Expression and Role of Response Regulating, Biosynthetic and Degrading Genes for Cytokinin Signaling during Clubroot Disease Development
Abstract
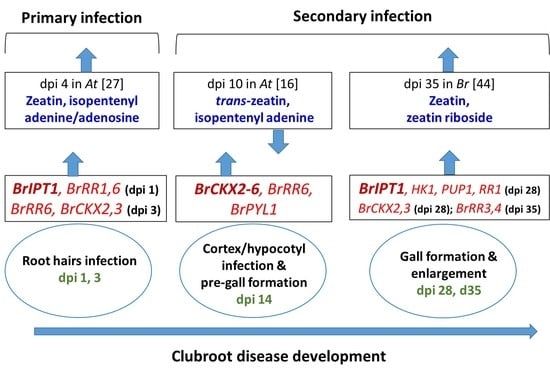
:1. Introduction
2. Results
2.1. Disease Severity Index of Four Korean Field Isolates
2.2. Properties of Cytokinin-Related Proteins
2.3. Expression Level Difference of Cytokinin-Related Genes in Leaf vs. Root
2.4. Expression Profiles of Response Regulator Genes
2.5. Expression Profiles of Other Cytokinin-Related Genes
2.6. Expression Profiles of Cytokinin Oxidase Genes
3. Discussion
3.1. Expression Level Difference in Leaf vs. Root Tissue
3.2. Expression Level Difference of BrIPT1 Gene
3.3. Expression Level Difference of BrRR Genes
3.4. Expression Level of CKX Genes
4. Materials and Methods
4.1. Preparation of Plant Materials
4.2. Gall Sample Collection
4.3. Spore Preparation from Galls and Infection of Plant Materials
4.4. Disease Scoring Criteria
4.5. Collection of Leaf and Root Samples from Plants Inoculated with Seosan Isolate
4.6. RNA Extraction and cDNA Synthesis
4.7. Identification of Cytokinin-Related Genes
4.8. qRT-PCR Expression Analysis
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dixon, G.R. The occurrence and economic impact of Plasmodiophora brassicae and clubroot disease. J. Plant Growth Regul. 2009, 28, 194–202. [Google Scholar] [CrossRef]
- Kim, H.; Jo, E.J.; Choi, Y.H.; Jang, K.S.; Choi, G.J. Pathotype Classification of Plasmodiophora brassicae Isolates Using Clubroot-Resistant Cultivars of Chinese Cabbage. Plant Pathol. J. 2016, 32, 423. [Google Scholar] [CrossRef] [Green Version]
- Williams, P.H. A system for the determination of races of Plasmodiophora brassicae that infect cabbage and rutabaga. Phytopathology 1966, 56, 624–626. [Google Scholar]
- Laila, R.; Robin, A.H.K.; Yang, K.; Choi, G.J.; Park, J.I.; Nou, I.S. Detection of Ribosomal DNA sequence polymorphisms in the protist Plasmodiophora brassicae for the identification of geographical isolates. Int. J. Mol. Sci. 2017, 18, 84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kageyama, K.; Asano, T. Life cycle of Plasmodiophora brassicae. J. Plant Growth Regul 2009, 28, 203. [Google Scholar] [CrossRef]
- Malinowski, R.; Smith, J.A.; Fleming, A.J.; Scholes, J.D.; Rolfe, S.A. Gall formation in clubroot-infected Arabidopsis results from an increase in existing meristematic activities of the host but is not essential for the completion of the pathogen life cycle. Plant J. 2012, 71, 226–238. [Google Scholar] [CrossRef]
- Donald, C.; Porter, I. Integrated control of clubroot. J. Plant Growth Regul. 2009, 28, 289. [Google Scholar] [CrossRef]
- Dixon, G.R. (2014). Clubroot (Plasmodiophora brassicae Woronin)–an agricultural and biological challenge worldwide. Can. J. Plant Path. 2009, 36 (Suppl. 1), 5–18. [Google Scholar] [CrossRef]
- Feng, J.; Hwang, S.F.; Strelkov, S.E. Studies into primary and secondary infection processes by Plasmodiophora brassicae on canola. Plant Path. 2013, 62, 177–183. [Google Scholar] [CrossRef]
