rs4143815-PDL1, a New Potential Immunogenetic Biomarker of Biochemical Recurrence in Locally Advanced Prostate Cancer after Radiotherapy
Abstract
:1. Introduction
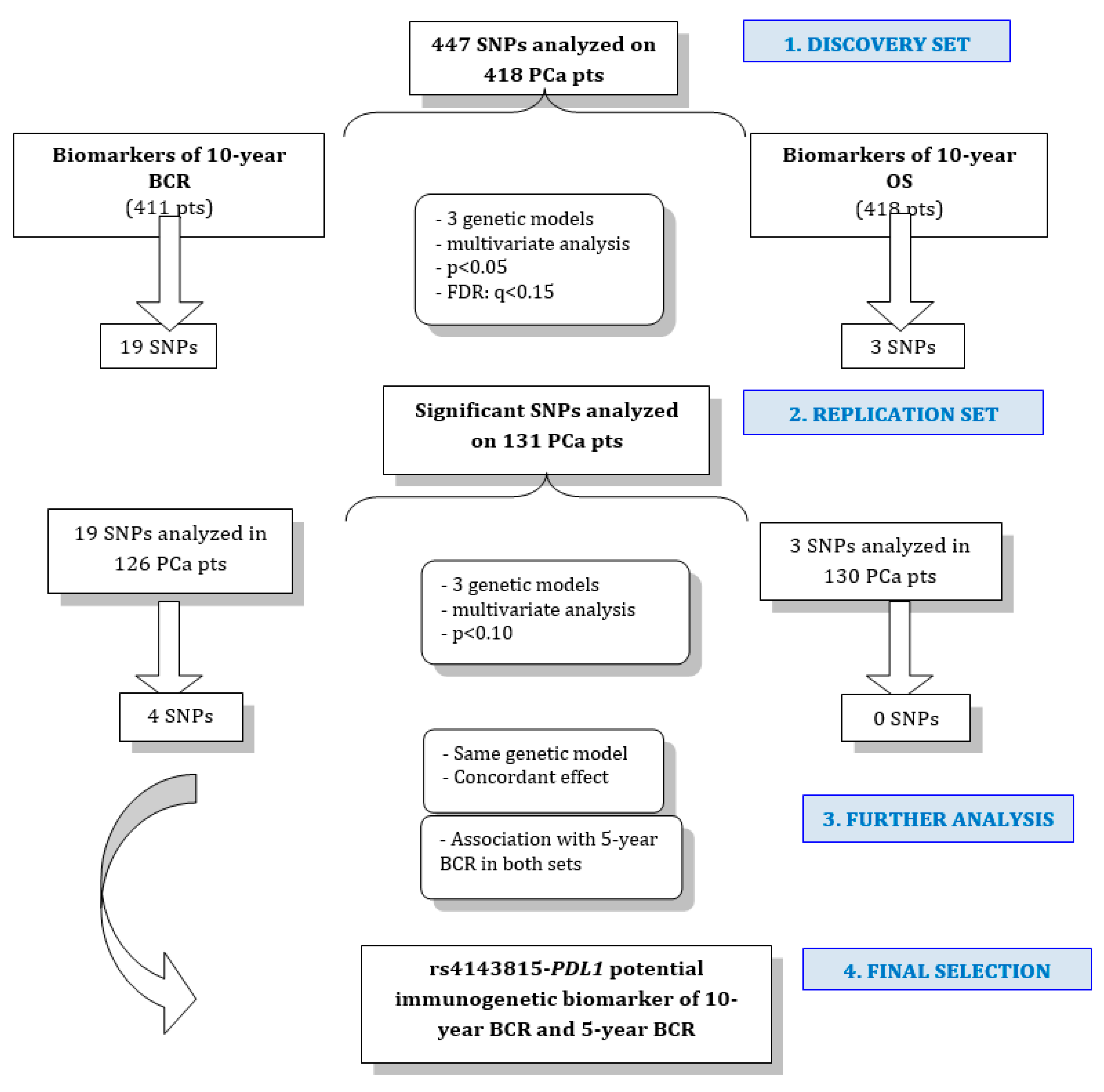
2. Results
2.1. Patient Characteristics and Genotyping
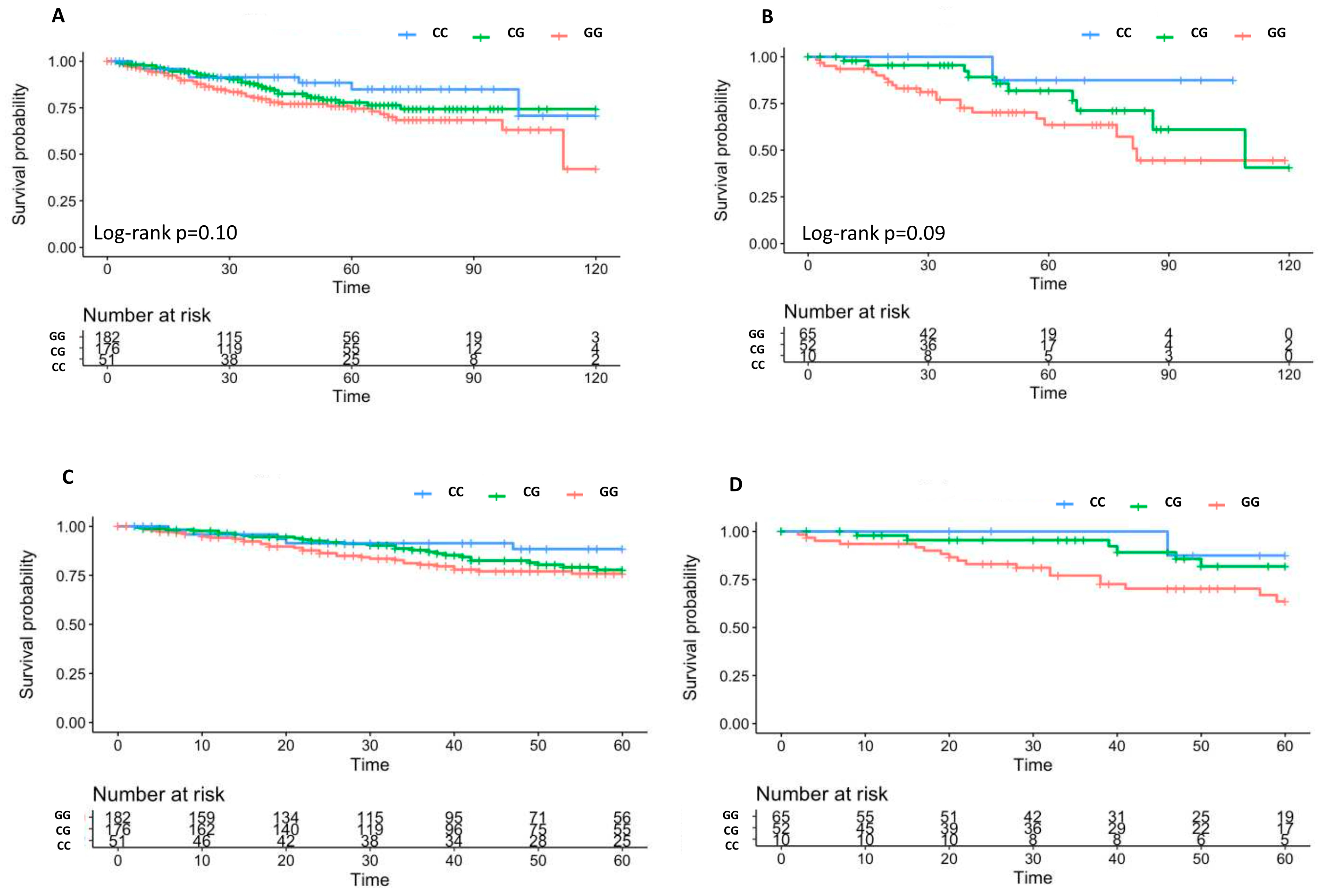
2.2. Associations of SNPs with 10 Year BCR
2.3. Associations of SNPs with 10 Year OS
3. Discussion
4. Materials and Methods
4.1. Patient Cohorts and Treatment
4.2. SNPs Selection
4.3. SNP Genotyping
4.4. Statistics: Identification of New Prognostic Biomarkers of BCR and OS
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Xie, W.; Regan, M.M.; Buyse, M.; Halabi, S.; Kantoff, P.W.; Sartor, O.; Soule, H.; Clarke, N.W.; Collette, L.; Dignam, J.J.; et al. Metastasis-Free Survival Is a Strong Surrogate of Overall Survival in Localized Prostate Cancer. J. Clin. Oncol. 2017, 35, 3097–3104. [Google Scholar] [CrossRef]
- Williams, S. Surrogate endpoints in early prostate cancer research. Transl. Androl. Urol. 2018, 7, 472–482. [Google Scholar] [CrossRef]
- De Langhe, S.; De Ruyck, K.; Ost, P.; Fonteyne, V.; Werbrouck, J.; De Meerleer, G.; De Neve, W.; Thierens, H. Acute radiation-induced nocturia in prostate cancer patients is associated with pretreatment symptoms, radical prostatectomy, and genetic markers in the TGFβ1 gene. Int. J. Radiat. Oncol. Biol. Phys. 2013, 85, 393–399. [Google Scholar] [CrossRef]
- Langsenlehner, T.; Thurner, E.M.; Renner, W.; Gerger, A.; Kapp, K.S.; Langsenlehner, U. Association of genetic variants in VEGF-A with clinical recurrence in prostate cancer patients treated with definitive radiotherapy. Strahlenther. Onkol. 2014, 190, 364–369. [Google Scholar] [CrossRef] [PubMed]
- Renner, W.; Langsenlehner, U.; Krenn-Pilko, S.; Eder, P.; Langsenlehner, T. BCL2 genotypes and prostate cancer survival. Strahlenther. Onkol. 2017, 193, 466–471. [Google Scholar] [CrossRef] [PubMed]
