. [Google Scholar] [CrossRef] [PubMed]
- Baudouin, E.; Hancock, J.T. Nitric oxide signaling in plants. Front. Plant Sci. 2014, 4, 553. [Google Scholar] [CrossRef] [PubMed]
- Polverari, A.; Molesini, B.; Pezzotti, M.; Buonaurio, R.; Marte, M.; Delledonne, M. Nitric oxide-mediated transcriptional changes in Arabidopsis thaliana. Mol. Plant-Microbe Interact. 2003, 16, 1094–1105. [Google Scholar] [CrossRef] [PubMed]
- Begara-Morales, J.C.; Sánchez-Calvo, B.; Luque, F.; Leyva-Pérez, M.O.; Leterrier, M.; Corpas, F.J.; Barroso, J.B. Differential transcriptomic analysis by RNA-seq of GSNO-responsive genes between Arabidopsis roots and leaves. Plant Cell Physiol. 2014, 55, 1080–1095. [Google Scholar] [CrossRef] [PubMed]
- Parani, M.; Rudrabhatla, S.; Myers, R.; Weirich, H.; Smith, B.; Leaman, D.W.; Goldman, S.L. Microarray analysis of nitric oxide responsive transcripts in Arabidopsis. Plant Biotechnol. J. 2004, 2, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Wei, H.; Li, L.; Yu, S. Transcriptome analysis of nitric oxide-responsive genes in upland cotton (Gossypium hirsutum). PLoS ONE 2018, 13, e0192367. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Gacio Mdel, C.; Matilla-Vázquez, M.A.; Matilla, A.J. Seed dormancy and ABA signaling: The breakthrough goes on. Plant Signal. Behav. 2009, 4, 1035–1049. [Google Scholar]
- Nambara, E.; Okamoto, M.; Tatematsu, K.; Yano, R.; Seo, M.; Kamiya, Y. Abscisic acid and the control of seed dormancy and germination. Seed Sci. Res. 2010, 20, 55–67. [Google Scholar] [CrossRef]
- Bethke, P.C.; Libourel, I.G.L.; Jones, R.L. Nitric oxide reduces seed dormancy in Arabidopsis. J. Exp. Bot. 2006, 57, 517–526. [Google Scholar] [CrossRef] [PubMed]
- Piterková, J.; Luhová, L.; Hofman, J.; Turecková, V.; Novák, O.; Petrivalsky, M.; Fellner, M. Nitric oxide is involved in light-specific responses of tomato during germination under normal and osmotic stress conditions. Ann. Bot. 2012, 110, 767–776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lefebvre, V.; North, H.; Frey, A.; Sotta, B.; Seo, M.; Okamoto, M.; Nambara, E.; Marion-Poll, A. Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J. 2006, 45, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Hussain, A.; Mun, B.-G.; Imran, Q.M.; Lee, S.-U.; Adamu, T.A.; Shahid, M.; Kim, K.-M.; Yun, B.-W. Nitric oxide mediated transcriptome profiling reveals activation of multiple regulatory pathways in Arabidopsis thaliana. Front. Plant Sci. 2016, 7, 975. [Google Scholar] [CrossRef] [PubMed]
- Hauser, F.; Waadt, R.; Schroeder, J.I. Evolution of abscisic acid synthesis and signaling mechanisms. Curr. Biol. 2011, 21, R346–R355. [Google Scholar] [CrossRef] [PubMed]
- Szostkiewicz, I.; Richter, K.; Kepka, M.; Demmel, S.; Ma, Y.; Korte, A.; Assaad, F.F.; Christmann, A.; Grill, E. Closely related receptor complexes differ in their ABA selectivity and sensitivity. Plant J. 2010, 61, 25–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freschi, L. Nitric oxide and phytohormone interactions: Current status and perspectives. Front. Plant Sci. 2013, 4, 398. [Google Scholar] [CrossRef] [PubMed]
- Jain, P.; Bhatla, S.C. Molecular mechanisms accompanying nitric oxide signalling through tyrosine nitration and S-nitrosylation of proteins in plants. Funct. Plant Biol. 2018, 45, 70. [Google Scholar] [CrossRef]
- Castillo, M.C.; Lozano-Juste, J.; González-Guzmán, M.; Rodriguez, L.; Rodriguez, P.L.; León, J. Inactivation of PYR/PYL/RCAR ABA receptors by tyrosine nitration may enable rapid inhibition of ABA signaling by nitric oxide in plants. Sci. Signal. 2015, 8, ra89. [Google Scholar] [CrossRef] [PubMed]