- McDonald, M.R.; Sharma, K.; Gossen, B.D.; Deora, A.; Feng, J.; Hwang, S.F. The role of primary and secondary infection in host response to Plasmodiophora brassicae. Phytopathology 2014, 104, 1078–1087. [Google Scholar] [CrossRef] [Green Version]
- Hwang, S.F.; Strelkov, S.E.; Feng, J.I.E.; Gossen, B.D.; Howard, R.J. Plasmodiophora brassicae: A review of an emerging pathogen of the Canadian canola (Brassica napus) crop. Mol. Plant Path. 2012, 13, 105–113. [Google Scholar] [CrossRef]
- Deora, A.; Gossen, B.D.; McDonald, M.R. Cytology of infection, development and expression of resistance to Plasmodiophora brassicae in canola. Ann. Appl. Biol. 2013, 163, 56–71. [Google Scholar] [CrossRef]
- Robin, A.H.K.; Hossain, M.R.; Kim, H.T.; Nou, I.S.; Park, J.I. Role of Cytokinins in Clubroot Disease Development. Plant Breed. Biotech. 2019, 7, 73–82. [Google Scholar] [CrossRef]
- Müller, P.; Hilgenberg, W. Isomers of zeatin and zeatin riboside in clubroot tissue: Evidence for trans-zeatin biosynthesis by Plasmodiophora brassicae. Physiologia Plantarum. 1986, 66, 245–250. [Google Scholar] [CrossRef]
- Dekhuijzen, H.M. The occurrence of free and bound cytokinins in plasmodia of Plasmodiophora brassicae isolated from tissue cultures of clubroots. Plant Cell Rep. 1981, 1, 18–20. [Google Scholar] [CrossRef] [PubMed]
- Malinowski, R.; Novák, O.; Borhan, M.H.; Spíchal, L.; Strnad, M.; Rolfe, S.A. The role of cytokinins in clubroot disease. Eur. J. Plant Path. 2016, 145, 543–557. [Google Scholar] [CrossRef] [Green Version]
- Prerostova, S.; Dobrev, P.I.; Konradyova, V.; Knirsch, V.; Gaudinova, A.; Kramna, B.; Kazda, J.; Ludwig-Müller, J.; Vankova, R. Hormonal responses to Plasmodiophora brassicae infection in Brassica napus cultivars differing in their pathogen resistance. Int. J. Mol. Sci. 2018, 19, 4024. [Google Scholar] [CrossRef] [Green Version]
- Dekhuijzen, H.M.; Overeem, J.C. The role of cytokinins in clubroot formation. Physiol. Plant Path. 1971, 1, 151–161. [Google Scholar] [CrossRef]
- Giron, D.; Kaiser, W.; Imbault, N.; Casas, J. Cytokinin-mediated leaf manipulation by a leaf miner caterpillar. Biol Lett. 2007, 3, 340–343. [Google Scholar] [CrossRef] [Green Version]
- Boivin, S.; Fonouni-Farde, C.; Frugier, F. How auxin and cytokinin phytohormones modulate root microbe interactions. Front. Plant Sci. 2016, 7, 1240. [Google Scholar] [CrossRef] [Green Version]
- Ludwig-Müller, J.; Auer, S.; Jülke, S.; Marschollek, S. Manipulation of Auxin and Cytokinin Balance during the Plasmodiophora brassicae–Arabidopsis Thaliana Interaction. In Auxins and Cytokinins in Plant Biology; Humana Press: New York, NY, USA, 2017; pp. 41–60. [Google Scholar]
- Ludwig-Müller, J.; Rolfe, S.A.; Scholes, J.D. Metabolism and plant hormone action during clubroot disease metabolism and plant hormone action during clubroot disease. J Plant Growth Regul. 2009, 28, 229. [Google Scholar] [CrossRef]
- Schuller, A.; Ludwig-Müller, J. A family of auxin conjugate hydrolases from Brassica rapa: Characterization and expression during clubroot disease. New Phytol. 2006, 171, 145–158. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Wei, X.; Yang, Y.; Yuan, Y.; Wei, F.; Zhao, Y.; Yang, S.; Yao, Q.; Wang, Z.; Tian, B.; et al. Root RNA-seq analysis reveals a distinct transcriptome landscape between clubroot-susceptible and clubroot-resistant Chinese cabbage lines after Plasmodiophora brassicae infection. Plant Soil. 2017, 421, 93–105. [Google Scholar] [CrossRef]