- Schoenfeld, J.D.; Margalit, D.N.; Kasperzyk, J.L.; Shui, I.M.; Rider, J.R.; Epstein, M.M.; Meisner, A.; Kenfield, S.A.; Martin, N.E.; Nguyen, P.L.; et al. A single nucleotide polymorphism in inflammatory gene RNASEL predicts outcome after radiation therapy for localized prostate cancer. Clin. Cancer Res. 2013, 19, 1612–1619. [Google Scholar] [CrossRef]
- Cushman, T.R.; Caetano, M.S.; Welsh, J.W.; Verma, V. Overview of ongoing clinical trials investigating combined radiotherapy and immunotherapy. Immunotherapy 2018, 10, 851–900. [Google Scholar] [CrossRef] [PubMed]
- Eze, C.; Manapov, F.; Gratzke, C.; Schmidt-Hegemann, N.S.; Jung, A.; Kirchner, T.; Heinemann, V.; Stief, C.G.; Belka, C.; Boeck, S. Concurrent radiotherapy and nivolumab in metachronous metastatic primary adenosquamous-cell carcinoma of the prostate. Eur. J. Cancer 2018, 95, 109–111. [Google Scholar] [CrossRef] [PubMed]
- Herbst, R.S.; Gordon, M.S.; Fine, G.D.; Sosman, J.A.; Soria, J.C.; Powderly, O.H.J.D.; Burris, H.A.; Mokatrin, A.; Kowanetz, M.; Leabman, M.; et al. A study of MPDL3280A, an engineered PD-L1 antibody in patients with locally advanced or metastatic tumors. JCO 2013, 31, 3000. [Google Scholar] [CrossRef]
- Calagua, C.; Russo, J.; Sun, Y.; Schaefer, R.; Lis, R.; Zhang, Z.; Mahoney, K.; Bubley, G.J.; Loda, M.; Taplin, M.E.; et al. Expression of PD-L1 in Hormone-naïve and Treated Prostate Cancer Patients Receiving Neoadjuvant Abiraterone Acetate plus Prednisone and Leuprolide. Clin. Cancer Res. 2017, 23, 6812–6822. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.S.; Mellman, I. Oncology meets immunology: the cancer-immunity cycle. Immunity 2013, 39, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Zou, W.; Chen, L. Inhibitory B7-family molecules in the tumour microenvironment. Nat. Rev. Immunol. 2008, 8, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Kwek, S.S.; Cha, E.; Fong, L. Unmasking the immune recognition of prostate cancer with CTLA4 blockade. Nat. Rev. Cancer 2012, 1, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Modena, A.; Ciccarese, C.; Iacovelli, R.; Brunelli, M.; Montironi, R.; Fiorentino, M.; Tortora, G.; Massari, F. Immune Checkpoint Inhibitors and Prostate Cancer: A New Frontier? Oncol. Rev. 2016, 10, 293. [Google Scholar] [CrossRef] [PubMed]
