Crosstalk between Androgen-ZIP9 Signaling and Notch Pathway in Rodent Sertoli Cells
Abstract
:1. Introduction
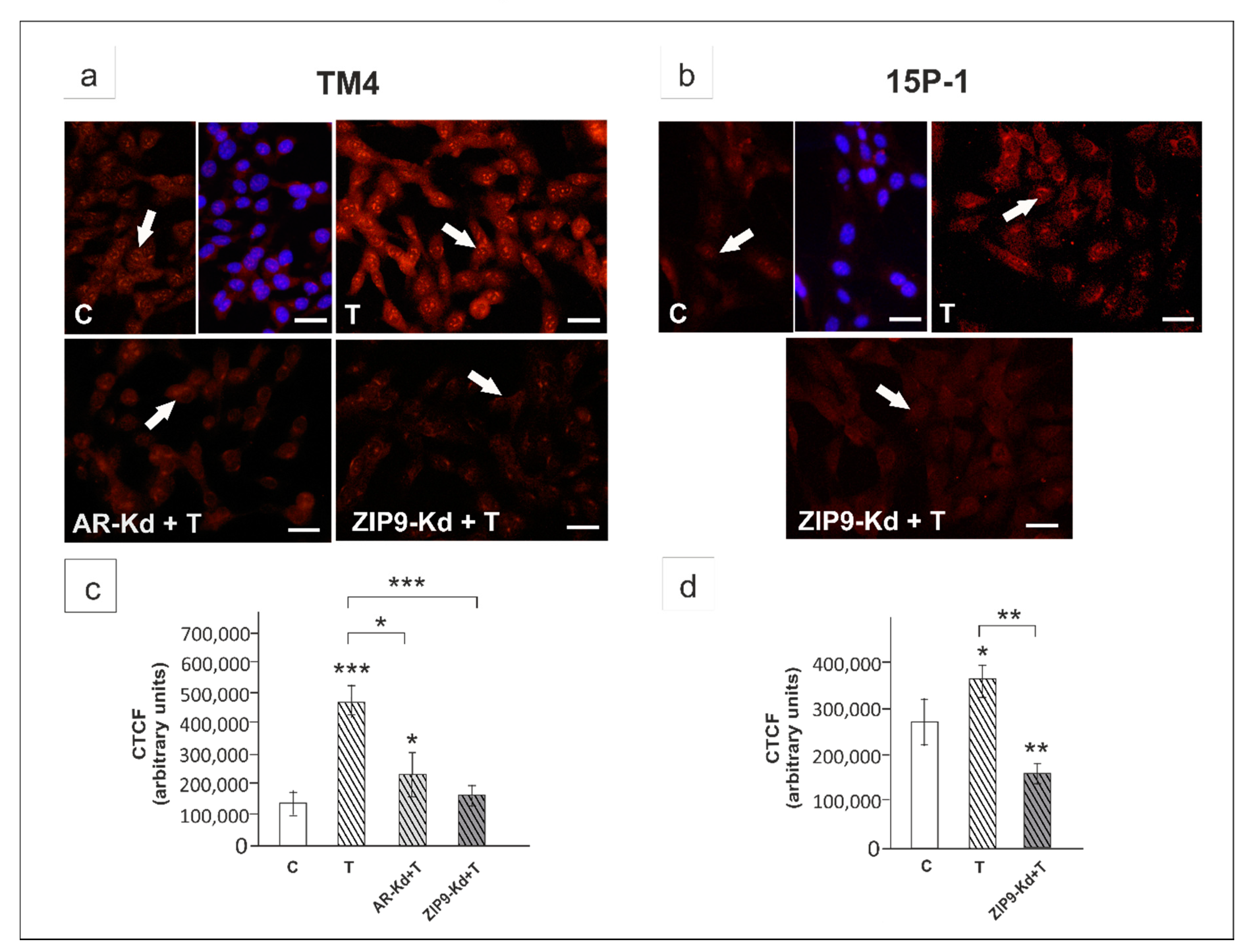
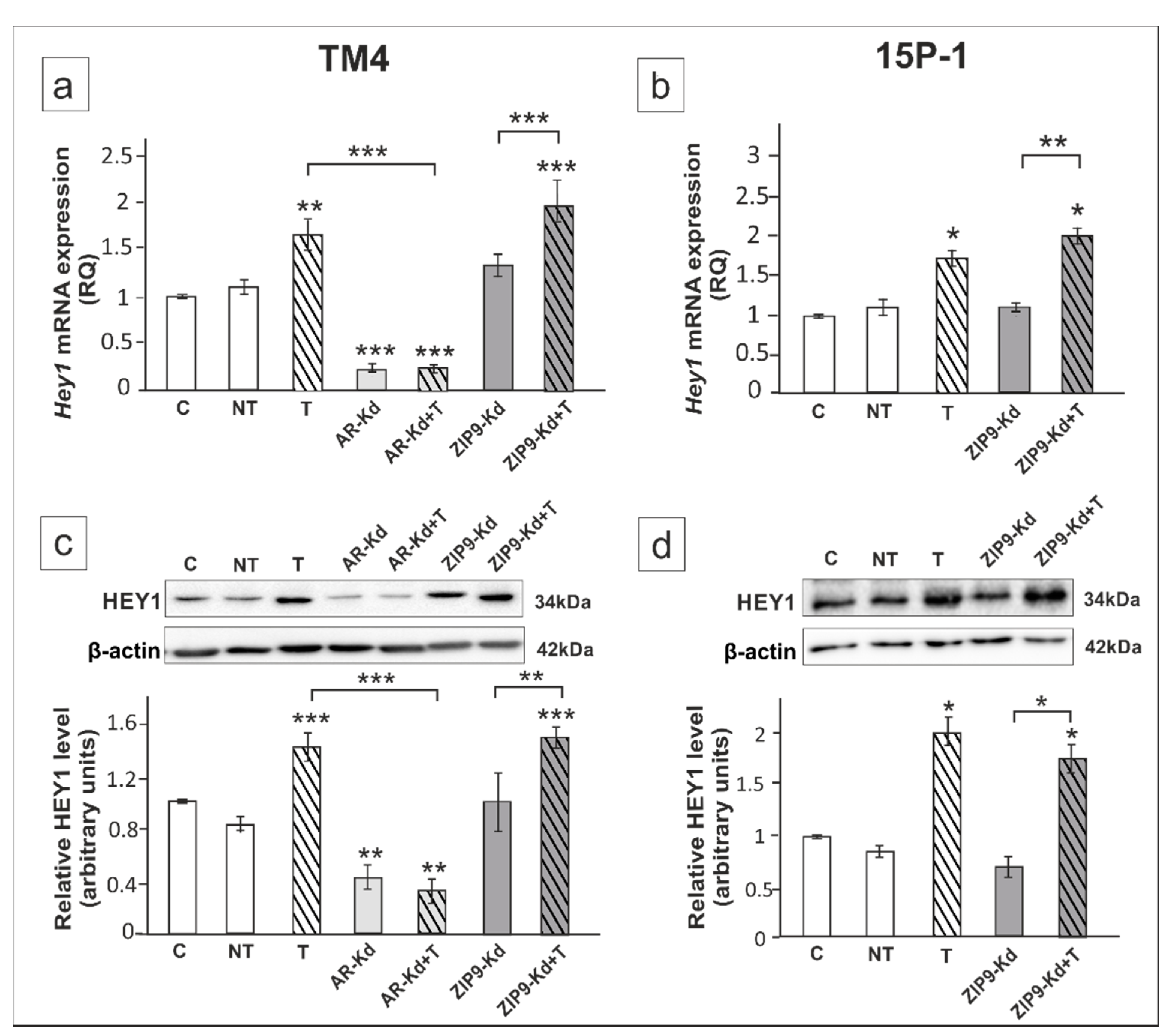
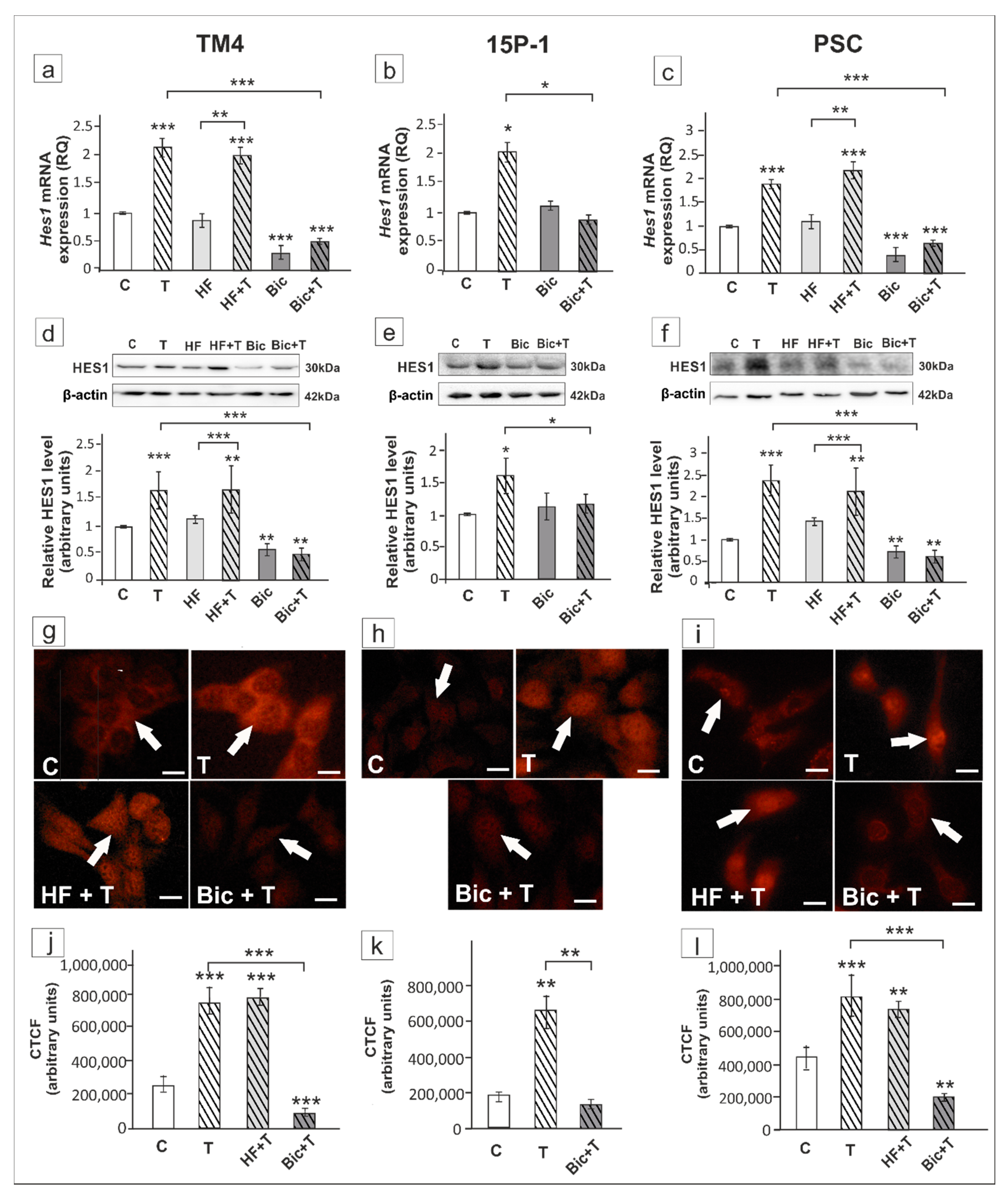
2. Results
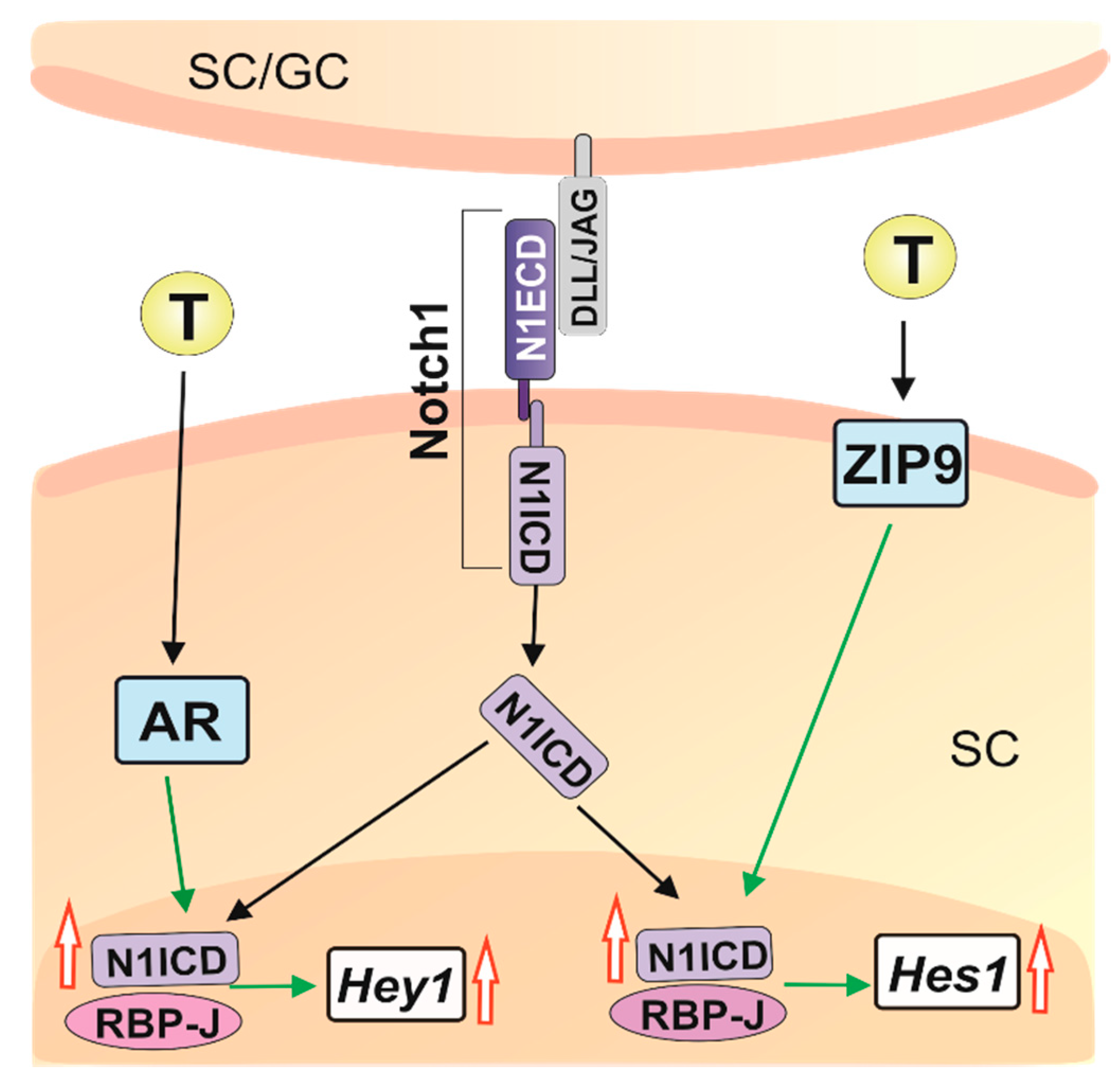
3. Discussion
4. Materials and Methods
4.1. Cell Line Cultures and Treatments
4.2. Transfection of TM4 or 15P-1 Cells with siRNA Duplexes
4.3. Reporter Assay
4.4. Primary Sertoli Cell (PSC) Culture and Treatments
4.5. RNA Isolation, Reverse Transcription and Real-Time Quantitative RT-PCR
4.6. Western Blot Analysis
4.7. Immunofluorescence
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AR | Nuclear androgen receptor |
| ATF-1 | Activating transcription factor 1 |
| Bic | Bicalutamide |
| CREB | cAMP-response element binding protein |
| DAPI | 4′,6-diamidino-2-phenylindole |
| DMEM | Dulbecco′s Modified Eagle Medium |
| DMSO | Dimethyl sulfoxide |
| EDTA | Ethylenediaminetetraacetic acid |
| ERK 1/2 | Extracellular signal-regulated protein kinase 1/2 |
| FOXA1 | Forkhead box protein A1 |
| GPRC6A | G protein-coupled receptor class C group 6 member A |
| Hes1/HES1 | Hairy/enhancer of split 1 |
| Hey1/HEY1 | Hes-related with YRPW motif 1 |
| HF | Hydroxyflutamide |
| N1ICD | Notch1 intracellular domain |
| PSC | Primary Sertoli cells |
| RBP-J | Recombination signal binding protein |
| RT-PCR | Reverse-transcriptase polymerase chain reaction |
| SDS-PAGE | Sodium dodecyl sulfate – poliacrylamide gel electrophoresis |
| TRPM8 | Transient receptor potential cation channel subfamily M member 8 |
| ZIP9 | Zrt- and Irt-like protein 9 |
References
- Bremner, W.J.; Millar, M.R.; Sharpe, R.M.; Saunders, P.T. Immunohistochemical localization of androgen receptors in the rat testis: Evidence for stage-dependent expression and regulation by androgens. Endocrinology 1994, 135, 1227–1234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rey, R.A.; Musse, M.; Venara, M.; Chemes, H.E. Ontogeny of the androgen receptor expression in the fetal and postnatal testis: Its relevance on Sertoli cell maturation and the onset of adult spermatogenesis. Microsc. Res. Tech. 2009, 72, 787–795. [Google Scholar] [CrossRef]
- De Gendt, K.; Swinnen, J.V.; Saunders, P.T.; Schoonjans, L.; Dewerchin, M.; Devos, A.; Tan, K.; Atanassova, N.; Claessens, F.; Lécureuil, C.; et al. A Sertoli cell-selective knockout of the androgen receptor causes spermatogenic arrest in meiosis. Proc. Natl. Acad. Sci. USA 2004, 101, 1327–1332. [Google Scholar] [CrossRef] [Green Version]
- Davey, R.A.; Grossmann, M. Androgen receptor structure, function and biology: From bench to bedside. Clin. Biochem. Rev. 2016, 37, 3–15. [Google Scholar]