- Ciaghi, S.; Schwelm, A.; Neuhauser, S. Transcriptomic response in symptomless roots of clubroot infected kohlrabi (Brassica oleracea var. gongylodes) mirrors resistant plants. BMC Plant Biol. 2019, 19, 288. [Google Scholar]
- Siemens, J.; González, M.C.; Wolf, S.; Hofmann, C.; Greiner, S.; Du, Y.; Rausch, T.; Roitsch, T.; Ludwig-Müller, J.U. Extracellular invertase is involved in the regulation of clubroot disease in Arabidopsis thaliana. Mol. Plant Path. 2011, 12, 247–262. [Google Scholar] [CrossRef]
- Devos, S.; Laukens, K.; Deckers, P.; Van Der Straeten, D.; Beeckman, T.; Inzé, D.; Van Onckelen, H.; Witters, E.; Prinsen, E. A hormone and proteome approach to picturing the initial metabolic events during Plasmodiophora brassicae infection on Arabidopsis. Mol Plant Microbe Interact. 2006, 19, 1431–1443. [Google Scholar] [CrossRef] [Green Version]
- Siemens, J.; Keller, I.; Sarx, J.; Kunz, S.; Schuller, A.; Nagel, W.; Schmülling, T.; Parniske, M.; Ludwig-Müller, J. Transcriptome analysis of Arabidopsis clubroots indicate a key role for cytokinins in disease development. Mol. Plant Microbe Interact. 2006, 19, 480–494. [Google Scholar] [CrossRef] [Green Version]
- Schuller, A.; Kehr, J.; Ludwig-Müller, J. Laser microdissection coupled to transcriptional profiling of Arabidopsis roots inoculated by Plasmodiophora brassicae indicates a role for brassinosteroids in clubroot formation. Plant Cell Physiol. 2013, 55, 392–411. [Google Scholar] [CrossRef]
- Agarwal, A.; Kaul, V.; Faggian, R.; Rookes, J.E.; Ludwig-Müller, J.; Cahill, D.M. Analysis of global host gene expression during the primary phase of the Arabidopsis thaliana–Plasmodiophora brassicae interaction. Functional Plant Biol. 2011, 38, 462–478. [Google Scholar] [CrossRef] [Green Version]
- Lan, M.; Li, G.; Hu, J.; Yang, H.; Zhang, L.; Xu, X.; Liu, J.; He, J.; Sun, R. iTRAQ-based quantitative analysis reveals proteomic changes in Chinese cabbage (Brassica rapa L.) in response to Plasmodiophora brassicae infection. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef]
- Gan, S.; Amasino, R.M. Inhibition of leaf senescence by autoregulated production of cytokinin. Science 1995, 270, 1986–1988. [Google Scholar] [CrossRef] [PubMed]
- Mok, D.W.; Mok, M.C. Cytokinin metabolism and action. Annu. Rev. Plant Biol. 2001, 52, 89–118. [Google Scholar] [CrossRef] [PubMed]
- Sakakibara, H. Cytokinins: Activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 2006, 57, 431–449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Werner, T.; Motyka, V.; Laucou, V.; Smets, R.; Van Onckelen, H.; Schmülling, T. Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 2003, 15, 2532–2550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Werner, T.; Motyka, V.; Strnad, M.; Schmülling, T. Regulation of plant growth by cytokinin. Proc. Natl. Acad. Sci. USA 2001, 98, 10487–10492. [Google Scholar] [CrossRef] [Green Version]