- Montironi, R.; Santoni, M.; Sotte, V.; Cheng, L.; Lopez-Beltran, A.; Massari, F.; Matrana, M.R.; Moch, H.; Berardi, R.; Scarpelli, M. Emerging Immunotargets and Immunotherapies in Prostate Cancer. Curr. Drug. Targets 2016, 17, 777–782. [Google Scholar] [CrossRef]
- Brahmer, J.R.; Drake, C.G.; Wollner, I.; Powderly, J.D.; Picus, J.; Sharfman, W.H.; Stankevich, E.; Pons, A.; Salay, T.M.; McMiller, T.L.; et al. Phase I study of single-agent anti-programmed death-1 (MDX-1106) in refractory solid tumors: safety, clinical activity, pharmacodynamics, and immunologic correlates. J. Clin. Oncol. 2010, 28, 3167–3175. [Google Scholar] [CrossRef] [PubMed]
- Topalian, S.L.; Hodi, F.S.; Brahmer, J.R.; Gettinger, S.N.; Smith, D.C.; McDermott, D.F.; Powderly, J.D.; Carvajal, R.D.; Sosman, J.A.; Atkins, M.B.; et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 2012, 366, 2443–2454. [Google Scholar] [CrossRef] [PubMed]
- Taube, J.M.; Klein, A.; Brahmer, J.R.; Xu, H.; Pan, X.; Kim, J.H.; Chen, L.; Pardoll, D.M.; Topalian, S.L.; Anders, R.A.; et al. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin. Cancer Res. 2014, 20, 5064–5074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Li, F.; Mao, Y.; Zhou, H.; Sun, J.; Li, R.; Liu, C.; Chen, W.; Hua, D.; Zhang, X. A miR-570 binding site polymorphism in the B7-H1 gene is associated with the risk of gastric adenocarcinoma. Hum. Genet. 2013, 132, 641–648. [Google Scholar] [CrossRef] [PubMed]
- Du, W.; Zhu, J.; Chen, Y.; Zeng, Y.; Shen, D.; Zhang, N.; Ning, W.; Liu, Z.; Huang, J.A. Variant SNPs at the microRNA complementary site in the B7-H1 3′-untranslated region increase the risk of non-small cell lung cancer. Mol. Med. Rep. 2017, 16, 2682–2690. [Google Scholar] [CrossRef] [Green Version]
- Yeo, M.K.; Choi, S.Y.; Seong, I.O.; Suh, K.S.; Kim, J.M.; Kim, K.H. Association of PD-L1 expression and PD-L1 gene polymorphism with poor prognosis in lung adenocarcinoma and squamous cell carcinoma. Hum. Pathol. 2017, 68, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Jung, D.K.; Choi, J.E.; Jin, C.C.; Hong, M.J.; Do, S.K.; Kang, H.G.; Lee, W.K.; Seok, Y.; Lee, E.B.; et al. Functional polymorphisms in PD-L1 gene are associated with the prognosis of patients with early stage non-small cell lung cancer. Gene 2017, 599, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Tao, L.H.; Zhou, X.R.; Li, F.C.; Chen, Q.; Meng, F.Y.; Mao, Y.; Li, R.; Hua, D.; Zhang, H.J.; Wang, W.P.; et al. A polymorphism in the promoter region of PD-L1 serves as a binding-site for SP1 and is associated with PD-L1 overexpression and increased occurrence of gastric cancer. Cancer Immunol. Immunother. 2017, 66, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Sun, J.; Li, F.; Li, R.; Gu, Y.; Liu, C.; Yang, P.; Zhu, M.; Chen, L.; Tian, W.; et al. A frequent somatic mutation in CD274 3′-UTR leads to protein over-expression in gastric cancer by disrupting miR-570 binding. Hum. Mutat. 2012, 33, 480–484. [Google Scholar] [CrossRef] [PubMed]
- Harada, K.; Ferdous, T.; Ueyama, Y. PD-L1 expression in malignant salivary gland tumors. BMC Cancer 2018, 18, 156. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.J.; Sun, W.P.; Peng, J.H.; Deng, Y.X.; Fang, Y.J.; Huang, J.; Zhang, H.; Wan, D.; Lin, J.; Pan, Z. Programmed death-ligand 1 expression correlates with diminished CD8+ T cell infiltration and predicts poor prognosis in anal squamous cell carcinoma patients. Cancer Manag. Res. 2018, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zanusso, C.; Bortolus, R.; Dreussi, E.; Polesel, J.; Montico, M.; Cecchin, E.; Gagno, S.; Rizzolio, F.; Arcicasa, M.; Novara, G.; et al. Impact of DNA repair gene polymorphisms on the risk of biochemical recurrence after radiotherapy and overall survival in prostate cancer. Oncotarget 2017, 8, 22863–22875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Characteristics | N | Training Set | Replication Set | Combined Datasets | Test Statistic |
|---|---|---|---|---|---|
| N = 418 | N = 131 | N = 549 | |||
| Basal parameters | |||||
| Age | 69.8 ± 5.6 | 70.6 ± 4.8 | 70.0 ± 5.5 | ||
| TNM Grade | 548 | χ24 = 3, p-value = 0.56 1 | |||
| 0 | 0.01 3⁄418 | 0.00 0⁄130 | 0.01 3⁄548 | ||
| 1 | 0.08 32⁄418 | 0.11 14⁄130 | 0.08 46⁄548 | ||
| 2 | 0.59 248⁄418 | 0.56 73⁄130 | 0.59 321⁄548 | ||
| 3 | 0.32 134⁄418 | 0.32 42⁄130 | 0.32 176⁄548 | ||
| 4 | 0.00 1⁄418 | 0.01 1⁄130 | 0.00 2⁄548 | ||
| Stage | 549 | χ23 = 6.8, p-value = 0.079 1 | |||
| A | 0.02 9⁄418 | 0.02 2⁄131 | 0.02 11⁄549 | ||
| B | 0.41 173⁄418 | 0.53 69⁄131 | 0.44 242⁄549 | ||
| C | 0.55 229⁄418 | 0.46 60⁄131 | 0.53 289⁄549 | ||
| D | 0.02 7⁄418 | 0.00 0⁄131 | 0.01 7⁄549 | ||
| Gleason score | 549 | χ22 = 0.69, p-value = 0.71 1 | |||
| 2–6 | 0.56 235⁄418 | 0.54 71⁄131 | 0.56 306⁄549 | ||
| 7 | 0.23 97⁄418 | 0.27 35⁄131 | 0.24 132⁄549 | ||
| 8–10 | 0.21 86⁄418 | 0.19 25⁄131 | 0.20 111⁄549 | ||
| D’Amico | 549 | χ22 = 2.6, p-value = 0.28 1 | |||
| 1 | 0.24 100⁄418 | 0.31 40⁄131 | 0.26 140⁄549 | ||
| 2 | 0.13 55⁄418 | 0.14 18⁄131 | 0.13 73⁄549 | ||
| 3 | 0.63 263⁄418 | 0.56 73⁄131 | 0.61 336⁄549 | ||
| PSA at diagnosis (ng/mL) | 549 | χ22 = 0.11, p-value = 0.95 1 | |||
| <4 | 0.06 27⁄418 | 0.07 9⁄131 | 0.07 36⁄549 | ||
| >10 | 0.43 180⁄418 | 0.44 58⁄131 | 0.43 238⁄549 | ||
| 4–10 | 0.50 211⁄418 | 0.49 64⁄131 | 0.50 275⁄549 | ||
| Treatment | |||||
| Hormone therapy | 549 | 0.86 359⁄418 | 0.87 114⁄131 | 0.86 473⁄549 | χ21 = 0.11, p-value = 0.74 1 |
| RT dose (cGy) | 549 | χ24 = 4.3, p-value = 0.36 1 | |||
| 6600 | 0.01 4⁄418 | 0.00 0⁄131 | 0.01 4⁄549 | ||
| 7000 | 0.09 36⁄418 | 0.05 7⁄131 | 0.08 43⁄549 | ||
| 7400 | 0.12 50⁄418 | 0.11 14⁄131 | 0.12 64⁄549 | ||
| 7600 | 0.74 311⁄418 | 0.82 107⁄131 | 0.76 418⁄549 | ||
| 8000 | 0.04 17⁄418 | 0.02 3⁄131 | 0.04 20⁄549 | ||
| Follow-up | |||||
| BCR | 549 | 0.19 80⁄418 | 0.24 32⁄131 | 0.20 112⁄549 | χ21 = 1.7, p-value = 0.19 1 |