- Brock, A.K.; Willmann, R.; Kolb, D.; Grefen, L.; Lajunen, H.M.; Bethke, G.; Lee, J.; Nürnberger, T.; Gust, A.A. The Arabidopsis mitogen-activated protein kinase phosphatase PP2C5 affects seed germination, stomatal aperture, and abscisic acid-inducible gene expression. Plant Physiol. 2010, 153, 1098–1111. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zhu, J.-K.; Lang, Z. Nitric oxide suppresses the inhibitory effect of abscisic acid on seed germination by S-nitrosylation of SnRK2 proteins. Plant Signal. Behav. 2015, 10, e1031939. [Google Scholar] [CrossRef] [PubMed]
- Oracz, K.; Karpiński, S. Phytohormones signaling pathways and ROS involvement in seed germination. Front. Plant Sci. 2016, 7, 864. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Juste, J.; Leon, J. Enhanced abscisic acid-mediated responses in nia1nia2noa1-2 triple mutant impaired in NIA/NR- and AtNOA1-dependent Nitric Oxide biosynthesis in Arabidopsis. Plant Physiol. 2010, 152, 891–903. [Google Scholar] [CrossRef] [PubMed]
- Albertos, P.; Romero-Puertas, M.C.; Tatematsu, K.; Mateos, I.; Sánchez-Vicente, I.; Nambara, E.; Lorenzo, O. S-nitrosylation triggers ABI5 degradation to promote seed germination and seedling growth. Nat. Commun. 2015, 6, 8669. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.R.; Wagner, R.L.; Verhey, S.D.; Walker-Simmons, M.K. The abscisic acid-responsive kinase PKABA1 interacts with a seed-specific abscisic acid response element-binding factor, TaABF, and phosphorylates TaABF peptide sequences. Plant Physiol. 2002, 130, 837–846. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Liu, B.; Liu, L.; Song, S. Jasmonate action in plant growth and development. J. Exp. Bot. 2017, 68, 1349–1359. [Google Scholar] [CrossRef] [PubMed]
- Yildiz, K.; Muradoglu, F.; Yilmaz, H. The effect of jasmonic acid on germination of dormant and nondormant pear (Pyrus communis L.) seeds. Seed Sci. Technol. 2008, 36, 569–574. [Google Scholar] [CrossRef]
- Ranjan, R.; Lewak, S. Jasmonic acid promotes germination and lipase activity in non-stratified apple embryos. Physiol. Plant. 1992, 86, 335–339. [Google Scholar] [CrossRef]
- Xu, Q.; Truong, T.T.; Barrero, J.M.; Jacobsen, J.V.; Hocart, C.H.; Gubler, F. A role for jasmonates in the release of dormancy by cold stratification in wheat. J. Exp. Bot. 2016, 67, 3497–3508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimmermann, G. The multigene family encoding Germin-Like proteins of barley. Regulation and function in basal host resistance. Plant Physiol. 2006, 142, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Han, G.-Z. Evolution of jasmonate biosynthesis and signaling mechanisms. J. Exp. Bot. 2016, 68, erw470. [Google Scholar] [CrossRef] [PubMed]
- Linkies, A.; Leubner-Metzger, G. Beyond gibberellins and abscisic acid: How ethylene and jasmonates control seed germination. Plant Cell Rep. 2012, 31, 253–270. [Google Scholar] [CrossRef] [PubMed]
- Chaki, M.; Valderrama, R.; Fernández-Ocaña, A.M.; Carreras, A.; López-Jaramillo, J.; Luque, F.; Palma, J.M.; Pedrajas, J.R.; Begara-Morales, J.C.; Sánchez-Calvo, B.; et al. Protein targets of tyrosine nitration in sunflower (Helianthus annuus L.) hypocotyls. J. Exp. Bot. 2009, 60, 4221–4234. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Arbaizar, A.; Regalado, J.J.; Lorenzo, O. Isolation and characterization of novel mutant loci suppressing the ABA hypersensitivity of the Arabidopsis coronatine insensitive 1-16 (coi1-16) mutant during germination and seedling growth. Plant Cell Physiol. 2012, 53, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Galland, M.; Rajjou, L. Regulation of mRNA translation controls seed germination and is critical for seedling vigor. Front. Plant Sci. 2015, 6, 284. [Google Scholar] [CrossRef] [PubMed]
