Molecular Regulation of the Polycomb Repressive-Deubiquitinase
Abstract
:1. Introduction
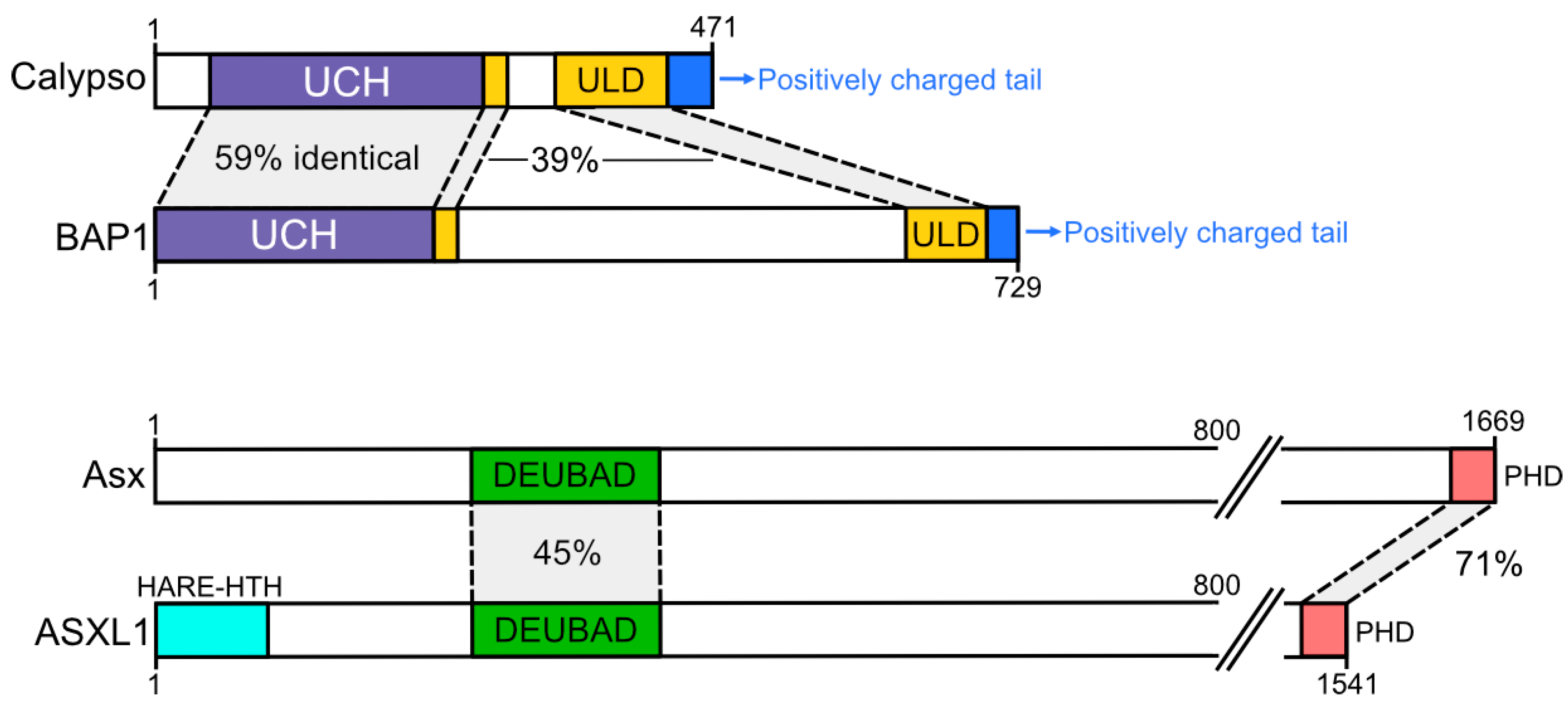
2. PR-DUB, the Polycomb-Repressive Deubiquitinase
3. Regulation of PR-DUB Catalytic Activity
4. Higher Order Complex Formation by the PR-DUB
5. PR-DUB Localisation and Recognition of H2AK119Ub
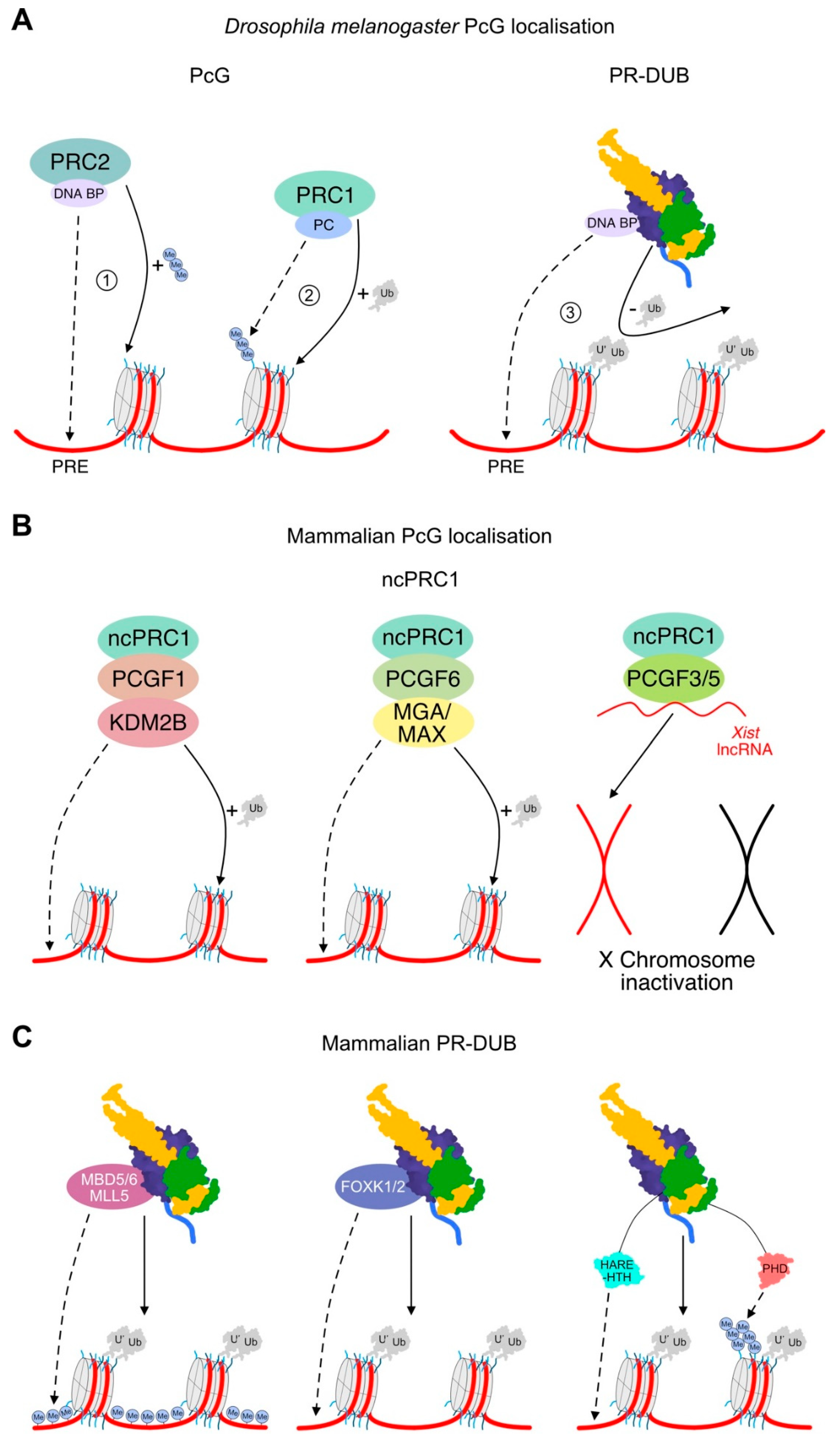
5.1. Relative Arrangement of PRC1 and PRC2
5.2. PR-DUB Localisation through Interaction Partners
6. PR-DUB and Human Disease
6.1. Consequences of PR-DUB Disruption
6.2. Mechanisms of PR-DUB Disruption through Mutation
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cao, J.; Yan, Q. Histone ubiquitination and deubiquitination in transcription, DNA damage response, and cancer. Front. Oncol. 2012, 2, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pickart, C.M. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 2001, 70, 503–533. [Google Scholar] [CrossRef] [PubMed]
- Schuettengruber, B.; Bourbon, H.M.; Di Croce, L.; Cavalli, G. Genome regulation by Polycomb and trithorax: 70 years and counting. Cell 2017, 171, 34–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohen, I.; Bar, C.; Ezhkova, E. Activity of PRC1 and histone H2AK119 monoubiquitination: Revising popular misconceptions. BioEssays 2020, 42, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Sahtoe, D.D.; Sixma, T.K. Layers of DUB regulation. Trends Biochem. Sci. 2015, 40, 456–467. [Google Scholar] [CrossRef] [PubMed]
- Scheuermann, J.C.; De Ayala Alonso, A.G.; Oktaba, K.; Ly-Hartig, N.; McGinty, R.K.; Fraterman, S.; Wilm, M.; Muir, T.W.; Müller, J. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature 2010, 465, 243–247. [Google Scholar] [CrossRef] [Green Version]
- Shukla, V.; Rao, M.; Zhang, H.; Beers, J.; Wangsa, D.; Wangsa, D.; Buishand, F.O.; Wang, Y.; Yu, Z.; Stevenson, H.S.; et al. ASXL3 is a novel pluripotency factor in human respiratory epithelial cells and a potential therapeutic target in small cell lung cancer. Cancer Res. 2017, 77, 6267–6281. [Google Scholar] [CrossRef] [Green Version]
- Foglizzo, M.; Middleton, A.J.; Burgess, A.E.; Crowther, J.M.; Dobson, R.C.J.; Murphy, J.M.; Day, C.L.; Mace, P.D. A bidentate Polycomb Repressive-Deubiquitinase complex is required for efficient activity on nucleosomes. Nat. Commun. 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Sahtoe, D.D.; Van Dijk, W.J.; Ekkebus, R.; Ovaa, H.; Sixma, T.K. BAP1/ASXL1 recruitment and activation for H2A deubiquitination. Nat. Commun. 2016, 7, 10292. [Google Scholar] [CrossRef] [Green Version]
- Daou, S.; Hammond-Martel, I.; Mashtalir, N.; Barbour, H.; Gagnon, J.; Iannantuono, N.V.G.; Nkwe, N.S.; Motorina, A.; Pak, H.; Yu, H.; et al. The BAP1/ASXL2 histone H2A deubiquitinase complex regulates cell proliferation and is disrupted in cancer. J. Biol. Chem. 2015, 290, 28643–28663. [Google Scholar] [CrossRef] [Green Version]