| Follow-up BCR | 538 | 22 (43–70) | 21 (46–68) | 22 (45–70) | F1 536 = 0.11, p-value = 0.74 2 |
| 47 ± 30 | 46 ± 31 | 47 ± 30 | |||
| Death | 549 | 0.18 76⁄418 | 0.19 25⁄131 | 0.18 101⁄549 | χ21 = 0.05, p-value = 0.82 1 |
| Follow-up OS | 549 | 45 (68–94) | 43 (65–90) | 44 (68–94) | F1 547 = 1.2, p-value = 0.26 2 |
| 70 ± 31 | 66 ± 32 | 69 ± 32 | |||
| Gene | SNP | Base Change | M | Discovery Cohort (n = 411) * | Replication Cohort (n = 126) * | |||
|---|---|---|---|---|---|---|---|---|
| HR (95% CI) | p-Value | q-Value | HR (95% CI) | p-Value | ||||
| IL2RB | rs84460 | C/T | R | 2.79 (1.34–5.81) | 0.006 | 0.121 | 0.96 (0.12–7.42) | 0.97 |
| SMAD3 | rs7162912 | G/T | A | 2.04 (1.43–2.92) | 0.0001 | 0.001 | 0.80 (0.44–1.45) | 0.46 |
| FOXO3 | rs7762395 | A/G | A | 1.91 (1.26–2.89) | 0.002 | 0.121 | 0.74 (0.31–1.79) | 0.51 |
| FOXO3 | rs2153960 | C/T | A | 1.83 (1.27–2.64) | 0.001 | 0.101 | 1.11 (0.63–1.98) | 0.71 |
| SMAD3 | rs9302242 | A/G | R | 2.03 (1.24–3.32) | 0.005 | 0.128 | 1.81 (0.82–4.02) | 0.14 |
| IL4R | rs1805011 | A/C | D | 2.28 (1.32–3.92) | 0.003 | 0.121 | 1.32 (0.51–3.42) | 0.57 |
| IL4R | rs3024586 | A/G | D | 2.56 (1.36–4.80) | 0.003 | 0.121 | 0.56 (0.15–2.15) | 0.40 |
| CCL5 | rs2280789 | C/T | R | 7.93 (2.31–27.15) | 0.001 | 0.101 | 0.70 (0.09–5.74) | 0.74 |
| TLR2 | rs3804099 | C/T | A | 1.71 (1.24–2.36) | 0.001 | 0.109 | 1.39 (0.74–2.61) | 0.30 |
| PDL1 | rs1411262 | A/G | A | 1.6 (1.15–2.28) | 0.005 | 0.123 | 0.48 (0.24–0.97) | 0.04 |
| PDL1 | rs10125854 | A/G | D | 3.01 (1.66–5.45) | 0.0003 | 0.065 | 0.54 (0.17–1.70) | 0.30 |
| PDL1 | rs4143815 | C/G | A | 0.58 (0.41–0.83) | 0.003 | 0.117 | 0.52 (0.26–1.04) | 0.06 |
| SMAD2 | rs1792666 | A/T | D | 2.09 (1.25–3.50) | 0.005 | 0.128 | 2.99 (1.10–8.12) | 0.03 |
| SMAD2 | rs4940086 | C/T | A | 1.65 (1.18–2.32) | 0.004 | 0.117 | 0.29 (0.14–0.59) | 0.0007 |
| STAT1 | rs16824035 | C/T | R | 3.97 (1.68–9.39) | 0.002 | 0.117 | 0 (0; inf) | 1.00 |
| VEGFR2 | rs12498529 | A/T | R | 4.26 (1.64–11.06) | 0.003 | 0.121 | 1.45 (0.31–6.80) | 0.64 |