- Bazin, J.; Langlade, N.; Vincourt, P.; Arribat, S.; Balzergue, S.; El-Maarouf-Bouteau, H.; Bailly, C. Targeted mRNA oxidation regulates sunflower seed dormancy alleviation during dry after-ripening. Plant Cell 2011, 23, 2196–2208. [Google Scholar] [CrossRef] [PubMed]
- Chomczynski, P.; Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef]






| Plant Material | Concentration of 8-NO2-G (pg µg−1 RNA) |
|---|---|
| C0 | 72.5 ± 3.2 a |
| C | 73.0 ± 3.9 a |
| NO | 84.2 ± 3.6 b |
| SIN-1 | 103 ± 4.1 c |
| Gene | Accession Number | Nucleotide Database | Forward Primer | Reverse Primer |
|---|---|---|---|---|
| NCED1 | XM_008380174.2 | NCBI | 5′- ATCGGCTCCTGCATGACA -3′ | 5′- CGAGAGCCAAATAAGTGAAC -3′ |
| NCED3 | XM_008384748.2 | NCBI | 5′- AGAAGCATATCTACGGTGAC -3′ | 5′- AGCGCTTATAAACGTTCCATG -3′ |
| CYP707A1 | AB593330.1 | NCBI | 5′- CTGGCATTGAAGCATTGGT -3′ | 5′- TCCTTGCCAAGAGAGCTT -3′ |
| CYP707A2 | AB593331.1 | NCBI | 5′- TGCAAGAGATGAAGAGGTATG -3′ | 5′- GAGAAGCTTCCTTGCCTTC -3′ |
| PYL1 | KX019762.1 | NCBI | 5′- GCACTTCATCCGGAGCTGT -3′ | 5′- GACAACAGTATCAGCGAAGAG -3′ |
| PYL2 | XM_008379295.2 | NCBI | 5′- CCTCACAGATACAAGCACT -3′ | 5′- CTCATTGACCGAAGTAACAG -3′ |
| RCAR3 | MDP0000191830 | GDR | 5′- CAAGTTGCTGAGAACAGTGA -3’ | 5′- CTATCAACGAATCTGTCAACTC -3′ |
| RCAR1 | MDP0000434532 | GDR | 5′- CTCTCGTCCGGTACATCA-3′ | 5′- GAAGAGTAGTTCCTAAGTCTGT-3′ |
| ABI1 | MDP0000893203 | GDR | 5′- TGTGGCGATTCGAGAGCA -3′ | 5′- CTGGATCAGGTATAATCCATGG -3′ |
| ABI2 | MDP0000647467 | GDR | 5′- GATGAATGCCTCGTATTAGC -3′ | 5′- GAGCTTTCAGATCCACCA-3′ |
| PP2C | XM_008358502.2 | NCBI | 5′- GATCGTCGTGGCTAACTGC -3′ | 5′- GTCGCTCGCCAGGATCAG -3′ |
| SNRK2 | KJ563286.1 | NCBI | 5′- AGATTGCAGATGTATGGTC -3′ | 5′- GCTTTGCATGGGTTGATC-3′ |
| AREB3 | MDP0000273211 | GDR | 5′- GTAGGTGCTGGAGCTATGAT -3′ | 5′- TGTATATGCCTGCTTCCTTG -3′ |
| ABF | MDP0000701734 | GDR | 5′- GAGCTGCAGAACACCATTG -3′ | 5′- TGTATATGCCTGCTTCCTTG -3′ |
| AOS1 | XM_008366758.2 | NCBI | 5′- CGCATCCAGAAATACCAGTCA -3′ | 5′′- GCTTCAGCTTGTCGTGCT -3′ |
| JMT | XM_008378987.2 | NCBI | 5′- GTTGCTCATCTGGACCAA - 3′ | 5′- TCAGTTGGTTGTAGAATGCC -3′ |
| JAR1 | XM_017327397.1 | NCBI | 5′- GTATTGCCATCTCTTGTGTG -3′ | 5′- ACAGCTGATCGGATTGATG - 3′ |
| COI1 | XM_008394693.2 | NCBI | 5′- GTGTCGTTGGTGTGCAAG -3′ | 5′- CAGATTGAACATCGCCGC -3′ |
| JAZ3 | MDP0000243322 | GDR | 5′- TGACTATTTCAACTGCTGATGC-3′ | 5′- GATTGGAGAACTGGAGAACTC-3′ |
| JAZ12 | KU179650.1 | NCBI | 5′- GAGACACTCTCTTCAGCG-3′ | 5′- TGAGTTTCTTCCTGAACCATG-3′ |
| MYC2 | NM_001328944.1 | NCBI | 5′- CGAACAAGAGTACCGCAAG -3′ | 5′-GTCGGAACGCAAACCATA - 3′ |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andryka-Dudek, P.; Ciacka, K.; Wiśniewska, A.; Bogatek, R.; Gniazdowska, A. Nitric Oxide-Induced Dormancy Removal of Apple Embryos Is Linked to Alterations in Expression of Genes Encoding ABA and JA Biosynthetic or Transduction Pathways and RNA Nitration. Int. J. Mol. Sci. 2019, 20, 1007. https://doi.org/10.3390/ijms20051007
Andryka-Dudek P, Ciacka K, Wiśniewska A, Bogatek R, Gniazdowska A. Nitric Oxide-Induced Dormancy Removal of Apple Embryos Is Linked to Alterations in Expression of Genes Encoding ABA and JA Biosynthetic or Transduction Pathways and RNA Nitration. International Journal of Molecular Sciences. 2019; 20(5):1007. https://doi.org/10.3390/ijms20051007
Chicago/Turabian StyleAndryka-Dudek, Paulina, Katarzyna Ciacka, Anita Wiśniewska, Renata Bogatek, and Agnieszka Gniazdowska. 2019. "Nitric Oxide-Induced Dormancy Removal of Apple Embryos Is Linked to Alterations in Expression of Genes Encoding ABA and JA Biosynthetic or Transduction Pathways and RNA Nitration" International Journal of Molecular Sciences 20, no. 5: 1007. https://doi.org/10.3390/ijms20051007