- Romanov, G.A.; Kieber, J.J.; Schmülling, T. A rapid cytokinin response assay in Arabidopsis indicates a role for phospholipase D in cytokinin signalling. FEBS Lett. 2002, 515, 39–43. [Google Scholar] [CrossRef] [Green Version]
- El-Showk, S.; Ruonala, R.; Helariutta, Y. Crossing paths: Cytokinin signalling and crosstalk. Development. 2013, 140, 1373–1383. [Google Scholar] [CrossRef] [Green Version]
- Rivero, R.M.; Shulaev, V.; Blumwald, E. Cytokinin-dependent photorespiration and the protection of photosynthesis during water deficit. Plant Physiol. 2009, 150, 1530–1540. [Google Scholar] [CrossRef] [Green Version]
- Qin, H.; Gu, Q.; Zhang, J.; Sun, L.; Kuppu, S.; Zhang, Y.; Burow, M.; Payton, P.; Blumwald, E.; Zhang, H. Regulated expression of an isopentenyltransferase gene (IPT) in peanut significantly improves drought tolerance and increases yield under field conditions. Plant Cell Physiol. 2011, 52, 1904–1914. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Lv, Y.; Zhang, M.; Liu, Y.; Kong, L.; Zou, M.; Lu, G.; Cao, J.; Yu, X. Identification, expression, and comparative genomic analysis of the IPT and CKX gene families in Chinese cabbage (Brassica rapa ssp. pekinensis). Bmc Genom. 2013, 14, 594. [Google Scholar] [CrossRef] [Green Version]
- Ando, S.; Asano, T.; Tsushima, S.; Kamachi, S.; Hagio, T.; Tabei, Y. Changes in gene expression of putative isopentenyltransferase during clubroot development in Chinese cabbage (Brassica rapa L.). Physiol. Mol. Plant Pathol. 2005, 67, 59–67. [Google Scholar] [CrossRef]
- Miyawaki, K.; Tarkowski, P.; Matsumoto-Kitano, M.; Kato, T.; Sato, S.; Tarkowska, D.; Tabata, S.; Sandberg, G.; Kakimoto, T. Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc. Nat. Acad. Sci. USA 2006, 103, 16598–16603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dekhuijzen, H.M. The occurrence of free and bound cytokinins in clubroots and Plasmodiophora brassicae infected turnip tissue cultures. Physiol. Plant. 1980, 49, 169–176. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, M.; Kong, L.; Lv, Y.; Zou, M.; Lu, G.; Cao, J.; Yu, X. Genome-wide identification, phylogeny, duplication, and expression analyses of two-component system genes in Chinese cabbage (Brassica rapa ssp. pekinensis). DNA Res. 2014, 21, 379–396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schaller, G.E.; Kieber, J.J.; Shiu, S.H. Two-component signaling elements and histidyl-aspartyl phosphorelays. Arabidopsis Book 2008, 6, e0112. [Google Scholar] [CrossRef] [Green Version]
- Zubo, Y.O.; Schaller, G.E. Role of the Cytokinin-Activated Type-B Response Regulators in Hormone Crosstalk. Plants 2020, 9, 166. [Google Scholar] [CrossRef] [Green Version]
- Hwang, I.; Sheen, J. Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 2001, 413, 383–389. [Google Scholar] [CrossRef]
- Sweere, U.; Eichenberg, K.; Lohrmann, J.; Mira-Rodado, V.; Bäurle, I.; Kudla, J.; Nagy, F.; Schäfer, E.; Harter, K. Interaction of the response regulator ARR4 with phytochrome B in modulating red light signaling. Science 2001, 294, 1108–1111. [Google Scholar] [CrossRef] [Green Version]