- Dey, A.; Seshasayee, D.; Noubade, R.; French, D.M.; Liu, J.; Chaurushiya, M.S.; Kirkpatrick, D.S.; Pham, V.C.; Lill, J.R.; Bakalarski, C.E.; et al. Loss of the tumor suppressor BAP1 causes myeloid transformation. Science 2012, 337, 1541–1546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Misaghi, S.; Ottosen, S.; Izrael-Tomasevic, A.; Arnott, D.; Lamkanfi, M.; Lee, J.; Liu, J.; O’Rourke, K.; Dixit, V.M.; Wilson, A.C. Association of C-terminal ubiquitin hydrolase BRCA1-associated protein 1 with cell cycle regulator host cell factor 1. Mol. Cell. Biol. 2009, 29, 2181–2192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, H.; Mashtalir, N.; Daou, S.; Hammond-Martel, I.; Ross, J.; Sui, G.; Hart, G.W.; Rauscher, F.J.; Drobetsky, E.; Milot, E.; et al. The ubiquitin carboxyl hydrolase BAP1 forms a ternary complex with YY1 and HCF-1 and is a critical regulator of gene expression. Mol. Cell. Biol. 2010, 30, 5071–5085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aravind, L.; Iyer, L.M. The HARE-HTH and associated domains: Novel modules in the coordination of epigenetic DNA and protein modifications. Cell Cycle 2012, 11, 119–131. [Google Scholar] [CrossRef] [Green Version]
- Johnston, S.C.; Riddle, S.M.; Cohen, R.E.; Hill, C.P. Structural basis for the specificity of ubiquitin C-terminal hydrolases. EMBO J. 1999, 18, 3877–3887. [Google Scholar] [CrossRef]
- Popp, M.W.; Artavanis-Tsakonas, K.; Ploegh, H.L. Substrate filtering by the active site crossover loop in UCHL3 revealed by sortagging and gain-of-function mutations. J. Biol. Chem. 2009, 284, 3593–3602. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.R.; Zhang, Y.H.; Liu, S.; Song, A.X.; Hu, H.Y. Length of the active-site crossover loop defines the substrate specificity of ubiquitin C-terminal hydrolases for ubiquitin chains. Biochem. J. 2012, 441, 143–149. [Google Scholar] [CrossRef]
- Mashtalir, N.; Daou, S.; Barbour, H.; Sen, N.N.; Gagnon, J.; Hammond-Martel, I.; Dar, H.H.; Therrien, M.; Affar, E.B. Autodeubiquitination protects the tumor suppressor BAP1 from cytoplasmic sequestration mediated by the atypical ubiquitin ligase UBE2O. Mol. Cell 2014, 54, 392–406. [Google Scholar] [CrossRef] [Green Version]
- Cohen, R.E.; Lam, Y.A.; Xu, W.; Demartinot, G.N. Editing of ubiquitin conjugates by an isopeptidase in the 265 proteasome. Nature 1997, 385, 737–740. [Google Scholar]
- Hölzl, H.; Kapelari, B.; Kellermann, J.; Seemüller, E.; Sümegi, M.; Udvardy, A.; Medalia, O.; Sperling, J.; Müller, S.A.; Engel, A.; et al. The regulatory complex of Drosophila melanogaster 26S proteasomes: Subunit composition and localization of a deubiquitylating enzyme. J. Cell Biol. 2000, 150, 119–129. [Google Scholar] [CrossRef] [Green Version]
