High Homology-Directed Repair Using Mitosis Phase and Nucleus Localizing Signal
Abstract
:1. Introduction
2. Results
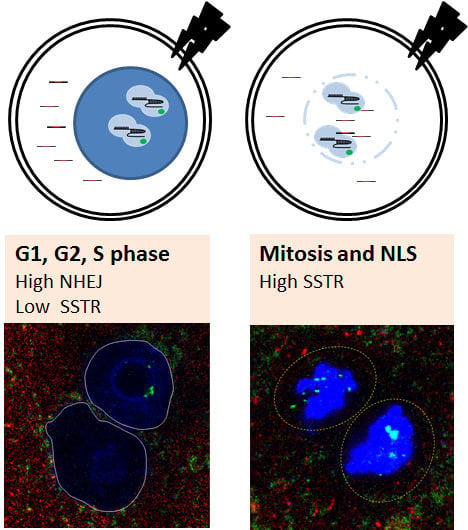
2.1. Embryonic Cell Cycle Analysis by Nuclear Envelope Morphology
2.2. Mitosis-Specific High SSTR Efficiency
2.3. The Nuclear Envelope as A Major Barrier to the Nuclear Import of ssODNs
2.4. High SSTR Efficiency in Mitotic Synchronous Embryos and Cells
2.5. Minimal Impact of ssODN Size and Cdk1 on SSTR Efficiency
2.6. Improving SSTR Efficiency Using NLS-tagged ssODNs
3. Discussion
4. Materials and Methods
4.1. Animal and Embryo Preparation
4.2. Embryonic Whole-Mount Immunostaining for Cell Cycle Determination
4.3. In silico sgRNA Design and Homologous Template Preparation
4.4. Electroporation-Mediated Ribonucleoprotein Transfection into Embryos and Genotyping
4.5. Nuclear Localization Analysis for Cas9 Protein and ssODNs
4.6. Establishing SpCas9 Overexpression NIH3T3 Cells
4.7. Preparation of Modified ssODNs and Cdk1
4.8. Cell Cycle Synchronization and Transfection of Cells by Electroporation
4.9. Targeted Deep Sequencing
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Richardson, C.D.; Kazane, K.R.; Feng, S.J.; Zelin, E.; Bray, N.L.; Schafer, A.J.; Floor, S.N.; Corn, J.E. CRISPR-Cas9 genome editing in human cells occurs via the Fanconi anemia pathway. Nat. Genet. 2018, 50, 1132–1139. [Google Scholar] [CrossRef] [PubMed]
- Renaud, J.B.; Boix, C.; Charpentier, M.; De Cian, A.; Cochennec, J.; Duvernois-Berthet, E.; Perrouault, L.; Tesson, L.; Edouard, J.; Thinard, R.; et al. Improved Genome Editing Efficiency and Flexibility Using Modified Oligonucleotides with TALEN and CRISPR-Cas9 Nucleases. Cell Rep. 2016, 14, 2263–2272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bertoni, C.; Rustagi, A.; Rando, T.A. Enhanced gene repair mediated by methyl-CpG-modified single-stranded oligonucleotides. Nucleic Acids Res. 2009, 37, 7468–7482. [Google Scholar] [CrossRef] [Green Version]
- Riesenberg, S.; Maricic, T. Targeting repair pathways with small molecules increases precise genome editing in pluripotent stem cells. Nat. Commun. 2018, 9, 2164. [Google Scholar] [CrossRef] [Green Version]
- Jang, D.E.; Lee, J.Y.; Lee, J.H.; Koo, O.J.; Bae, H.S.; Jung, M.H.; Bae, J.H.; Hwang, W.S.; Chang, Y.J.; Lee, Y.H.; et al. Multiple sgRNAs with overlapping sequences enhance CRISPR/Cas9-mediated knock-in efficiency. Exp. Mol. Med. 2018, 50, 16. [Google Scholar] [CrossRef] [Green Version]
- Rothkamm, K.; Kruger, I.; Thompson, L.H.; Lobrich, M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol. Cell. Biol. 2003, 23, 5706–5715. [Google Scholar] [CrossRef] [Green Version]
- Arnoult, N.; Correia, A.; Ma, J.; Merlo, A.; Garcia-Gomez, S.; Maric, M.; Tognetti, M.; Benner, C.W.; Boulton, S.J.; Saghatelian, A.; et al. Regulation of DNA repair pathway choice in S and G2 phases by the NHEJ inhibitor CYREN. Nature 2017, 549, 548–552. [Google Scholar] [CrossRef] [Green Version]
- Lin, S.; Staahl, B.T.; Alla, R.K.; Doudna, J.A. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. Elife 2014, 3, e04766. [Google Scholar] [CrossRef]