- Mok, M.C.; Martin, R.C.; Mok, D.W. Cytokinins: Biosynthesis metabolism and perception. In Vitro Cell. Dev. Biol. Plant 2000, 36, 102–107. [Google Scholar] [CrossRef]
- Werner, T.; Schmülling, T. Cytokinin action in plant development. Curr. Opin. Plant Biol. 2009, 12, 527–538. [Google Scholar] [CrossRef]
- Mrízová, K.; Jiskrová, E.; Vyroubalová, Š.; Novák, O.; Ohnoutková, L.; Pospíšilová, H.; Frébort, I.; Harwood, W.A.; Galuszka, P. Overexpression of cytokinin dehydrogenase genes in barley (Hordeum vulgare cv. Golden Promise) fundamentally affects morphology and fertility. PLoS ONE 2013, 8, e79029. [Google Scholar]
- Liu, P.; Zhang, C.; Ma, J.Q.; Zhang, L.Y.; Yang, B.; Tang, X.Y.; Huang, L.; Zhou, X.T.; Lu, K.; Li, J.N. Genome-wide identification and expression profiling of cytokinin oxidase/dehydrogenase (CKX) genes reveal likely roles in pod development and stress responses in oilseed rape (Brassica napus L.). Genes 2018, 9, 168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jo, E.J.; Jang, K.S.; Choi, Y.H.; Ahn, K.G.; Choi, G.J. Resistance of Cabbage Plants to Isolates of Plasmodiophora brassicae. Korean J. Hortic. Sci. Technol. 2016, 34, 442–452. [Google Scholar]
- Laila, R.; Park, J.I.; Robin, A.H.; Natarajan, S.; Vijayakumar, H.; Shirasawa, K.; Isobe, S.; Kim, H.T.; Nou, I.S. Mapping of a novel clubroot resistance QTL using ddRAD-seq in Chinese cabbage (Brassica rapa L.). BMC Plant Biol. 2019, 19, 13. [Google Scholar] [CrossRef]
- Cheng, F.; Liu, S.; Wu, J.; Fang, L.; Sun, S.; Liu, B.; Li, P.; Hua, W.; Wang, X. BRAD, the genetics and genomics database for Brassica plants. BMC Plant Biol. 2011, 11, 136. [Google Scholar] [CrossRef] [Green Version]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy—the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [Green Version]
- Robin, A.H.; Yi, G.E.; Laila, R.; Yang, K.; Park, J.I.; Kim, H.R.; Nou, I.S. Expression profiling of glucosinolate biosynthetic genes in Brassica oleracea L. var. capitata inbred lines reveals their association with glucosinolate content. Molecules 2016, 21, 787. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]









| Gene Name | BRAD ID | Chromosomal Location | Start Codon | Stop Codon | Strand | Gene Identity | (aa) | Pi | MW (kDa) | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|---|---|
| BrRR1 | Bra023972 | A03 | 28566478 | 28568797 | − | Two-component response regulator | 518 | 5.92 | 57.96 | Nuclear |
| BrHK1 | Bra024849 | A06 | 22745808 | 22750254 | + | Histidine kinase (CRE1/AHK4) | 1040 | 6.21 | 115.0 | Endoplasmic reticulum |
| BrRR2 | Bra018084 | A06 | 9832893 | 9834443 | + | Response regulator 5 (ARR5) | 179 | 6.18 | 20.30 | Nuclear |
| BrRR2.1 | Bra033773 | A01 | 13811201 | 13812750 | − | 180 | 5.54 | 20.404 | Nuclear | |
| BrRR3 | Bra036963 | Scaffold000123 | 487890 | 489100 | − | Response regulator 4 (ARR4) | 261 | 4.62 | 28.525 | Nuclear |
| BrRR3.1 | Bra031714 | A09 | 36858902 | 36860112 | + | 261 | 4.62 | 28.525 | Nuclear | |
| BrRR3.2 | Bra019932 | A06 | 3644746 | 3645865 | − | 253 | 4.63 | 27.682 | Nuclear | |
| BrRR3.3 | Bra018439 | A05 | 8234767 | 8235847 | + | 251 | 4.55 | 27.317 | Nuclear | |
| BrRR4 | Bra015885 | A07 | 23646646 | 23647740 | − | Response regulator 15 (ARR15) | 185 | 5.94 | 20.55 | Nuclear |
| BrRR4.1 | Bra003782 | A07 | 18316602 | 18317690 | + | 197 | 5.06 | 21.850 | Nuclear | |
| BrRR4.2 | Bra025708 | A06 | 7115317 | 7116482 | − | 213 | 5.54 | 23.480 | Nuclear | |
| BrPUP1 | Bra010890 | A08 | 16197869 | 16199402 | + | Purine permease (PUP1) | 302 | 8.72 | 38.77 | Nuclear |
| BrADA1 | Bra040578 | Scaffold000219 | 46992 | 49506 | − | Transcriptional adaptor (ADA2b) | 486 | 6.51 | 56.07 | Nuclear |
| BrADA1.1 | Bra012721 | A03 | 22587864 | 22590255 | − | 469 | 7.53 | 54.33 | Nuclear | |
| BrIPT1 | Bra001737 | A03 | 18198110 | 18199443 | − | Isopentenyl transferase (IPT8) | 327 | 9.26 | 37.03 | Chloroplast |
| BrRR5 | Bra003265 | A07 | 15543847 | 15545066 | + | Response regulator 9 (ARR9) | 240 | 5.07 | 26.847 | Nuclear |
| BrRR5.1 | Bra014649 | A04 | 2014110 | 2015202 | − | 234 | 5.20 | 25.932 | Nuclear | |
| BrRR6 | Bra016526 | A08 | 18109570 | 18110672 | + | Response regulator 7 (ARR7) | 207 | 5.62 | 23.04 | Nuclear |
| BrRR6.1 | Bra025708 | A06 | 7115317 | 7116482 | − | 213 | 5.54 | 23.48 | Plasma membrane | |
| BrCKX2 | Bra036719 | A09 | 5855383 | 5858908 | − | Cytokinin oxidase | 502 | 5.96 | 55.48 | ER |
| BrCKX2.1 | Bra040677 | Scaffold000232 | 36707 | 41121 | − | 505 | 5.62 | 55.60 | ER | |
| BrCKX3 | Bra035640 | A02 | 6020617 | 6023613 | + | 518 | 5.62 | 58.89 | Vacuole/ER | |
| BrCKX3.1 | Bra002777 | A10 | 7709311 | 7712297 | − | 366 | 5.63 | 40.97 | Vacuole/ER | |
| BrCKX4 | Bra024135 | A03 | 27423204 | 27426573 | + | 524 | 5.71 | 57.99 | Extracellular | |
| BrCKX5 | Bra015842 | A07 | 23842777 | 23845621 | − | 524 | 5.62 | 58.83 | Extracellular | |
| BrCKX6 | Bra007743 | A09 | 32193279 | 32195072 | + | 449 | 7.30 | 50.498 | Extracellular | |
| BrCRR1 | Bra007202 | A09 | 29452438 | 29453460 | − | Protein kinase family | 340 | 5.71 | 37.562 | Plasma membrane |
| BrPYL1 | Bra009113 | A10 | 15189844 | 15190458 | − | ABA binding protein | 204 | 6.02 | 22.777 | Peroxisomal |
| BrPYL1.1 | Bra005853 | A03 | 930207 | 930818 | + | 203 | 5.72 | 22.72 | Peroxisomal | |
| BrPYL11.2 | Bra028758 | A02 | 798022 | 798519 | + | 165 | 5.52 | 18.37 | Peroxisomal |
| Sources of Variation | df | Genes | MS | F Statistic | p Value | Genes | MS | F Statistic | p Value |
|---|---|---|---|---|---|---|---|---|---|
| Time-point (Tm) | 4 | BrRR1 | 1.84 | 253.7 | <0.01 | BrADA1 | 1.05 | 62.5 | <0.01 |
| Treatment (Tr) | 1 | 1.74 | 239.4 | <0.01 | 0.0003 | 0.02 | 0.891 | ||
| Tm x Tr | 4 | 0.88 | 121.1 | <0.01 | 0.559 | 33.06 | <0.01 | ||
| Leaf vs. Root (Tm Tr) | 10 | 1.01 | 139 | <0.01 | 0.56 | 33.13 | <0.01 | ||
| Error | 40 | 0.033 | 0.016 | ||||||
| Time-point | 4 | BrRR2 | 7.64 | 1305.2 | <0.01 | BrIPT1 | 7.52 | 671.2 | <0.01 |
| Treatment | 1 | 2.46 | 420.9 | <0.01 | 12.2 | 1089.8 | <0.01 | ||
| Tm x Tr | 4 | 2.67 | 457.9 | <0.01 | 5.58 | 498.3 | <0.01 | ||
| Leaf vs. Root (Tm Tr) | 10 | 2.97 | 507.1 | <0.01 | 5.63 | 502.3 | <0.01 | ||