| VEGFR2 | rs2034965 | A/G | A | 1.60 (1.14–2.27) | 0.007 | 0.154 | 0.74 (0.41–1.33) | 0.32 |
| VEGFR2 | rs4576072 | C/T | R | 4.67 (1.59–13.74) | 0.005 | 0.128 | 1.76 (0.38–8.22) | 0.47 |
| AKT2/miR641 | rs11880261 | C/T | A | 0.64 (0.44–0.92) | 0.02 | 0.128 | 1.33 (0.74–2.36) | 0.34 |
| Gene | SNP | Base Change | Discovery Cohort (n = 418) * | Replication Cohort (n = 130) * | ||||
|---|---|---|---|---|---|---|---|---|
| M | HR (95% CI) | p-Value | q-Value | HR (95% CI) | p-Value | |||
| MMP9 | rs3918262 | A/G | R | 4.31 (1.81–10.26) | 0.001 | 0.14 | 0 (0–Inf) | 1.00 |
| VEGFR2 | rs7692791 | C/T | A | 1.82 (1.31–2.53) | 0.0003 | 0.14 | 1.09 (0.58–2.03) | 0.79 |
| VEGFR2 | rs2034967 | C/T | D | 0.45 (0.28–0.72) | 0.00000008 | 0.14 | 0.68 (0.30–1.56) | 0.36 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zanusso, C.; Dreussi, E.; Bortolus, R.; Romualdi, C.; Gagno, S.; De Mattia, E.; Romanato, L.; Sartor, F.; Quartuccio, L.; Cecchin, E.; et al. rs4143815-PDL1, a New Potential Immunogenetic Biomarker of Biochemical Recurrence in Locally Advanced Prostate Cancer after Radiotherapy. Int. J. Mol. Sci. 2019, 20, 2082. https://doi.org/10.3390/ijms20092082
Zanusso C, Dreussi E, Bortolus R, Romualdi C, Gagno S, De Mattia E, Romanato L, Sartor F, Quartuccio L, Cecchin E, et al. rs4143815-PDL1, a New Potential Immunogenetic Biomarker of Biochemical Recurrence in Locally Advanced Prostate Cancer after Radiotherapy. International Journal of Molecular Sciences. 2019; 20(9):2082. https://doi.org/10.3390/ijms20092082
Chicago/Turabian StyleZanusso, Chiara, Eva Dreussi, Roberto Bortolus, Chiara Romualdi, Sara Gagno, Elena De Mattia, Loredana Romanato, Franca Sartor, Luca Quartuccio, Erika Cecchin, and et al. 2019. "rs4143815-PDL1, a New Potential Immunogenetic Biomarker of Biochemical Recurrence in Locally Advanced Prostate Cancer after Radiotherapy" International Journal of Molecular Sciences 20, no. 9: 2082. https://doi.org/10.3390/ijms20092082





