Protective Effect of l-Hexaguluroic Acid Hexasodium Salt on UVA-Induced Photo-Aging in HaCaT Cells
Abstract
:1. Introduction
2. Results
2.1. G6 Inhibits UVA-Irradiated Cytotoxicity in HaCaT Cells
2.2. Effect of G6 on Mitochondrial Membrane Potential (MMP)
2.3. G6 Co-Localize with Mitochondria
2.4. Effect of G6 on ROS Production, ATP Content, and NAD+/NADH Ratio Change
2.5. Effect of G6 on Mitochondrial Functions
2.6. Molecular Docking Predicted the Possible Interaction between G6 and SIRT1 Protein
2.7. Effect of G6 on Key Protein Expressions of SIRT1 Signaling Pathway
2.8. Effects of G6 on the mRNA Expression of SIRT1 Pathways
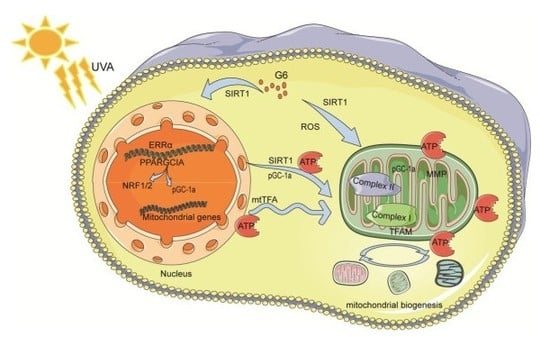
3. Discussion
4. Materials and Methods
4.1. Materials and Reagents
4.2. Cell Culture and Treatments
4.3. MTT Assay
4.4. Measurement of Mitochondrial Membrane Potential (MMP, ΔΨm)
4.5. Measurement of Reactive Oxygen Species (ROS) Generation
4.6. Determination of ATP Content
4.7. NAD+/NADH Ratio Change
4.8. Activity of Mitochondrial Complexes I and II
4.9. Western Blot Analysis
4.10. Real-Time RT-PCR Assay
4.11. Preparation of FITC-Labeled G6
4.12. Intracellular Localization of G6
4.13. Molecule Docking Studies
4.14. SIRT1 Inhibitor Interferes with SIRT1 Protein Expression
4.15. Statistics
Supplementary Materials
Supplementary File 1Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| G6 | l-Hexaguluroic acid hexasodium salt |
| pGC-1α | peroxisome proliferator-activated receptor gamma coactivator-1 alpha |
| SIRT1 | sirtuin 1 |
| ROS | reactive oxygen species |
| MMP | mitochondrial membrane potential |
| FITC | fluorescin isothiocyanate |
| ddH2O | double-distilled water |
| NADH | nicotinamide adenine dinucleotide |
| PVDF | polyvinylidene fluoride |
| JC-1 | J-Aggregate forming lipophilic cation 55′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolcarbocyanine iodide |
| BSA | bovine serum albumin |
References
- Janjetovic, Z.; Nahmias, Z.P.; Hanna, S.; Jarrett, S.G.; Kim, T.K.; Reiter, R.J.; Slominski, A.T. Melatonin and its metabolites ameliorate ultraviolet B-induced damage in human epidermal keratinocytes. J. Pineal Res. 2014, 57, 90–102. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-W.; Chi, M.-C.; Peng, K.-T.; Chiang, Y.-C.; Hsu, L.-F.; Yan, Y.-L.; Li, H.-Y.; Chen, M.-C.; Lee, I.-T.; Lai, C.-H. Water-Soluble Fullerenol C60 (OH) 36 toward Effective Anti-Air Pollution Induced by Urban Particulate Matter in HaCaT Cell. Int. J. Mol. Sci. 2019, 20, 4259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsumura, Y.; Ananthaswamy, H.N. Toxic effects of ultraviolet radiation on the skin. Toxicol. Appl. Pharmacol. 2004, 195, 298–308. [Google Scholar] [CrossRef] [PubMed]
- Debacq-Chainiaux, F.; Leduc, C.; Verbeke, A.; Toussaint, O. UV, stress and aging. Derm. Endocrinol. 2012, 4, 236–240. [Google Scholar] [CrossRef] [Green Version]