| Error | 40 | 0.055 | 0.0112 | ||||||
| Time-point | 4 | BrRR3 | 7.98 | 241.5 | <0.01 | BrPYL1 | 85.1 | 642.1 | <0.01 |
| Treatment | 1 | 0.068 | 2.07 | 0.158 | 79.3 | 598.1 | <0.01 | ||
| Tm x Tr | 4 | 4.04 | 122.4 | <0.01 | 106.4 | 802.7 | <0.01 | ||
| Leaf vs. Root (Tm Tr) | 10 | 12.8 | 389.9 | <0.01 | 115.9 | 873.8 | <0.01 | ||
| Error | 40 | 0.033 | 0.133 | ||||||
| Time-point | 4 | BrRR4 | 4.48 | 242.5 | <0.01 | BrCKX2 | 114.1 | 562.9 | <0.01 |
| Treatment | 1 | 0.073 | 3.95 | 0.054 | 41.4 | 204.5 | <0.01 | ||
| Tm x Tr | 4 | 4.41 | 238.6 | <0.01 | 32.8 | 161.9 | <0.01 | ||
| Leaf vs. Root (Tm Tr) | 10 | 9.74 | 526.8 | <0.01 | 53.8 | 265.8 | <0.01 | ||
| Error | 40 | 0.018 | 0.203 | ||||||
| Time-point | 4 | BrRR5 | 376.1 | 191.9 | <0.01 | BrCKX3 | 131.6 | 586.8 | <0.01 |
| Treatment | 1 | 925.2 | 472.2 | <0.01 | 53.9 | 240.4 | <0.01 | ||
| Tm x Tr | 4 | 235.2 | 120.1 | <0.01 | 139.8 | 623.2 | <0.01 | ||
| Leaf vs. Root (Tm Tr) | 10 | 461.6 | 235.6 | <0.01 | 135.2 | 602.5 | <0.01 | ||
| Error | 40 | 1.959 | 0.224 | ||||||
| Time-point | 4 | BrRR6 | 121.8 | 479.8 | <0.01 | BrCKX4 | 1248.8 | 2545.6 | <0.01 |
| Treatment | 1 | 75.4 | 297.0 | <0.01 | 70.6 | 144.1 | <0.01 | ||
| Tm x Tr | 4 | 33.4 | 131.5 | <0.01 | 78.8 | 160.6 | <0.01 | ||
| Leaf vs. Root (Tm Tr) | 10 | 161.9 | 637.8 | <0.01 | 1074.3 | 2189.9 | <0.01 | ||
| Error | 40 | 0.254 | 0.49 | ||||||
| Time-point | 4 | BrHK1 | 32.04 | 351.1 | <0.01 | BrCKX5 | 112,4 | 2488.7 | <0.01 |
| Treatment | 1 | 0.05 | 0.56 | 0.458 | 75.6 | 1675.7 | <0.01 | ||
| Tm x Tr | 4 | 12.55 | 137.6 | <0.01 | 4.89 | 108.4 | <0.01 | ||
| Leaf vs. Root (Tm Tr) | 10 | 72.8 | 798.7 | <0.01 | 87.3 | 1933.2 | <0.01 | ||
| Error | 40 | 0.09 | 0.045 | ||||||
| Time-point | 4 | BrCRR1 | 3.19 | 968.4 | <0.01 | BrCKX6 | 1618.7 | 2676.6 | <0.01 |
| Treatment | 1 | 0.47 | 145.3 | <0.01 | 431.9 | 714.2 | <0.01 | ||
| Tm x Tr | 4 | 1.11 | 336.8 | <0.01 | 157.1 | 259.8 | <0.01 | ||
| Leaf vs. Root (Tm Tr) | 10 | 1.74 | 530.6 | <0.01 | 1558.9 | 2577.8 | <0.01 | ||
| Error | 40 | 0.003 | 0.60 | ||||||
| Time-point | 4 | BrPUP1 | 3.43 | 354.4 | <0.01 | ||||
| Treatment | 1 | 2.21 | 227.4 | <0.01 | |||||
| Tm x Tr | 4 | 0.769 | 79.2 | <0.01 | |||||
| Leaf vs. Root (Tm Tr) | 10 | 4.38 | 451.8 | <0.01 | |||||
| Error | 40 | 0.009 |
| Sample ID | Treatment | Time-Point | Sample Type |
|---|---|---|---|
| 1 | Mock | Day 1 | Leaf |
| 2 | Mock | Day 1 | Root |
| 3 | Seosan-inoculated | Day 1 | Leaf |
| 4 | Seosan-inoculated | Day 1 | Root |
| 5 | Mock | Day 3 | Leaf |
| 6 | Mock | Day 3 | Root |
| 7 | Seosan-inoculated | Day 3 | Leaf |
| 8 | Seosan-inoculated | Day 3 | Root |
| 9 | Mock | Day 14 | Leaf |
| 10 | Mock | Day 14 | Root |
| 11 | Seosan-inoculated | Day 14 | Leaf |
| 12 | Seosan-inoculated | Day 14 | Root |
| 13 | Mock | Day 28 | Leaf |
| 14 | Mock | Day 28 | Root |
| 15 | Seosan-inoculated | Day 28 | Leaf |
| 16 | Seosan-inoculated | Day 28 | Root |
| 17 | Mock | Day 35 | Leaf |
| 18 | Mock | Day 35 | Root |
| 19 | Seosan-inoculated | Day 35 | Leaf |
| 20 | Seosan-inoculated | Day 35 | Root |