- Lam, Y.A.; DeMartino, G.N.; Pickart, C.M.; Cohen, R.E. Specificity of the ubiquitin isopeptidase in the PA700 regulatory complex of 26 S proteasomes. J. Biol. Chem. 1997, 272, 28438–28446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, T.; Naqvi, N.I.; Yang, H.; Teo, T.S. Identification of a 26S proteasome-associated UCH in fission yeast. Biochem. Biophys. Res. Commun. 2000, 272, 270–275. [Google Scholar] [CrossRef] [PubMed]
- De, I.; Chittock, E.C.; Grötsch, H.; Miller, T.C.R.; McCarthy, A.A.; Müller, C.W. Structural basis for the activation of the deubiquitinase Calypso by the Polycomb protein ASX. Structure 2019, 27, 528–536.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sahtoe, D.D.; van Dijk, W.J.; ElOualid, F.; Ekkebus, R.; Ovaa, H.; Sixma, T.K. Mechanism of UCH-L5 activation and inhibition by DEUBAD domains in RPN13 and INO80G. Mol. Cell 2015, 57, 887–900. [Google Scholar] [CrossRef] [Green Version]
- Linden, R.T.V.; Hemmis, C.W.; Schmitt, B.; Ndoja, A.; Whitby, F.G.; Robinson, H.; Cohen, R.E.; Yao, T.; Hill, C.P. Structural basis for the activation and inhibition of the UCH37 deubiquitylase. Mol. Cell 2015, 57, 901–911. [Google Scholar] [CrossRef] [Green Version]
- Maiti, T.K.; Permaul, M.; Boudreaux, D.A.; Mahanic, C.; Mauney, S.; Das, C. Crystal structure of the catalytic domain of UCHL5, a proteasome-associated human deubiquitinating enzyme, reveals an unproductive form of the enzyme. FEBS J. 2011, 278, 4917–4926. [Google Scholar] [CrossRef]
- Nishio, K.; Kim, S.W.; Kawai, K.; Mizushima, T.; Yamane, T.; Hamazaki, J.; Murata, S.; Tanaka, K.; Morimoto, Y. Crystal structure of the de-ubiquitinating enzyme UCH37 (human UCH-L5) catalytic domain. Biochem. Biophys. Res. Commun. 2009, 390, 855–860. [Google Scholar] [CrossRef]
- Burgie, S.E.; Bingman, C.A.; Soni, A.B.; Phillips, G.N. Structural characterization of human Uch37. Proteins 2012, 80, 649–654. [Google Scholar] [CrossRef] [Green Version]
- Blackledge, N.P.; Fursova, N.A.; Kelley, J.R.; Huseyin, M.K.; Feldmann, A.; Klose, R.J. PRC1 catalytic activity is central to Polycomb system function. Mol. Cell 2020, 77, 857–874.e9. [Google Scholar] [CrossRef] [Green Version]
- Fursova, N.A.; Blackledge, N.P.; Nakayama, M.; Ito, S.; Koseki, Y.; Farcas, A.M.; King, H.W.; Koseki, H.; Klose, R.J. Synergy between variant PRC1 complexes defines Polycomb-Mediated gene repression. Mol. Cell 2019, 74, 1020–1036.e8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamburri, S.; Lavarone, E.; Fernández-Pérez, D.; Conway, E.; Zanotti, M.; Manganaro, D.; Pasini, D. Histone H2AK119 mono-ubiquitination is essential for Polycomb-Mediated transcriptional repression. Mol. Cell 2020, 77, 840–856.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Müller, J.; Kassis, J.A. Polycomb response elements and targeting of Polycomb group proteins in Drosophila. Curr. Opin. Genet. Dev. 2006, 16, 476–484. [Google Scholar] [CrossRef] [PubMed]