- Berneburg, M.; Plettenberg, H.; Krutmann, J. Photoaging of human skin. Photodermatol. Photoimmunol. Photomed. Rev. Artic. 2000, 16, 239–244. [Google Scholar] [CrossRef]
- Puizina-Ivic, N. Skin aging. Acta Dermatovenerol. Alp. Panon. Adriat. 2008, 17, 47. [Google Scholar]
- Yang, T.-H.; Lai, Y.-H.; Lin, T.-P.; Liu, W.-S.; Kuan, L.-C.; Liu, C.-C. Chronic exposure to Rhodobacter sphaeroides extract Lycogen™ prevents UVA-induced malondialdehyde accumulation and procollagen I down-regulation in human dermal fibroblasts. Int. J. Mol. Sci. 2014, 15, 1686–1699. [Google Scholar] [CrossRef] [Green Version]
- Xie, H.; Liu, F.; Liu, L.; Dan, J.; Luo, Y.; Yi, Y.; Chen, X.; Li, J. Protective role of AQP3 in UVA-induced NHSFs apoptosis via Bcl2 up-regulation. Arch. Dermatol. Res. 2013, 305, 397–406. [Google Scholar] [CrossRef]
- Ichihashi, M.; Ueda, M.; Budiyanto, A.; Bito, T.; Oka, M.; Fukunaga, M.; Tsuru, K.; Horikawa, T. UV-induced skin damage. Toxicology 2003, 189, 21–39. [Google Scholar] [CrossRef]
- Fan, J.; Zhuang, Y.; Li, B. Effects of collagen and collagen hydrolysate from jellyfish umbrella on histological and immunity changes of mice photoaging. Nutrients 2013, 5, 223–233. [Google Scholar] [CrossRef] [Green Version]
- Ryu, J.; Park, S.-J.; Kim, I.-H.; Choi, Y.H.; Nam, T.-J. Protective effect of porphyra-334 on UVA-induced photoaging in human skin fibroblasts. Int. J. Mol. Med. 2014, 34, 796–803. [Google Scholar] [CrossRef] [Green Version]
- Kwon, O.S.; Jung, S.H.; Yang, B.S. Topical administration of manuka oil prevents UV-B irradiation-induced cutaneous photoaging in mice. Evid. Based Complementary Altern. Med. 2013, 2013, 930857. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Fiskum, G.; Schubert, D. Generation of reactive oxygen species by the mitochondrial electron transport chain. J. Neurochem. 2002, 80, 780–787. [Google Scholar] [CrossRef]
- Andreyev, A.Y.; Kushnareva, Y.E.; Starkov, A. Mitochondrial metabolism of reactive oxygen species. Biochem. Mosc. 2005, 70, 200–214. [Google Scholar] [CrossRef]
- Harman, D. The biologic clock: The mitochondria? J. Am. Geriatr. Soc. 1972, 20, 145–147. [Google Scholar] [CrossRef]
- Trounce, I.; Byrne, E.; Marzuki, S. Decline in skeletal muscle mitochondrial respiratory chain function: Possible factor in ageing. Lancet 1989, 333, 637–639. [Google Scholar] [CrossRef]
- Yen, T.-C.; Chen, Y.-S.; King, K.-L.; Yeh, S.-H.; Wei, Y.-H. Liver mitochondrial respiratory functions decline with age. Biochem. Biophys. Res. Commun. 1989, 165, 994–1003. [Google Scholar] [CrossRef]
- Frenzei, H.; Feimann, J. Age-dependent structural changes in the myocardium of rats. A quantitative light-and electron-microscopic study on the right and left chamber wall. Mech. Ageing Dev. 1984, 27, 29–41. [Google Scholar] [CrossRef]
- Cottrell, D.A.; Turnbull, D.M. Mitochondria and ageing. Curr. Opin. Clin. Nutr. