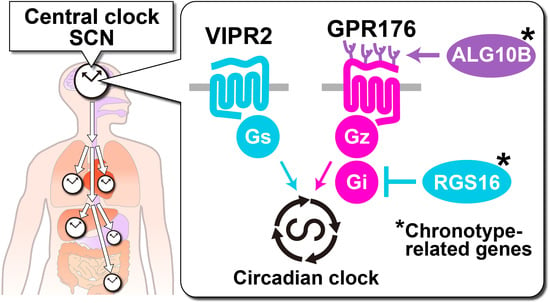
Time-Restricted G-Protein Signaling Pathways via GPR176, Gz, and RGS16 Set the Pace of the Master Circadian Clock in the Suprachiasmatic Nucleus
Abstract
:1. Introduction
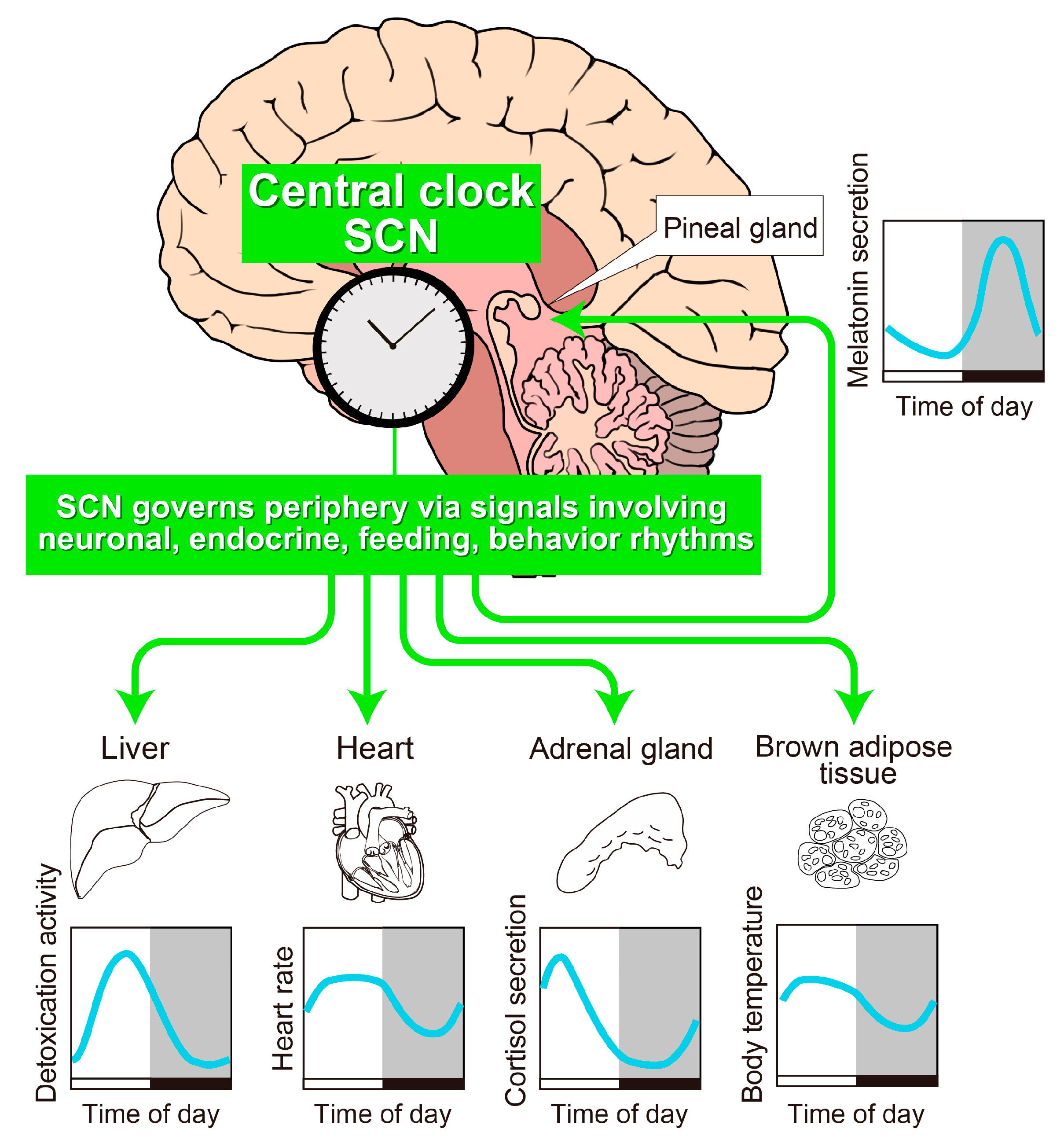
2. The SCN Is the Principal Circadian Pacemaker in Mammals
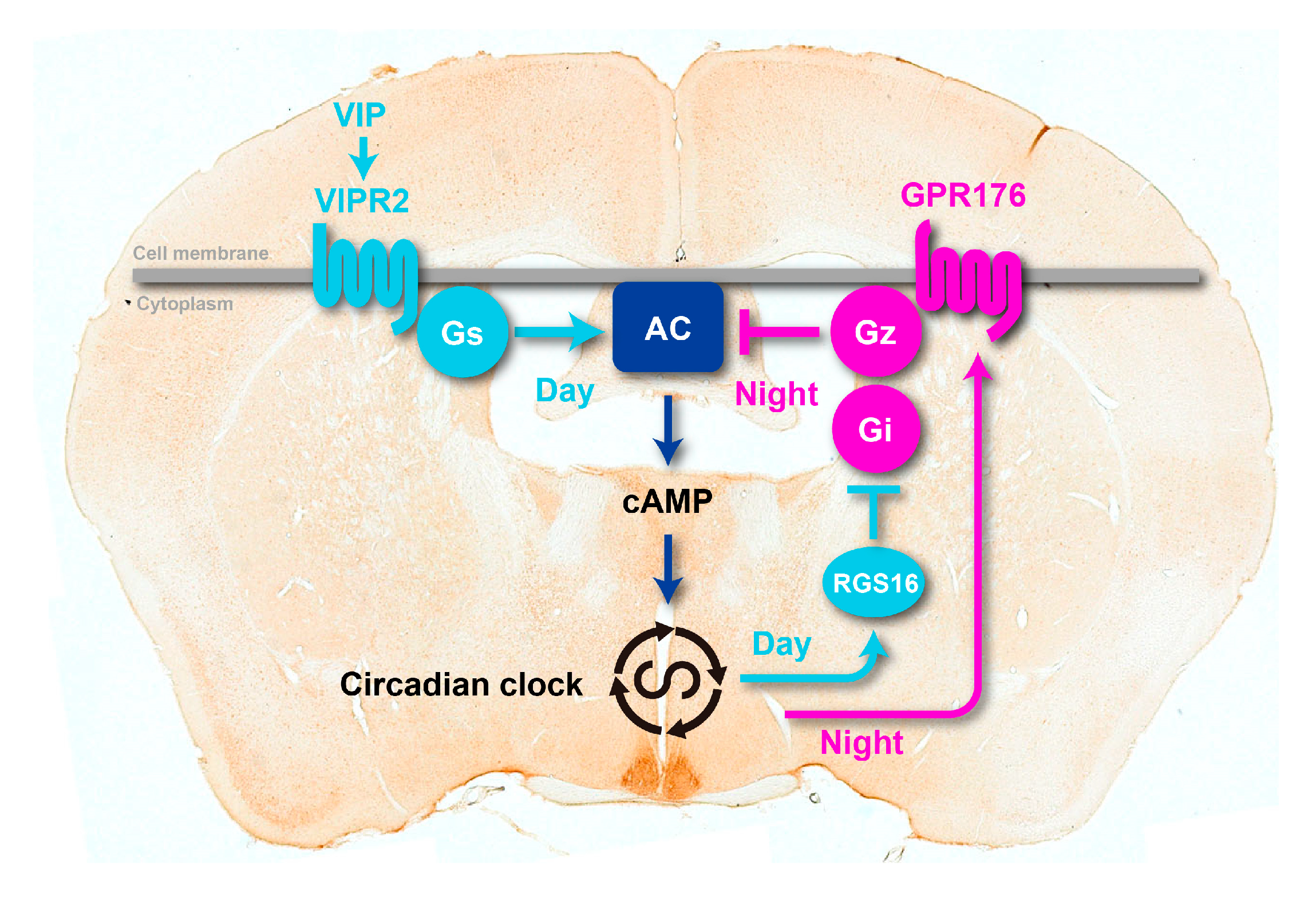
3. GPR176 Is a Gz-Linked Orphan GPCR that Sets the Pace of the SCN Clock
3.1. GPR176 Colocalizes with VIPR2 and Displays Oscillatory Abundance
3.2. Agonist-Independent Activity of GPR176 Counteracts VIPR2-cAMP Signaling
3.3. The Unique G-Protein Subclass Gz Is Required for GPR176 Basal Activity
3.4. GPR176 Is an N-Glycosylated GPCR
4. RGS16 Is a Regulator of G-Protein-cAMP Signaling in the SCN
5. Advantage of Focusing on GPCRs in the SCN
6. Deorphanizing GPR176
7. Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Santos, R.; Ursu, O.; Gaulton, A.; Bento, A.P.; Donadi, R.S.; Bologa, C.G.; Karlsson, A.; Al-Lazikani, B.; Hersey, A.; Oprea, T.I.; et al. A comprehensive map of molecular drug targets. Nat. Rev. Drug Discov. 2017, 16, 19–34. [Google Scholar] [CrossRef] [PubMed]
- Hauser, A.S.; Attwood, M.M.; Rask-Andersen, M.; Schioth, H.B.; Gloriam, D.E. Trends in GPCR drug discovery: New agents, targets and indications. Nat. Rev. Drug Discov. 2017, 16, 829–842. [Google Scholar] [CrossRef] [PubMed]
- Wacker, D.; Stevens, R.C.; Roth, B.L. How Ligands Illuminate GPCR Molecular Pharmacology. Cell 2017, 170, 414–427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doi, M.; Takahashi, Y.; Komatsu, R.; Yamazaki, F.; Yamada, H.; Haraguchi, S.; Emoto, N.; Okuno, Y.; Tsujimoto, G.; Kanematsu, A.; et al. Salt-sensitive hypertension in circadian clock-deficient Cry-null mice involves dysregulated adrenal Hsd3b6. Nat. Med. 2010, 16, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Doi, M. Circadian clock-deficient mice as a tool for exploring disease etiology. Biol. Pharm. Bull. 2012, 35, 1385–1391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cederroth, C.R.; Albrecht, U.; Bass, J.; Brown, S.A.; Dyhrfjeld-Johnsen, J.; Gachon, F.; Green, C.B.; Hastings, M.H.; Helfrich-Forster, C.; Hogenesch, J.B.; et al. Medicine in the Fourth Dimension. Cell Metab. 2019, 30, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Doi, M.; Murai, I.; Kunisue, S.; Setsu, G.; Uchio, N.; Tanaka, R.; Kobayashi, S.; Shimatani, H.; Hayashi, H.; Chao, H.W.; et al. Gpr176 is a Gz-linked orphan G-protein-coupled receptor that sets the pace of circadian behaviour. Nat. Commun. 2016, 7, 10583. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Nakagawa, S.; Miyake, T.; Setsu, G.; Kunisue, S.; Goto, K.; Hirasawa, A.; Okamura, H.; Yamaguchi, Y.; Doi, M. Identification and functional characterisation of N-linked glycosylation of the orphan G protein-coupled receptor Gpr176. Sci. Rep. 2020, 10, 4429. [Google Scholar] [CrossRef]
- Martin, A.L.; Steurer, M.A.; Aronstam, R.S. Constitutive Activity among Orphan Class-A G Protein Coupled Receptors. PLoS ONE 2015, 10, e0138463. [Google Scholar] [CrossRef] [Green Version]
- Doi, M.; Ishida, A.; Miyake, A.; Sato, M.; Komatsu, R.; Yamazaki, F.; Kimura, I.; Tsuchiya, S.; Kori, H.; Seo, K.; et al. Circadian regulation of intracellular G-protein signalling mediates intercellular synchrony and rhythmicity in the suprachiasmatic nucleus. Nat. Commun. 2011, 2, 327. [Google Scholar] [CrossRef] [Green Version]
- Hayasaka, N.; Aoki, K.; Kinoshita, S.; Yamaguchi, S.; Wakefield, J.K.; Tsuji-Kawahara, S.; Horikawa, K.; Ikegami, H.; Wakana, S.; Murakami, T.; et al. Attenuated food anticipatory activity and abnormal circadian locomotor rhythms in Rgs16 knockdown mice. PLoS ONE 2011, 6, e17655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Y.; Shmygelska, A.; Tran, D.; Eriksson, N.; Tung, J.Y.; Hinds, D.A. GWAS of 89,283 individuals identifies genetic variants associated with self-reporting of being a morning person. Nat. Commun. 2016, 7, 10448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lane, J.M.; Vlasac, I.; Anderson, S.G.; Kyle, S.D.; Dixon, W.G.; Bechtold, D.A.; Gill, S.; Little, M.A.; Luik, A.; Loudon, A.; et al. Genome-wide association analysis identifies novel loci for chronotype in 100,420 individuals from the UK Biobank. Nat. Commun. 2016, 7, 10889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, S.E.; Tyrrell, J.; Wood, A.R.; Beaumont, R.N.; Ruth, K.S.; Tuke, M.A.; Yaghootkar, H.; Hu, Y.; Teder-Laving, M.; Hayward, C.; et al. Genome-Wide Association Analyses in 128,266 Individuals Identifies New Morningness and Sleep Duration Loci. PLoS Genet. 2016, 12, e1006125. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.E.; Lane, J.M.; Wood, A.R.; van Hees, V.T.; Tyrrell, J.; Beaumont, R.N.; Jeffries, A.R.; Dashti, H.S.; Hillsdon, M.; Ruth, K.S.; et al. Genome-wide association analyses of chronotype in 697,828 individuals provides insights into circadian rhythms. Nat. Commun. 2019, 10, 343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herzog, E.D.; Hermanstyne, T.; Smyllie, N.J.; Hastings, M.H. Regulating the Suprachiasmatic Nucleus (SCN) Circadian Clockwork: Interplay between Cell-Autonomous and Circuit-Level Mechanisms. Cold Spring Harb. Perspect Biol. 2017, 9, a027706. [Google Scholar] [CrossRef] [PubMed]
- Lehman, M.N.; Silver, R.; Gladstone, W.R.; Kahn, R.M.; Gibson, M.; Bittman, E.L. Circadian rhythmicity restored by neural transplant. Immunocytochemical characterization of the graft and its integration with the host brain. J. Neurosci. 1987, 7, 1626–1638. [Google Scholar] [CrossRef] [Green Version]
- Ralph, M.R.; Foster, R.G.; Davis, F.C.; Menaker, M. Transplanted suprachiasmatic nucleus determines circadian period. Science 1990, 247, 975–978. [Google Scholar] [CrossRef] [Green Version]
- Sujino, M.; Masumoto, K.H.; Yamaguchi, S.; van der Horst, G.T.; Okamura, H.; Inouye, S.T. Suprachiasmatic nucleus grafts restore circadian behavioral rhythms of genetically arrhythmic mice. Curr. Biol. 2003, 13, 664–668. [Google Scholar] [CrossRef] [Green Version]
- Lee, I.T.; Chang, A.S.; Manandhar, M.; Shan, Y.; Fan, J.; Izumo, M.; Ikeda, Y.; Motoike, T.; Dixon, S.; Seinfeld, J.E.; et al. Neuromedin s-producing neurons act as essential pacemakers in the suprachiasmatic nucleus to couple clock neurons and dictate circadian rhythms. Neuron 2015, 85, 1086–1102. [Google Scholar] [CrossRef] [Green Version]
- Mieda, M.; Ono, D.; Hasegawa, E.; Okamoto, H.; Honma, K.; Honma, S.; Sakurai, T. Cellular clocks in AVP neurons of the SCN are critical for interneuronal coupling regulating circadian behavior rhythm. Neuron 2015, 85, 1103–1116. [Google Scholar] [CrossRef] [Green Version]
- Smyllie, N.J.; Chesham, J.E.; Hamnett, R.; Maywood, E.S.; Hastings, M.H. Temporally chimeric mice reveal flexibility of circadian period-setting in the suprachiasmatic nucleus. Proc. Natl. Acad. Sci. USA 2016, 113, 3657–3662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyake, T.; Doi, M. Reconstitution of organismal liver clock function requires light. Trends Endocrinol. Metab. 2019, 30, 569–571. [Google Scholar] [CrossRef] [PubMed]
- Koronowski, K.B.; Kinouchi, K.; Welz, P.S.; Smith, J.G.; Zinna, V.M.; Shi, J.; Samad, M.; Chen, S.; Magnan, C.N.; Kinchen, J.M.; et al. Defining the Independence of the Liver Circadian Clock. Cell 2019, 177, 1448–1462.e14. [Google Scholar] [CrossRef] [PubMed]
- Welz, P.S.; Zinna, V.M.; Symeonidi, A.; Koronowski, K.B.; Kinouchi, K.; Smith, J.G.; Guillen, I.M.; Castellanos, A.; Crainiciuc, G.; Prats, N.; et al. BMAL1-driven tissue clocks respond independently to light to maintain homeostasis. Cell 2019, 177, 1436–1447.e12. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, J.S. Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 2017, 18, 164–179. [Google Scholar] [CrossRef] [Green Version]
- O’Neill, J.S.; Reddy, A.B. Circadian clocks in human red blood cells. Nature 2011, 469, 498–503. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.S.; Green, E.W.; Zhao, Y.; van Ooijen, G.; Olmedo, M.; Qin, X.; Xu, Y.; Pan, M.; Valekunja, U.K.; Feeney, K.A.; et al. Peroxiredoxins are conserved markers of circadian rhythms. Nature 2012, 485, 459–464. [Google Scholar] [CrossRef] [Green Version]
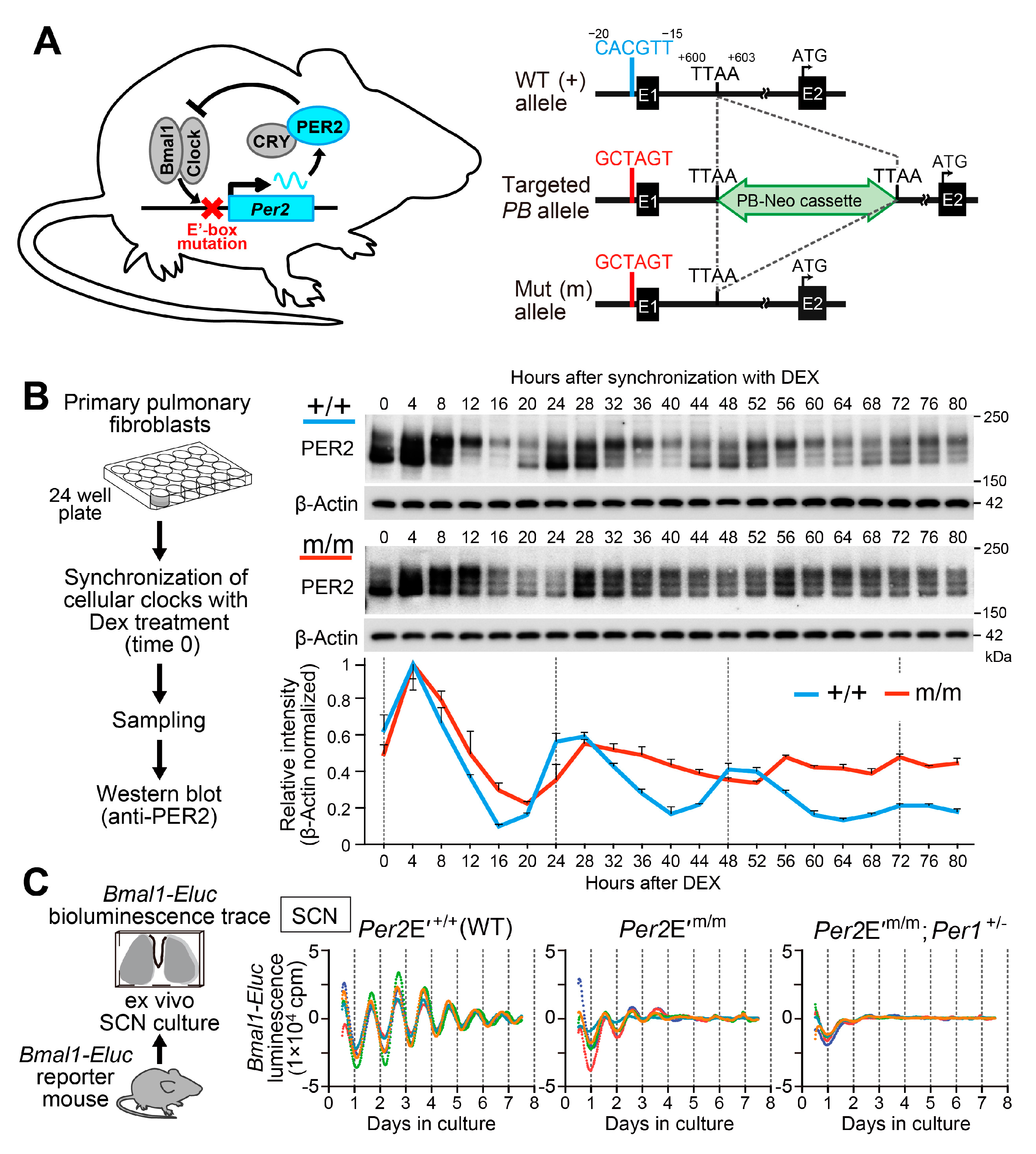
- Doi, M.; Shimatani, H.; Atobe, Y.; Murai, I.; Hayashi, H.; Takahashi, Y.; Fustin, J.M.; Yamaguchi, Y.; Kiyonari, H.; Koike, N.; et al. Non-coding cis-element of Period2 is essential for maintaining organismal circadian behaviour and body temperature rhythmicity. Nat. Commun. 2019, 10, 2563. [Google Scholar] [CrossRef]
- Mermet, J.; Yeung, J.; Hurni, C.; Mauvoisin, D.; Gustafson, K.; Jouffe, C.; Nicolas, D.; Emmenegger, Y.; Gobet, C.; Franken, P.; et al. Clock-dependent chromatin topology modulates circadian transcription and behavior. Genes Dev. 2018, 32, 347–358. [Google Scholar] [CrossRef] [Green Version]
- Hastings, M.H.; Maywood, E.S.; Brancaccio, M. Generation of circadian rhythms in the suprachiasmatic nucleus. Nat. Rev. Neurosci. 2018, 19, 453–469. [Google Scholar] [CrossRef] [PubMed]
- Maywood, E.S.; Chesham, J.E.; O’Brien, J.A.; Hastings, M.H. A diversity of paracrine signals sustains molecular circadian cycling in suprachiasmatic nucleus circuits. Proc. Natl. Acad. Sci. USA 2011, 108, 14306–14311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aton, S.J.; Colwell, C.S.; Harmar, A.J.; Waschek, J.; Herzog, E.D. Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nat. Neurosci. 2005, 8, 476–483. [Google Scholar] [CrossRef]
- Goda, T.; Doi, M.; Umezaki, Y.; Murai, I.; Shimatani, H.; Chu, M.L.; Nguyen, V.H.; Okamura, H.; Hamada, F.N. Calcitonin receptors are ancient modulators for rhythms of preferential temperature in insects and body temperature in mammals. Genes Dev. 2018, 32, 140–155. [Google Scholar] [CrossRef]
- Goto, K.; Doi, M.; Wang, T.; Kunisue, S.; Murai, I.; Okamura, H. G-protein-coupled receptor signaling through Gpr176, Gz, and RGS16 tunes time in the center of the circadian clock [Review]. Endocr. J. 2017, 64, 571–579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goda, T.; Hamada, F.N. Drosophila Temperature Preference Rhythms: An Innovative Model to Understand Body Temperature Rhythms. Int. J. Mol. Sci. 2019, 20, 1988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, S.; Tsai, C.; Ronecker, J.; Bayly, A.; Herzog, E.D. Spatiotemporal distribution of vasoactive intestinal polypeptide receptor 2 in mouse suprachiasmatic nucleus. J. Comp. Neurol. 2012, 520, 2730–2741. [Google Scholar] [CrossRef] [Green Version]
- O’Neill, J.S.; Maywood, E.S.; Chesham, J.E.; Takahashi, J.S.; Hastings, M.H. cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science 2008, 320, 949–953. [Google Scholar] [CrossRef] [Green Version]
- Ho, M.K.; Wong, Y.H. G(z) signaling: Emerging divergence from G(i) signaling. Oncogene 2001, 20, 1615–1625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graziano, M.P.; Freissmuth, M.; Gilman, A.G. Expression of Gs alpha in Escherichia coli. Purification and properties of two forms of the protein. J. Biol. Chem. 1989, 264, 409–418. [Google Scholar]
- Linder, M.E.; Ewald, D.A.; Miller, R.J.; Gilman, A.G. Purification and characterization of Go alpha and three types of Gi alpha after expression in Escherichia coli. J. Biol. Chem. 1990, 265, 8243–8251. [Google Scholar] [PubMed]
- Casey, P.J.; Fong, H.K.; Simon, M.I.; Gilman, A.G. Gz, a guanine nucleotide-binding protein with unique biochemical properties. J. Biol. Chem. 1990, 265, 2383–2390. [Google Scholar] [PubMed]
- Vancura, P.; Abdelhadi, S.; Csicsely, E.; Baba, K.; Tosini, G.; Iuvone, P.M.; Spessert, R. Gnaz couples the circadian and dopaminergic system to G protein-mediated signaling in mouse photoreceptors. PLoS ONE 2017, 12, e0187411. [Google Scholar] [CrossRef] [Green Version]
- Barr, A.J.; Brass, L.F.; Manning, D.R. Reconstitution of receptors and GTP-binding regulatory proteins (G proteins) in Sf9 cells. A direct evaluation of selectivity in receptor.G protein coupling. J. Biol. Chem. 1997, 272, 2223–2229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serres, F.; Li, Q.; Garcia, F.; Raap, D.K.; Battaglia, G.; Muma, N.A.; Van de Kar, L.D. Evidence that G(z)-proteins couple to hypothalamic 5-HT(1A) receptors in vivo. J. Neurosci. 2000, 20, 3095–3103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yung, L.Y.; Tsim, S.T.; Wong, Y.H. Stimulation of cAMP accumulation by the cloned Xenopus melatonin receptor through Gi and Gz proteins. FEBS Lett. 1995, 372, 99–102. [Google Scholar] [CrossRef] [Green Version]
- Chan, A.S.; Lai, F.P.; Lo, R.K.; Voyno-Yasenetskaya, T.A.; Stanbridge, E.J.; Wong, Y.H. Melatonin mt1 and MT2 receptors stimulate c-Jun N-terminal kinase via pertussis toxin-sensitive and -insensitive G proteins. Cell Signal. 2002, 14, 249–257. [Google Scholar] [CrossRef]
- Tansky, M.F.; Pothoulakis, C.; Leeman, S.E. Functional consequences of alteration of N-linked glycosylation sites on the neurokinin 1 receptor. Proc. Natl. Acad. Sci. USA 2007, 104, 10691–10696. [Google Scholar] [CrossRef] [Green Version]
- Kozielewicz, P.; Alomar, H.; Yusof, S.; Grafton, G.; Cooper, A.J.; Curnow, S.J.; Ironside, J.W.; Pall, H.; Barnes, N.M. N-glycosylation and expression in human tissues of the orphan GPR61 receptor. FEBS Open Bio 2017, 7, 1982–1993. [Google Scholar] [CrossRef]
- Lutz, E.M.; Sheward, W.J.; West, K.M.; Morrow, J.A.; Fink, G.; Harmar, A.J. The VIP2 receptor: Molecular characterisation of a cDNA encoding a novel receptor for vasoactive intestinal peptide. FEBS Lett. 1993, 334, 3–8. [Google Scholar] [CrossRef] [Green Version]
- Hawtin, S.R.; Davies, A.R.; Matthews, G.; Wheatley, M. Identification of the glycosylation sites utilized on the V1a vasopressin receptor and assessment of their role in receptor signalling and expression. Biochem. J. 2001, 357, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, Y.; Suzuki, T.; Mizoro, Y.; Kori, H.; Okada, K.; Chen, Y.; Fustin, J.M.; Yamazaki, F.; Mizuguchi, N.; Zhang, J.; et al. Mice genetically deficient in vasopressin V1a and V1b receptors are resistant to jet lag. Science 2013, 342, 85–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lichnerova, K.; Kaniakova, M.; Park, S.P.; Skrenkova, K.; Wang, Y.X.; Petralia, R.S.; Suh, Y.H.; Horak, M. Two N-glycosylation Sites in the GluN1 Subunit Are Essential for Releasing N-methyl-d-aspartate (NMDA) Receptors from the Endoplasmic Reticulum. J. Biol. Chem. 2015, 290, 18379–18390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kandel, M.B.; Yamamoto, S.; Midorikawa, R.; Morise, J.; Wakazono, Y.; Oka, S.; Takamiya, K. N-glycosylation of the AMPA-type glutamate receptor regulates cell surface expression and tetramer formation affecting channel function. J. Neurochem. 2018, 147, 730–747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bansal, G.; Druey, K.M.; Xie, Z. R4 RGS proteins: Regulation of G-protein signaling and beyond. Pharmacol. Ther. 2007, 116, 473–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mure, L.S.; Le, H.D.; Benegiamo, G.; Chang, M.W.; Rios, L.; Jillani, N.; Ngotho, M.; Kariuki, T.; Dkhissi-Benyahya, O.; Cooper, H.M.; et al. Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science 2018, 359, eaao0318. [Google Scholar] [CrossRef] [Green Version]
- Yamazaki, S.; Numano, R.; Abe, M.; Hida, A.; Takahashi, R.; Ueda, M.; Block, G.D.; Sakaki, Y.; Menaker, M.; Tei, H. Resetting central and peripheral circadian oscillators in transgenic rats. Science 2000, 288, 682–685. [Google Scholar] [CrossRef] [Green Version]
- Foster, S.R.; Hauser, A.S.; Vedel, L.; Strachan, R.T.; Huang, X.P.; Gavin, A.C.; Shah, S.D.; Nayak, A.P.; Haugaard-Kedstrom, L.M.; Penn, R.B.; et al. Discovery of human signaling systems: Pairing peptides to G Protein-Coupled receptors. Cell 2019, 179, 895–908.e21. [Google Scholar] [CrossRef] [Green Version]
- Kroeze, W.K.; Sassano, M.F.; Huang, X.P.; Lansu, K.; McCorvy, J.D.; Giguere, P.M.; Sciaky, N.; Roth, B.L. PRESTO-Tango as an open-source resource for interrogation of the druggable human GPCRome. Nat. Struct. Mol. Biol. 2015, 22, 362–369. [Google Scholar] [CrossRef] [Green Version]
- Colosimo, D.A.; Kohn, J.A.; Luo, P.M.; Piscotta, F.J.; Han, S.M.; Pickard, A.J.; Rao, A.; Cross, J.R.; Cohen, L.J.; Brady, S.F. Mapping interactions of microbial metabolites with human G-Protein-Coupled receptors. Cell Host Microbe 2019, 26, 273–282.e7. [Google Scholar] [CrossRef] [Green Version]
- Cohen, L.J.; Esterhazy, D.; Kim, S.H.; Lemetre, C.; Aguilar, R.R.; Gordon, E.A.; Pickard, A.J.; Cross, J.R.; Emiliano, A.B.; Han, S.M.; et al. Commensal bacteria make GPCR ligands that mimic human signalling molecules. Nature 2017, 549, 48–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Subtype | kcat (min–1) | Source | Ref |
|---|---|---|---|
| Gαs | 4.5 | bovine, recombinant, purified from E. coli | [40] |
| Gαi1 | 2.4 | rat, recombinant, purified from E. coli | [41] |
| Gαi2 | 2.7 | rat, recombinant, purified from E. coli | [41] |
| Gαi3 | 1.8 | rat, recombinant, purified from E. coli | [41] |
| Gαo | 2.2 | rat, recombinant, purified from E. coli | [41] |
| Gαz | 0.05 | human, recombinant, purified from E. coli | [42] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nakagawa, S.; Nguyen Pham, K.T.; Shao, X.; Doi, M. Time-Restricted G-Protein Signaling Pathways via GPR176, Gz, and RGS16 Set the Pace of the Master Circadian Clock in the Suprachiasmatic Nucleus. Int. J. Mol. Sci. 2020, 21, 5055. https://doi.org/10.3390/ijms21145055
Nakagawa S, Nguyen Pham KT, Shao X, Doi M. Time-Restricted G-Protein Signaling Pathways via GPR176, Gz, and RGS16 Set the Pace of the Master Circadian Clock in the Suprachiasmatic Nucleus. International Journal of Molecular Sciences. 2020; 21(14):5055. https://doi.org/10.3390/ijms21145055
Chicago/Turabian StyleNakagawa, Shumpei, Khanh Tien Nguyen Pham, Xinyan Shao, and Masao Doi. 2020. "Time-Restricted G-Protein Signaling Pathways via GPR176, Gz, and RGS16 Set the Pace of the Master Circadian Clock in the Suprachiasmatic Nucleus" International Journal of Molecular Sciences 21, no. 14: 5055. https://doi.org/10.3390/ijms21145055