- Bauer, M.; Trupke, J.; Ringrose, L. The quest for mammalian Polycomb response elements: Are we there yet? Chromosoma 2016, 125, 471–496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Isono, K.; Endo, T.A.; Ku, M.; Yamada, D.; Suzuki, R.; Sharif, J.; Ishikura, T.; Toyoda, T.; Bernstein, B.E.; Koseki, H. SAM domain polymerization links subnuclear clustering of PRC1 to gene silencing. Dev. Cell 2013, 26, 565–577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blackledge, N.P.; Farcas, A.M.; Kondo, T.; King, H.W.; McGouran, J.F.; Hanssen, L.L.P.; Ito, S.; Cooper, S.; Kondo, K.; Koseki, Y.; et al. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell 2014, 157, 1445–1459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farcas, A.M.; Blackledge, N.P.; Sudbery, I.; Long, H.K.; McGouran, J.F.; Rose, N.R.; Lee, S.; Sims, D.; Cerase, A.; Sheahan, T.W.; et al. KDM2B links the polycomb repressive complex 1 (PRC1) to recognition of CpG islands. Elife 2012, 2012, e00205. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Zhang, J.; Bonasio, R.; Strino, F.; Sawai, A.; Parisi, F.; Kluger, Y.; Reinberg, D. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol. Cell 2012, 45, 344–356. [Google Scholar] [CrossRef] [Green Version]
- Stielow, B.; Finkernagel, F.; Stiewe, T.; Nist, A.; Suske, G. MGA, L3MBTL2 and E2F6 determine genomic binding of the non-canonical Polycomb repressive complex PRC1.6. PLoS Genet. 2018, 14, e1007193. [Google Scholar] [CrossRef]
- Palmer, W.H.; Obbard, D.J. Variation and evolution in the glutamine-rich repeat region of Drosophila Argonaute-2. G3 Genes Genomes Genet. 2016, 6, 2563–2572. [Google Scholar] [CrossRef] [Green Version]
- Hauri, S.; Comoglio, F.; Seimiya, M.; Gerstung, M.; Glatter, T.; Hansen, K.; Aebersold, R.; Paro, R.; Gstaiger, M.; Beisel, C. A high-density map for navigating the human Polycomb complexome. Cell Rep. 2016, 17, 583–595. [Google Scholar] [CrossRef] [Green Version]
- Ji, Z.; Mohammed, H.; Webber, A.; Ridsdale, J.; Han, N.; Carroll, J.S.; Sharrocks, A.D. The forkhead transcription factor FOXK2 acts as a chromatin targeting factor for the BAP1-containing histone deubiquitinase complex. Nucleic Acids Res. 2014, 42, 6232–6242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, K.L.; Huang, C.Y.; Chang, C.H.; Sun, Y.J.; Chuang, W.J.; Hsiao, C.D. Crystal structure of the human FOXK1a-DNA complex and its implications on the diverse binding specificity of winged helix/forkhead proteins. J. Biol. Chem. 2006, 281, 17400–17409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolovos, P.; Nishimura, K.; Sankar, A.; Sidoli, S.; Cloos, P.A.; Helin, K.; Christensen, J. PR-DUB maintains the expression of critical genes through FOXK1/2- and ASXL1/2/3-dependent recruitment to chromatin and H2AK119ub1 deubiquitination. Genome Res. 2020, 30, 1119–1130. [Google Scholar] [CrossRef]