- Yang, D.; Scavuzzo, M.A.; Chmielowiec, J.; Sharp, R.; Bajic, A.; Borowiak, M. Enrichment of G2/M cell cycle phase in human pluripotent stem cells enhances HDR-mediated gene repair with customizable endonucleases. Sci. Rep. 2016, 6, 21264. [Google Scholar] [CrossRef] [Green Version]
- Chazaud, C.; Yamanaka, Y. Lineage specification in the mouse preimplantation embryo. Development 2016, 143, 1063–1074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Zhao, H.; Dan, J.; Kim, S.; Hardikar, S.; Hollowell, D.; Lin, K.; Lu, Y.; Takata, Y.; Shen, J.; et al. Maternal Setdb1 Is Required for Meiotic Progression and Preimplantation Development in Mouse. PLoS Genet. 2016, 12, e1005970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wossidlo, M.; Nakamura, T.; Lepikhov, K.; Marques, C.J.; Zakhartchenko, V.; Boiani, M.; Arand, J.; Nakano, T.; Reik, W.; Walter, J. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat. Commun. 2011, 2, 241. [Google Scholar] [CrossRef] [Green Version]
- Kang, E.; Wu, G.; Ma, H.; Li, Y.; Tippner-Hedges, R.; Tachibana, M.; Sparman, M.; Wolf, D.P.; Scholer, H.R.; Mitalipov, S. Nuclear reprogramming by interphase cytoplasm of two-cell mouse embryos. Nature 2014, 509, 101–104. [Google Scholar] [CrossRef] [Green Version]
- Schwinn, M.K.; Machleidt, T.; Zimmerman, K.; Eggers, C.T.; Dixon, A.S.; Hurst, R.; Hall, M.P.; Encell, L.P.; Binkowski, B.F.; Wood, K.V. CRISPR-Mediated Tagging of Endogenous Proteins with a Luminescent Peptide. ACS Chem. Biol. 2018, 13, 467–474. [Google Scholar] [CrossRef]
- Signoretto, E.; Honisch, S.; Briglia, M.; Faggio, C.; Castagna, M.; Lang, F. Nocodazole Induced Suicidal Death of Human Erythrocytes. Cell Physiol. Biochem. 2016, 38, 379–392. [Google Scholar] [CrossRef]
- Gavet, O.; Pines, J. Activation of cyclin B1-Cdk1 synchronizes events in the nucleus and the cytoplasm at mitosis. J. Cell Biol. 2010, 189, 247–259. [Google Scholar] [CrossRef]
- Adli, M. The CRISPR tool kit for genome editing and beyond. Nat. Commun. 2018, 9, 1911. [Google Scholar] [CrossRef]
- Wiernert, B.; Feng, S.J.; Locke, M.; Nguyen, D.N.; Wyman, S.K.; Kazane, K.R.; Marson, A.; Richardson, C.D.; Corn, J.E. Timed inhibition of CDC7 increases CRISPR-Cas9 mediated templated repair. bioRxiv 2018. [Google Scholar] [CrossRef]
- Rodriguez, A.; D’Andrea, A. Fanconi anemia pathway. Curr. Biol. 2017, 27, R986–R988. [Google Scholar] [CrossRef] [Green Version]
- Nalepa, G.; Clapp, D.W. Fanconi anemia and the cell cycle: New perspectives on aneuploidy. F1000Prime Rep 2014, 6, 23. [Google Scholar] [CrossRef] [PubMed]
- Kee, Y.; Kim, J.M.; D’Andrea, A.D. Regulated degradation of FANCM in the Fanconi anemia pathway during mitosis. Genes Dev. 2009, 23, 555–560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giraud, G.; Stadhouders, R.; Conidi, A.; Dekkers, D.H.; Huylebroeck, D.; Demmers, J.A.; Soler, E.; Grosveld, F.G. NLS-tagging: An alternative strategy to tag nuclear proteins. Nucleic Acids Res. 2014, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanimoto, M.; Kamiya, H.; Minakawa, N.; Matsuda, A.; Harashima, H. No enhancement of nuclear entry by direct conjugation of a nuclear localization signal peptide to linearized DNA. Bioconjug Chem. 2003, 14, 1197–1202. [Google Scholar] [CrossRef] [PubMed]
- Lou, C.; Martos-Maldonado, M.C.; Madsen, C.S.; Thomsen, R.P.; Midtgaard, S.R.; Christensen, N.J.; Kjems, J.; Thulstrup, P.W.; Wengel, J.; Jensen, K.J. Peptide-oligonucleotide conjugates as nanoscale building blocks for assembly of an artificial three-helix protein mimic. Nat. Commun. 2016, 7, 12294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, B.; Posfai, E.; Rossant, J. Efficient generation of targeted large insertions by microinjection into two-cell-stage mouse embryos. Nat. Biotechnol. 2018, 36, 632–637. [Google Scholar] [CrossRef]
- Aird, E.J.; Lovendahl, K.N.; St Martin, A.; Harris, R.S.; Gordon, W.R. Increasing Cas9-mediated homology-directed repair efficiency through covalent tethering of DNA repair template. Commun. Biol. 2018, 1, 54. [Google Scholar] [CrossRef] [Green Version]
- Savic, N.; Ringnalda, F.C.; Lindsay, H.; Berk, C.; Bargsten, K.; Li, Y.; Neri, D.; Robinson, M.D.; Ciaudo, C.; Hall, J.; et al. Covalent linkage of the DNA repair template to the CRISPR-Cas9 nuclease enhances homology-directed repair. Elife 2018, 7. [Google Scholar] [CrossRef]
- Wang, D.; Tai, P.W.L.; Gao, G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat. Rev. Drug Discov. 2019. [Google Scholar] [CrossRef]
- Zhu, J.Y.; Wan, S.S.; Zheng, D.W.; Lei, Q.; Zhuo, R.X.; Feng, J.; Zhang, X.Z. Propelled Transnuclear Gene Transport Achieved through Intracellularly Redox-Responsive and Acidity-Accelerative Decomposition of Supramolecular Florescence-Quenchable Vectors. ACS Appl. Mater. Interfaces 2017, 9, 255–265. [Google Scholar] [CrossRef]
- Hashimoto, M.; Yamashita, Y.; Takemoto, T. Electroporation of Cas9 protein/sgRNA into early pronuclear zygotes generates non-mosaic mutants in the mouse. Dev. Biol. 2016, 418, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Lim, K.; Kim, J.S.; Bae, S. Cas-analyzer: An online tool for assessing genome editing results using NGS data. Bioinformatics 2016. [Google Scholar] [CrossRef] [PubMed] [Green Version]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, J.P.; Chang, Y.J.; Song, D.W.; Choi, B.S.; Koo, O.J.; Yi, S.Y.; Park, T.S.; Yeom, S.C. High Homology-Directed Repair Using Mitosis Phase and Nucleus Localizing Signal. Int. J. Mol. Sci. 2020, 21, 3747. https://doi.org/10.3390/ijms21113747
Han JP, Chang YJ, Song DW, Choi BS, Koo OJ, Yi SY, Park TS, Yeom SC. High Homology-Directed Repair Using Mitosis Phase and Nucleus Localizing Signal. International Journal of Molecular Sciences. 2020; 21(11):3747. https://doi.org/10.3390/ijms21113747
Chicago/Turabian StyleHan, Jeong Pil, Yoo Jin Chang, Dong Woo Song, Beom Seok Choi, Ok Jae Koo, Seung Youn Yi, Tae Sub Park, and Su Cheong Yeom. 2020. "High Homology-Directed Repair Using Mitosis Phase and Nucleus Localizing Signal" International Journal of Molecular Sciences 21, no. 11: 3747. https://doi.org/10.3390/ijms21113747