- Walker, W.H. Non-classical actions of testosterone and spermatogenesis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2010, 365, 1557–1569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Hara, L.; Smith, L.B. Androgen receptor roles in spermatogenesis and infertility. Best Pract. Res. Clin. Endocrinol. Metab. 2015, 29, 595–605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berg, A.H.; Rice, C.D.; Rahman, M.S.; Dong, J.; Thomas, P. Identification and characterization of membrane androgen receptors in the ZIP9 zinc transporter subfamily: I. Discovery in female atlantic croaker and evidence ZIP9 mediates testosterone-induced apoptosis of ovarian follicle cells. Endocrinology 2014, 155, 4237–4249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bulldan, A.; Dietze, R.; Shihan, M.; Scheiner-Bobis, G. Non-classical testosterone signaling mediated through ZIP9 stimulates claudin expression and tight junction formation in Sertoli cells. Cell Signal. 2016, 28, 1075–1085. [Google Scholar] [CrossRef]
- Kamińska, A.; Pardyak, L.; Marek, S.; Wróbel, K.; Kotula-Balak, M.; Bilińska, B.; Hejmej, A. Notch signaling regulates nuclear androgen receptor AR and membrane androgen receptor ZIP9 in mouse Sertoli cells. Andrology 2020, 8, 457–472. [Google Scholar] [CrossRef] [PubMed]
- Profaska-Szymik, M.; Galuszka, A.; Korzekwa, A.J.; Hejmej, A.; Gorowska-Wojtowicz, E.; Pawlicki, P.; Kotula-Balak, M.; Tarasiuk, K.; Tuz, R. Implication of membrane androgen receptor (ZIP9) in cell senescence in regressed testes of the bank vole. Int. J. Mol. Sci. 2020, 21, 6888. [Google Scholar] [CrossRef]
- Bulldan, A.; Shihan, M.; Goericke-Pesch, S.; Scheiner-Bobis, G. Signaling events associated with gonadotropin releasing hormone-agonist-induced hormonal castration and its reversal in canines. Mol. Reprod. Dev. 2016, 83, 1092–1101. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.; Pang, Y.; Dong, J.; Berg, A.H. Identification and characterization of membrane androgen receptors in the ZIP9 zinc transporter subfamily: II. Role of human ZIP9 in testosterone-induced prostate and breast cancer cell apoptosis. Endocrinology 2014, 155, 4250–4265. [Google Scholar] [CrossRef] [Green Version]
- Bulldan, A.; Bartsch, J.W.; Konrad, L.; Scheiner-Bobis, G. ZIP9 but not the androgen receptor mediates testosterone-induced migratory activity of metastatic prostate cancer cells. Biochim. Biophys. Acta Mol. Cell Res. 2018, 1865, 1857–1868. [Google Scholar] [CrossRef] [PubMed]