- Baymaz, H.I.; Fournier, A.; Laget, S.; Ji, Z.; Jansen, P.W.T.C.; Smits, A.H.; Ferry, L.; Mensinga, A.; Poser, I.; Sharrocks, A.; et al. MBD5 and MBD6 interact with the human PR-DUB complex through their methyl-CpG-binding domain. Proteomics 2014, 14, 2179–2189. [Google Scholar] [CrossRef]
- Zhou, P.; Wang, Z.; Yuan, X.; Zhou, C.; Liu, L.; Wan, X.; Zhang, F.; Ding, X.; Wang, C.; Xiong, S.; et al. Mixed Lineage Leukemia 5 (MLL5) protein regulates cell cycle progression and E2F1-responsive gene expression via association with Host Cell Factor-1 (HCF-1). J. Biol. Chem. 2013, 288, 17532–17543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inoue, D.; Fujino, T.; Sheridan, P.; Zhang, Y.Z.; Nagase, R.; Horikawa, S.; Li, Z.; Matsui, H.; Kanai, A.; Saika, M.; et al. A novel ASXL1-OGT axis plays roles in H3K4 methylation and tumor suppression in myeloid malignancies. Leukemia 2018, 32, 1327–1337. [Google Scholar] [CrossRef]
- Fujiki, R.; Hashiba, W.; Sekine, H.; Yokoyama, A.; Chikanishi, T.; Ito, S.; Imai, Y.; Kim, J.; He, H.H.; Igarashi, K.; et al. GlcNAcylation of histone H2B facilitates its monoubiquitination. Nature 2011, 480, 557–560. [Google Scholar] [CrossRef]
- Park, U.H.; Kang, M.R.; Kim, E.J.; Kwon, Y.S.; Hur, W.; Yoon, S.K.; Song, B.J.; Park, J.H.; Hwang, J.T.; Jeong, J.C.; et al. ASXL2 promotes proliferation of breast cancer cells by linking ERα to histone methylation. Oncogene 2016, 35, 3742–3752. [Google Scholar] [CrossRef]
- Abdel-Wahab, O.; Adli, M.; LaFave, L.M.; Gao, J.; Hricik, T.; Shih, A.H.; Pandey, S.; Patel, J.P.; Chung, Y.R.; Koche, R.; et al. ASXL1 mutations promote myeloid transformation through loss of PRC2-mediated gene repression. Cancer Cell 2012, 22, 180–193. [Google Scholar] [CrossRef] [Green Version]
- Lai, H.L.; Wang, Q.T. Additional sex combs-like 2 is required for Polycomb repressive complex 2 binding at select targets. PLoS ONE 2013, 8, e73983. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Kurtenbach, S.; Guo, Y.; Lohse, I.; Durante, M.A.; Li, J.; Li, Z.; Al-Ali, H.; Li, L.; Chen, Z.; et al. Gain of function of ASXL1 truncating protein in the pathogenesis of myeloid malignancies. Blood 2018, 131, 328–341. [Google Scholar] [CrossRef] [PubMed]
- Campagne, A.; Lee, M.K.; Zielinski, D.; Michaud, A.; Le Corre, S.; Dingli, F.; Chen, H.; Shahidian, L.Z.; Vassilev, I.; Servant, N.; et al. BAP1 complex promotes transcription by opposing PRC1-mediated H2A ubiquitylation. Nat. Commun. 2019, 10, 348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carbone, M.; Yang, H.; Pass, H.I.; Krausz, T.; Testa, J.R.; Gaudino, G. BAP1 and cancer. Nat. Rev. Cancer 2013, 13, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Wiesner, T.; Obenauf, A.C.; Murali, R.; Fried, I.; Griewank, K.G.; Ulz, P.; Windpassinger, C.; Wackernagel, W.; Loy, S.; Wolf, I.; et al. Germline mutations in BAP1 predispose to melanocytic tumors. Nat. Genet. 2011, 43, 1018–1022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Testa, J.R.; Cheung, M.; Pei, J.; Below, J.E.; Tan, Y.; Sementino, E.; Cox, N.J.; Dogan, A.U.; Pass, H.I.; Trusa, S.; et al. Germline BAP1 mutations predispose to malignant mesothelioma. Nat. Genet. 2012, 43, 1022–1025. [Google Scholar] [CrossRef] [Green Version]