| Arabidopsis Homolog (Accession Number) | Gene Name | BRAD ID | cDNA Size (bp) | Primer Forward (F) and Reverse(R) | Product Size (bp) |
|---|---|---|---|---|---|
| At4g31920 | BrRR1 | Bra023972 | 1557 | F:AGCTCAAGAACATATGGCAA | 191 |
| R:TGGATCATCGTTCTCATTCC | |||||
| At2g01830 | BrHK1 | Bra024849 | 3123 | F:AGCTCTGAAGAAGTTTGGAG | 200 |
| R:GTAAATGCCATTCCAGCTTC | |||||
| At3g48100 | BrRR2 | Bra018084 | 540 | F:TACTCAGAGTCTCTTCGTGT | 171 |
| R:CTTCTTGAGTAGTTCATATC | |||||
| At1g10470 | BrRR3 | Bra036963 | 786 | F:GTTCAGAGATGATGAGGGTC | 208 |
| R:GCAGATGCTTTCTCGTTGTC | |||||
| At1g74890 | BrRR4 | Bra015885 | 558 | F:TCTACCTCGGAGTTACATGT | 161 |
| R:CCAGAAGATCCTTTGTCTCC | |||||
| At3g55950 | BrCRR1 | Bra007202 | 1023 | F:GATTACTATGGGTGAGCTGG | 167 |
| R:CTCTCTCCAAATTTCCGACA | |||||
| At1g28230 | BrPUP1 | Bra010890 | 1059 | F:TAGCTTTCAACGCTCTCTTT | 159 |
| R:CAACCACATACTCCTTGTGA | |||||
| At4g16420 | BrADA1 | Bra040578 | 1461 | F:ATGAAATGCTTCTCCTGGAG | 178 |
| R:TTCTTGTTCTTCCCTGCTAC | |||||
| At3g19160 | BrIPT1 | Bra001737 | 984 | F:TTCTGAGCTGAGGTACGATT | 194 |
| R:ACTCCGGTACTCCTATAGCC | |||||
| At3g57040 | BrRR5 | Bra003265 | 723 | F:GGCATGACTGGTTATGATTT | 237 |
| R:AAGAGATTGAAGCACCTTCA | |||||
| At1g19050 | BrRR6 | Bra016526 | 624 | F:TTAGTTCGCCTGACCTACAT | 163 |
| R:TCAGAATCTCCTGTGTTTCC | |||||
| At5g05440 | BrPYL1 | Bra009113 | 615 | F:GAGAGGCTCGAGATCCTGGAC | 237 |
| R:ACAGCTTCTAGCCAGAGACTG | |||||
| At2g19500 | BrCKX2 | Bra036719 | 1509 | F:TTAGTGAAGAGCCACGGTAT | 210 |
| R:CCATAAACCCAAAGATCTGA | |||||
| At5g56970 | BrCKX3 | Bra035640 | 1557 | F:GGGAGAGCTAAACCTGAAAT | 238 |
| R:TAAGAACCCTACCGCATAAA | |||||
| At4g29740 | BrCKX4 | Bra024135 | 1575 | F:GCTGGTGGATCACATAACTT | 170 |
| R:ATTTTCCTCCTCGAGAAATC | |||||
| At1g75450 | BrCKX5 | Bra015842 | 1575 | F:GTATCTTTCCGTTGGTGGTA | 200 |
| R:CTCGTGTAATGATCCCAAAT | |||||
| At3g63440 | BrCKX6 | Bra007743 | 1350 | F:GGTTATCCTTCACACCAGAA | 225 |
| R:AGACATGTACCCTGTCCAAG | |||||
| XM_009147610.2 | ACTIN1 | 1371 | F: CAACCAATCGTCTGTGACAA | 105 | |
| R: ATGTCTTGGCCTACCAACAA | |||||
| FJ969844.1 | ACTIN2 | 491 | F: AATGGTACCGGAATGGTCAA | 119 | |
| R: TCCTTCTGGTTCATCCCAAC |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Laila, R.; Robin, A.H.K.; Park, J.-I.; Saha, G.; Kim, H.-T.; Kayum, M.A.; Nou, I.-S. Expression and Role of Response Regulating, Biosynthetic and Degrading Genes for Cytokinin Signaling during Clubroot Disease Development. Int. J. Mol. Sci. 2020, 21, 3896. https://doi.org/10.3390/ijms21113896
Laila R, Robin AHK, Park J-I, Saha G, Kim H-T, Kayum MA, Nou I-S. Expression and Role of Response Regulating, Biosynthetic and Degrading Genes for Cytokinin Signaling during Clubroot Disease Development. International Journal of Molecular Sciences. 2020; 21(11):3896. https://doi.org/10.3390/ijms21113896
Chicago/Turabian StyleLaila, Rawnak, Arif Hasan Khan Robin, Jong-In Park, Gopal Saha, Hoy-Taek Kim, Md. Abdul Kayum, and Ill-Sup Nou. 2020. "Expression and Role of Response Regulating, Biosynthetic and Degrading Genes for Cytokinin Signaling during Clubroot Disease Development" International Journal of Molecular Sciences 21, no. 11: 3896. https://doi.org/10.3390/ijms21113896