- Kalyvianaki, K.; Panagiotopoulos, A.A.; Malamos, P.; Moustou, E.; Tzardi, M.; Stathopoulos, E.N.; Ioannidis, G.S.; Marias, K.; Notas, G.; Theodoropoulos, P.A.; et al. Membrane androgen receptors (Oxer1, Gprc6a and ZIP9) in prostate and breast cancer: A comparative study of their expression. Steroids 2019, 142, 100–108. [Google Scholar] [CrossRef]
- Griswold, M.D. 50 years of spermatogenesis: Sertoli cells and their interactions with germ cells. Biol. Reprod. 2018, 99, 87–100. [Google Scholar] [CrossRef] [PubMed]
- Garcia, T.X.; DeFalco, T.; Capel, B.; Hofmann, M.C. Constitutive activation of Notch1 signaling in Sertoli cells causes gonocyte exit from quiescence. Dev. Biol. 2013, 377, 188–201. [Google Scholar] [CrossRef] [Green Version]
- Garcia, T.X.; Parekh, P.; Gandhi, P.; Sinha, K.; Hofmann, M.C. The Notch Ligand JAG1 Regulates GDNF Expression in Sertoli Cells. Stem Cells Dev. 2017, 26, 585–598. [Google Scholar] [CrossRef] [Green Version]
- Parekh, P.A.; Garcia, T.X.; Waheeb, R.; Jain, V.; Gandhi, P.; Meistrich, M.L.; Shetty, G.; Hofmann, M.C. Undifferentiated spermatogonia regulate Cyp26b1 expression through NOTCH signaling and drive germ cell differentiation. FASEB J. 2019, 33, 8423–8435. [Google Scholar] [CrossRef]
- Murta, D.; Batista, M.; Trindade, A.; Silva, E.; Henrique, D.; Duarte, A.; Lopes-da-Costa, L. In vivo notch signaling blockade induces abnormal spermatogenesis in the mouse. PLoS ONE 2014, 9, e113365. [Google Scholar] [CrossRef]
- Falo-Sanjuan, J.; Bray, S.J. Decoding the Notch signal. Dev. Growth Differ. 2020, 62, 4–14. [Google Scholar] [CrossRef] [Green Version]
- Iso, T.; Kedes, L.; Hamamori, Y. HES and HERP families: Multiple effectors of the Notch signaling pathway. J. Cell Physiol. 2003, 194, 237–255. [Google Scholar] [CrossRef]
- Bray, S.J. Notch signalling in context. Nat. Rev. Mol. Cell Biol. 2016, 17, 722–735. [Google Scholar] [CrossRef] [PubMed]
- Krejcí, A.; Bernard, F.; Housden, B.E.; Collins, S.; Bray, S.J. Direct response to Notch activation: Signaling crosstalk and incoherent logic. Sci. Signal. 2009, 2. [Google Scholar] [CrossRef]
- Bertrand, F.E. The cross-talk of NOTCH and GSK-3 signaling in colon and other cancers. Biochim. Biophys. Acta Mol. Cell Res. 2020, 1867, 118738. [Google Scholar] [CrossRef] [PubMed]
- Kamakura, S.; Oishi, K.; Yoshimatsu, T.; Nakafuku, M.; Masuyama, N.; Gotoh, Y. Hes binding to STAT3 mediates crosstalk between Notch and JAK-STAT signalling. Nat. Cell Biol. 2004, 6, 547–554. [Google Scholar] [CrossRef]
- Li, N.; Zhang, L.; Li, Q.; Du, Y.; Liu, H.; Liu, Y.; Xiong, W. Notch activity mediates oestrogen-induced stromal cell invasion in endometriosis. Reproduction 2018, 157, 371–381. [Google Scholar] [CrossRef] [Green Version]
- Piccirilli, D.; Baldini, E.; Massimiani, M.; Camaioni, A.; Salustri, A.; Bernardini, R.; Centanni, M.; Ulisse, S.; Moretti, C.; Campagnolo, L. Thyroid hormone regulates protease expression and activation of Notch signaling in implantation and embryo development. J. Endocrinol. 2018, 236, 1–12. [Google Scholar] [CrossRef]
- Sinha-Hikim, I.; Cornford, M.; Gaytan, H.; Lee, M.L.; Bhasin, S. Effects of testosterone supplementation on skeletal muscle fiber hypertrophy and satellite cells in community-dwelling older men. J. Clin. Endocrinol. Metab. 2006, 91, 3024–3033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nantermet, P.V.; Xu, J.; Yu, Y.; Hodor, P.; Holder, D.; Adamski, S.; Gentile, M.A.; Kimmel, D.B.; Harada, S.; Gerhold, D.; et al. Identification of genetic pathways activated by the androgen receptor during the induction of proliferation in the ventral prostate gland. J. Biol. Chem. 2003, 23. [Google Scholar] [CrossRef] [Green Version]
- Aldahl, J.; Yu, E.J.; He, Y.; Hooker, E.; Wong, M.; Le, V.; Olson, A.; Lee, D.H.; Kim, W.K.; Murtaugh, C.L.; et al. A pivotal role of androgen signaling in Notch-responsive cells in prostate development, maturation, and regeneration. Differentiation 2019, 107, 1–10. [Google Scholar] [CrossRef]
- Defalco, T.; Saraswathula, A.; Briot, A.; Iruela-Arispe, M.L.; Capel, B. Testosterone levels influence mouse fetal Leydig cell progenitors through notch signaling. Biol. Reprod. 2013, 88, 91. [Google Scholar] [CrossRef]
- Willems, A.; Batlouni, S.R.; Esnal, A.; Swinnen, J.V.; Saunders, P.T.; Sharpe, R.M.; França, L.R.; De Gendt, K.; Verhoeven, G. Selective ablation of the androgen receptor in mouse sertoli cells affects sertoli cell maturation, barrier formation and cytoskeletal development. PLoS ONE 2010, 5, e14168. [Google Scholar] [CrossRef] [Green Version]
- Kamińska, A.; Marek, S.; Pardyak, L.; Brzoskwinia, M.; Pawlicki, P.; Bilińska, B.; Hejmej, A. Disruption of androgen signaling during puberty affects Notch pathway in rat seminiferous epithelium. Reprod. Biol. Endocrinol. 2020, 18, 30. [Google Scholar] [CrossRef]
- Handelsman, D.J.; Jimenez, M.; Singh, G.K.; Spaliviero, J.; Desai, R.; Walters, K.A. Measurement of testosterone by immunoassays and mass spectrometry in mouse serum, testicular, and ovarian extracts. Endocrinology 2015, 156, 400–405. [Google Scholar] [CrossRef] [Green Version]
- Oduwole, O.O.; Vydra, N.; Wood, N.E.; Samanta, L.; Owen, L.; Keevil, B.; Donaldson, M.; Naresh, K.; Huhtaniemi, I.T. Overlapping dose responses of spermatogenic and extragonadal testosterone actions jeopardize the principle of hormonal male contraception. FASEB J. 2014, 28, 2566–2576. [Google Scholar] [CrossRef] [PubMed]
- Rato, L.; Alves, M.G.; Dias, T.R.; Lopes, G.; Cavaco, J.E.; Socorro, S.; Oliveira, P.F. High-energy diets may induce a pre-diabetic state altering testicular glycolytic metabolic profile and male reproductive parameters. Andrology 2013, 1, 495–504. [Google Scholar] [CrossRef]
- Gatson, J.W.; Singh, M. Activation of a membrane-associated androgen receptor promotes cell death in primary cortical astrocytes. Endocrinology 2007, 148, 2458–2464. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Kang, L.; Zhang, Y.; Feng, B.; Du, J.; Cui, H. Detecting the presence of hippocampus membrane androgen receptors in male SAMP8 mice and their induced synaptic plasticity. Mol. Cell. Endocrinol. 2015, 414, 82–90. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, S.; Chen, H.; Chen, M.; Mi, S.; Ma, J.; Wang, C.; Sun, H.; Liu, X.; Cui, H. Non-genomic mechanisms mediate androgen-induced PSD95 expression. Aging 2019, 11, 2281–2294. [Google Scholar] [CrossRef] [PubMed]
- Bulldan, A.; Malviya, V.N.; Upmanyu, N.; Konrad, L.; Scheiner-Bobis, G. Testosterone/bicalutamide antagonism at the predicted extracellular androgen binding site of ZIP9. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 2402–2414. [Google Scholar] [CrossRef]
- Aguirre-Portolés, C.; Payne, R.; Trautz, A.; Foskett, J.K.; Natale, C.A.; Ridky, T.W. Testosterone signaling through ZIP9 renders melanoma more aggressive in males than in females. bioRxiv 2020, 2020, 989160. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.H.; Han, G.C.; Seo, J.Y.; Park, I.; Park, W.; Jeong, H.W.; Lee, S.H.; Bae, S.H.; Seong, J.; Yum, M.K.; et al. Sex hormones establish a reserve pool of adult muscle stem cells. Nat. Cell Biol. 2016, 18, 930–940. [Google Scholar] [CrossRef]
- Sharma, N.L.; Massie, C.E.; Ramos-Montoya, A.; Zecchini, V.; Scott, H.E.; Lamb, A.D.; MacArthur, S.; Stark, R.; Warren, A.Y.; Mills, I.G.; et al. The androgen receptor induces a distinct transcriptional program in castration-resistant prostate cancer in man. Cancer Cell 2013, 23, 35–47. [Google Scholar] [CrossRef] [Green Version]
- Olsen, J.R.; Azeem, W.; Hellem, M.R.; Marvyin, K.; Hua, Y.; Qu, Y.; Li, L.; Lin, B.; Ke, X.; Øyan, A.M.; et al. Context dependent regulatory patterns of the androgen receptor and androgen receptor target genes. BMC Cancer 2016, 16, 377. [Google Scholar] [CrossRef] [Green Version]
- Converse, A.; Zhang, C.; Thomas, P. Membrane androgen receptor ZIP9 induces croaker ovarian cell apoptosis via stimulatory G protein alpha subunit and MAP kinase signaling. Endocrinology 2017, 158, 3015–3029. [Google Scholar] [CrossRef]
- Shihan, M.; Chan, K.H.; Konrad, L.; Scheiner-Bobis, G. Non-classical testosterone signaling in spermatogenic GC-2 cells is mediated through ZIP9 interacting with Gnα11. Cell Signal. 2015, 27, 2077–2086. [Google Scholar] [CrossRef]
- Belandia, B.; Powell, S.M.; García-Pedrero, J.M.; Walker, M.M.; Bevan, C.L.; Parker, M.G. Hey1, a mediator of notch signaling, is an androgen receptor corepressor. Mol. Cell Biol. 2005, 25, 1425–1436. [Google Scholar] [CrossRef] [Green Version]
- Asuthkar, S.; Velpula, K.K.; Elustondo, P.A.; Demirkhanyan, L.; Zakharian, E. TRPM8 channel as a novel molecular target in androgen-regulated prostate cancer cells. Oncotarget 2015, 6, 17221–17236. [Google Scholar] [CrossRef] [Green Version]
- Grolez, G.P.; Gordiendko, D.V.; Clarisse, M.; Hammadi, M.; Desruelles, E.; Fromont, G.; Prevarskaya, N.; Slomianny, C.; Gkika, D. TRPM8-androgen receptor association within lipid rafts promotes prostate cancer cell migration. Cell Death Dis. 2019, 10, 652. [Google Scholar] [CrossRef] [Green Version]
- Ye, R.; Pi, M.; Nooh, M.M.; Bahout, S.W.; Quarles, L.D. Human GPRC6A mediates testosterone-induced mitogen-activated protein kinases and mTORC1 signaling in prostate cancer cells. Mol. Pharmacol. 2019, 95, 563–572. [Google Scholar] [CrossRef] [Green Version]
- Borowiec, A.S.; Sion, B.; Chalmel, F.; Rolland, A.D.; Lemonnier, L.; De Clerck, T.; Bokhobza, A.; Derouiche, S.; Dewailly, E.; Slomianny, C.; et al. Cold/menthol TRPM8 receptors initiate the cold-shock response and protect germ cells from cold-shock-induced oxidation. FASEB J. 2016, 30, 3155–3170. [Google Scholar] [CrossRef] [Green Version]
- Pi, M.; Chen, L.; Huang, M.Z.; Zhu, W.; Ringhofer, B.; Luo, J.; Christenson, L.; Li, B.; Zhang, J.; Jackson, P.D.; et al. GPRC6A null mice exhibit osteopenia, feminization and metabolic syndrome. PLoS ONE 2008, 3, e3858. [Google Scholar] [CrossRef]
- Qiu, M.; Bao, W.; Wang, J.; Yang, T.; He, X.; Liao, Y.; Wan, X. FOXA1 promotes tumor cell proliferation through AR involving the Notch pathway in endometrial cancer. BMC Cancer 2014, 14, 78. [Google Scholar] [CrossRef]
- Chen, H.; Libertini, S.J.; George, M.; Dandekar, S.; Tepper, C.G.; Al-Bataina, B.; Kung, H.J.; Ghosh, P.M.; Mudryj, M. Genome-wide analysis of androgen receptor binding and gene regulation in two CWR22-derived prostate cancer cell lines. Endocr. Relat. Cancer 2010, 17, 857–873. [Google Scholar] [CrossRef] [Green Version]
- Clocchiatti, A.; Ghosh, S.; Procopio, M.G.; Mazzeo, L.; Bordignon, P.; Ostano, P.; Goruppi, S.; Bottoni, G.; Katarkar, A.; Levesque, M.; et al. Androgen receptor functions as transcriptional repressor of cancer-associated fibroblast activation. J. Clin. Investig. 2018, 128, 5531–5548. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Zou, J.; Zhao, B.; Johannsen, E.; Ashworth, T.; Wong, H.; Pear, W.S.; Schug, J.; Blacklow, S.C.; Arnett, K.L.; et al. Genome-wide analysis reveals conserved and divergent features of Notch1/RBPJ binding in human and murine T-lymphoblastic leukemia cells. Proc. Natl. Acad. Sci. USA 2011, 108, 14908–14913. [Google Scholar] [CrossRef] [Green Version]
- Lake, R.J.; Tsai, P.F.; Choi, I.; Won, K.J.; Fan, H.Y. RBPJ, the major transcriptional effector of Notch signaling, remains associated with chromatin throughout mitosis, suggesting a role in mitotic bookmarking. PLoS Genet. 2014, 10, e1004204. [Google Scholar] [CrossRef]
- Ghosh, S.; Thakur, M.K. PS1 expression is downregulated by gonadal steroids in adult mouse brain. Neurochem. Res. 2008, 33, 365–369. [Google Scholar] [CrossRef] [PubMed]
- Drummond, E.S.; Martins, R.N.; Handelsman, D.J.; Harvey, A.R. Altered expression of Alzheimer’s disease-related proteins in male hypogonadal mice. Endocrinology 2012, 153, 2789–2799. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.H.; Wu, Y.; Yao, S.; Levine, A.C.; Kirschenbaum, A.; Collier, L.; Bauman, W.A.; Cardozo, C.P. Androgens up-regulate transcription of the Notch inhibitor Numb in C2C12 myoblasts via Wnt/β-catenin signaling to T cell factor elements in the Numb promoter. J. Biol. Chem. 2013, 288, 17990–17998. [Google Scholar] [CrossRef] [Green Version]
- Curry, C.L.; Reed, L.L.; Nickoloff, B.J.; Miele, L.; Foreman, K.E. Notch-independent regulation of Hes-1 expression by c-Jun N-terminal kinase signaling in human endothelial cells. Lab. Invest. 2006, 86, 842–852. [Google Scholar] [CrossRef]
- Mruk, D.D.; Cheng, C.Y. An in vitro system to study Sertoli cell blood-testis barrier dynamics. Methods Mol. Biol. 2011, 763, 237–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chojnacka, K.; Zarzycka, M.; Hejmej, A.; Mruk, D.D.; Gorowska, E.; Kotula-Balak, M.; Klimek, M.; Bilinska, B. Hydroxyflutamide affects connexin 43 via the activation of PI3K/Akt-dependent pathway but has no effect on the crosstalk between PI3K/Akt and ERK1/2 pathways at the Raf-1 kinase level in primary rat Sertoli cells. Toxicol. In Vitro 2016, 31, 146–157. [Google Scholar] [CrossRef]
- Galdieri, M.; Ziparo, E.; Palombi, F.; Russo, M.A.; Stefanini, M. Pure Sertoli cell cultures: A new model for the study of somatic—germ cell interactions. J. Androl. 1981, 2, 249–254. [Google Scholar] [CrossRef] [Green Version]
- Svec, D.; Tichopad, A.; Novosadova, V.; Pfaffl, M.W.; Kubista, M. How good is a PCR efficiency estimate: Recommendations for precise and robust qPCR efficiency assessments. Biomol. Detect. Quantif. 2015, 3, 9–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)). Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Bilinska, B.; Hejmej, A.; Kotula-Balak, M. Preparation of testicular samples for histology and immunohistochemistry. Methods Mol. Biol. 2018, 1748, 17–36. [Google Scholar] [CrossRef]
- Jakic, B.; Buszko, M.; Cappellano, G.; Wick, G. Elevated sodium leads to the increased expression of HSP60 and induces apoptosis in HUVECs. PLoS ONE 2017, 12, e0179383. [Google Scholar] [CrossRef] [Green Version]







| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| Mouse | ||
| Actb | CTGGAACGGTGAAGGTGACA | AAGGGGACTTCCTGTAACAATGCA |
| B2m | TTCTGGTGCTTGTCTCACTCA | CAGTATGTTCGGCTTCCCATTC |
| Gapdh | GGAGATTGTTGCCATCAACG | GGAGATTGTTGCCATCAACG |
| Hes1 | ACCTTCCAGTGGCTCCTC | TTTAGTGTCCGTCAGAAGAGAG |
| Hey1 | GCCGAAGTTGCCCGTTATCTG | GCCGAAGTTGCCCGTTATCTG |
| Notch1 | GATGCCACCTGAACAACTGC | TGACAACAGCAACAGCAAGG |
| Rn18S | CTCTGGTTGCTCTGTGCAGT | GGCTCCTTGTAGGGGTTCTC |
| Rat | ||
| Actb | AAGTACCCCATTGAACACGG | ATCACAATGCCAGTGGTACG |
| B2m | GGACTGGTCTTTCTATATCCTGGC | GATCACATGTCTCGATCCCAGTAG |
| Gapdh | GGAGATTGTTGCCATCAACG | CACAATGCCAAAGTTGTCA |
| Hes1 | GGCAGGCGCACCCCGCCTTG | GCAGCCAGGCTGGAGAGGCT |
| Hey1 | AAAGACGGAGAGGCATCATCG | GCAGTGTGCAGCATTTTCAGG |
| Notch1 | GCAGCCACAGAACTTACAAATCCAG | TAAATGCCTCTGGAATGTGGGTGAT |
| Rn18S | GCCGCGGTAATTCCAGCTCCA | CCCGCCCGCTCCCAAGATC |
| Antibody | Host Species | Vendor | Cat. Number | Dilution |
|---|---|---|---|---|
| Anti-actin | Mouse | Sigma-Aldrich | A2228 | 1:3000 (WB) |
| Anti-HES1 | Rabbit | Thermo Fisher | PA5-28802 | 1:1000 (WB); 1:100 (IF) |
| Anti-HEY1 | Rabbit | Thermo Fisher | PA5-40553 | 1:2000 (WB), 1:100 (IF) |
| Anti-N1ICD | Rabbit | Abcam | ab8925 | 1:1000 (WB), 1:200 (IF) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kamińska, A.; Marek, S.; Pardyak, L.; Brzoskwinia, M.; Bilinska, B.; Hejmej, A. Crosstalk between Androgen-ZIP9 Signaling and Notch Pathway in Rodent Sertoli Cells. Int. J. Mol. Sci. 2020, 21, 8275. https://doi.org/10.3390/ijms21218275
Kamińska A, Marek S, Pardyak L, Brzoskwinia M, Bilinska B, Hejmej A. Crosstalk between Androgen-ZIP9 Signaling and Notch Pathway in Rodent Sertoli Cells. International Journal of Molecular Sciences. 2020; 21(21):8275. https://doi.org/10.3390/ijms21218275
Chicago/Turabian StyleKamińska, Alicja, Sylwia Marek, Laura Pardyak, Małgorzata Brzoskwinia, Barbara Bilinska, and Anna Hejmej. 2020. "Crosstalk between Androgen-ZIP9 Signaling and Notch Pathway in Rodent Sertoli Cells" International Journal of Molecular Sciences 21, no. 21: 8275. https://doi.org/10.3390/ijms21218275