- Bott, M.; Brevet, M.; Taylor, B.S.; Shimizu, S.; Ito, T.; Wang, L.; Creaney, J.; Lake, R.A.; Zakowski, M.F.; Reva, B.; et al. The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nat. Genet. 2011, 43, 668–672. [Google Scholar] [CrossRef]
- Kapur, P.; Peña-Llopis, S.; Christie, A.; Zhrebker, L.; Pavía-Jiménez, A.; Rathmell, W.K.; Xie, X.-J.; Brugarolas, J. Effects on survival of BAP1 and PBRM1 mutations in sporadic clear-cell renal-cell carcinoma: A retrospective analysis with independent validation. Lancet. Oncol. 2013, 14, 159–167. [Google Scholar] [CrossRef] [Green Version]
- Napolitano, A.; Pellegrini, L.; Dey, A.; Larson, D.; Tanji, M.; Flores, E.G.; Kendrick, B.; Lapid, D.; Powers, A.; Kanodia, S.; et al. Minimal asbestos exposure in germline BAP1 heterozygous mice is associated with deregulated inflammatory response and increased risk of mesothelioma. Oncogene 2016, 35, 1996–2002. [Google Scholar] [CrossRef]
- He, M.; Chaurushiya, M.S.; Webster, J.D.; Kummerfeld, S.; Reja, R.; Chaudhuri, S.; Chen, Y.J.; Modrusan, Z.; Haley, B.; Dugger, D.L.; et al. Intrinsic apoptosis shapes the tumor spectrum linked to inactivation of the deubiquitinase BAP1. Science 2019, 364, 283–285. [Google Scholar] [CrossRef]
- Micol, J.B.; Pastore, A.; Inoue, D.; Duployez, N.; Kim, E.; Lee, S.C.W.; Durham, B.H.; Chung, Y.R.; Cho, H.; Zhang, X.J.; et al. ASXL2 is essential for haematopoiesis and acts as a haploinsufficient tumour suppressor in leukemia. Nat. Commun. 2017, 8, 15429. [Google Scholar] [CrossRef] [Green Version]
- Balasubramani, A.; Larjo, A.; Bassein, J.A.; Chang, X.; Hastie, R.B.; Togher, S.M.; Lähdesmäki, H.; Rao, A. Cancer-associated ASXL1 mutations may act as gain-of-function mutations of the ASXL1-BAP1 complex. Nat. Commun. 2015, 6, 7307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdel-Wahab, O.; Dey, A. The ASXL-BAP1 axis: New factors in myelopoiesis, cancer and epigenetics. Leukemia 2013, 27, 10–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gelsi-Boyer, V.; Brecqueville, M.; Devillier, R.; Murati, A.; Mozziconacci, M.J.; Birnbaum, D. Mutations in ASXL1 are associated with poor prognosis across the spectrum of malignant myeloid diseases. J. Hematol. Oncol. 2012, 5, 12. [Google Scholar] [CrossRef] [Green Version]
- Asada, S.; Goyama, S.; Inoue, D.; Shikata, S.; Takeda, R.; Fukushima, T.; Yonezawa, T.; Fujino, T.; Hayashi, Y.; Kawabata, K.C.; et al. Mutant ASXL1 cooperates with BAP1 to promote myeloid leukaemogenesis. Nat. Commun. 2018, 9, 2733. [Google Scholar] [CrossRef] [Green Version]
- Kobrinski, D.A.; Yang, H.; Kittaneh, M. BAP1: Role in carcinogenesis and clinical implications. Transl. Lung Cancer Res. 2020, 9, S60–S66. [Google Scholar] [CrossRef]
- Okonska, A.; Felley-Bosco, E. BAP1 missense mutations in cancer: Friend or foe? Trends Cancer 2019, 5, 659–662. [Google Scholar] [CrossRef]
- Dinan, A.M.; Atkins, J.F.; Firth, A.E. ASXL gain-of-function truncation mutants: Defective and dysregulated forms of a natural ribosomal frameshifting product? Biol. Direct 2017, 12, 24. [Google Scholar] [CrossRef] [PubMed]
- Hoischen, A.; Van Bon, B.W.M.; Rodríguez-Santiago, B.; Gilissen, C.; Vissers, L.E.L.M.; De Vries, P.; Janssen, I.; Van Lier, B.; Hastings, R.; Smithson, S.F.; et al. De novo nonsense mutations in ASXL1 cause Bohring-Opitz syndrome. Nat. Genet. 2011, 43, 729–731. [Google Scholar] [CrossRef] [PubMed]
- Shashi, V.; Pena, L.D.M.; Kim, K.; Burton, B.; Hempel, M.; Schoch, K.; Walkiewicz, M.; McLaughlin, H.M.; Cho, M.; Stong, N.; et al. De novo truncating variants in ASXL2 are associated with a unique and recognizable clinical phenotype. Am. J. Hum. Genet. 2016, 99, 991–999, Erratum in Am. J. Hum. Genet. 2017, 100, 179, 10.1016/j.ajhg.2016.12.004. [Google Scholar] [CrossRef] [Green Version]
- Bainbridge, M.N.; Hu, H.; Muzny, D.M.; Musante, L.; Lupski, J.R.; Graham, B.H.; Chen, W.; Gripp, K.W.; Jenny, K.; Wienker, T.F.; et al. De novo truncating mutations in ASXL3 are associated with a novel clinical phenotype with similarities to Bohring-Opitz syndrome. Genome Med. 2013, 5, 11. [Google Scholar] [CrossRef] [PubMed]
- Ventii, K.H.; Devi, N.S.; Friedrich, K.L.; Chernova, T.A.; Tighiouart, M.; Van Meir, E.G.; Wilkinson, K.D. BRCA1-associated protein-1 is a tumor suppressor that requires deubiquitinating activity and nuclear localization. Cancer Res. 2008, 68, 6953–6962. [Google Scholar] [CrossRef] [Green Version]
- Gelsi-Boyer, V.; Trouplin, V.; Adélaïde, J.; Bonansea, J.; Cervera, N.; Carbuccia, N.; Lagarde, A.; Prebet, T.; Nezri, M.; Sainty, D.; et al. Mutations of polycomb-associated gene ASXL1 in myelodysplastic syndromes and chronic myelomonocytic leukaemia. Br. J. Haematol. 2009, 145, 788–800. [Google Scholar] [CrossRef] [PubMed]
- Inoue, D.; Matsumoto, M.; Nagase, R.; Saika, M.; Fujino, T.; Nakayama, K.I.; Kitamura, T. Truncation mutants of ASXL1 observed in myeloid malignancies are expressed at detectable protein levels. Exp. Hematol. 2016, 44, 172–176. [Google Scholar] [CrossRef] [PubMed]
- Inoue, D.; Kitaura, J.; Togami, K.; Nishimura, K.; Enomoto, Y.; Uchida, T.; Kagiyama, Y.; Kawabata, K.C.; Nakahara, F.; Izawa, K.; et al. Myelodysplastic syndromes are induced by histone methylation altering ASXL1 mutations. J. Clin. Invest. 2013, 123, 4627–4640. [Google Scholar] [CrossRef]
- Nagase, R.; Inoue, D.; Pastore, A.; Fujino, T.; Hou, H.A.; Yamasaki, N.; Goyama, S.; Saika, M.; Kanai, A.; Sera, Y.; et al. Expression of mutant Asxl1 perturbs hematopoiesis and promotes susceptibility to leukemic transformation. J. Exp. Med. 2018, 215, 1729–1747. [Google Scholar] [CrossRef] [Green Version]
- Daou, S.; Barbour, H.; Ahmed, O.; Masclef, L.; Baril, C.; Nkwe, N.S.; Tchelougou, D.; Uriarte, M.; Bonneil, E.; Ceccarelli, D.; et al. Monoubiquitination of ASXLs controls the deubiquitinase activity of the tumor suppressor BAP1. Nat. Commun. 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Xia, Y.K.; Zeng, Y.R.; Zhang, M.L.; Liu, P.; Liu, F.; Zhang, H.; He, C.X.; Sun, Y.P.; Zhang, J.Y.; Zhang, C.; et al. Tumor-derived neomorphic mutations in ASXL1 impairs the BAP1-ASXL1-FOXK1/K2 transcription network. Protein Cell 2020. [Google Scholar] [CrossRef]
- Chiang, C.-M. Brd4 engagement from chromatin targeting to transcriptional regulation: Selective contact with acetylated histone H3 and H4. F1000 Biol. Rep. 2009, 7, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szczepanski, A.P.; Zhao, Z.; Sosnowski, T.; Goo, Y.A.; Bartom, E.T.; Wang, L. ASXL3 bridges BRD4 to BAP1 complex and governs enhancer activity in small cell lung cancer. Genome Med. 2020, 12, 63. [Google Scholar] [CrossRef] [PubMed]
- Saika, M.; Inoue, D.; Nagase, R.; Sato, N.; Tsuchiya, A.; Yabushita, T.; Kitamura, T.; Goyama, S. ASXL1 and SETBP1 mutations promote leukaemogenesis by repressing TGFβ pathway genes through histone deacetylation. Sci. Rep. 2018, 8, 15873. [Google Scholar] [CrossRef]
- Luchini, C.; Veronese, N.; Yachida, S.; Cheng, L.; Nottegar, A.; Stubbs, B.; Solmi, M.; Capelli, P.; Pea, A.; Barbareschi, M.; et al. Different prognostic roles of tumor suppressor gene BAP1 in cancer: A systematic review with meta-analysis. Genes Chromosom. Cancer 2016, 55, 741–749. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Wang, Y.; Wei, P.; Du, X. BRCA1-associated protein 1 deficiency in lung adenocarcinoma predicts poor outcome and increased tumor invasion. BMC Cancer 2016, 16, 670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.Y.; Wang, Z.; Huang, J.B.; Ren, X.D.; Ye, D.; Zhu, W.W.; Qin, L.X. Tissue-specific significance of BAP1 gene mutation in prognostic prediction and molecular taxonomy among different types of cancer. Tumor Biol. 2017, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reddington, C.J.; Fellner, M.; Burgess, A.E.; Mace, P.D. Molecular Regulation of the Polycomb Repressive-Deubiquitinase. Int. J. Mol. Sci. 2020, 21, 7837. https://doi.org/10.3390/ijms21217837
Reddington CJ, Fellner M, Burgess AE, Mace PD. Molecular Regulation of the Polycomb Repressive-Deubiquitinase. International Journal of Molecular Sciences. 2020; 21(21):7837. https://doi.org/10.3390/ijms21217837
Chicago/Turabian StyleReddington, Cameron J., Matthias Fellner, Abigail E. Burgess, and Peter D. Mace. 2020. "Molecular Regulation of the Polycomb Repressive-Deubiquitinase" International Journal of Molecular Sciences 21, no. 21: 7837. https://doi.org/10.3390/ijms21217837





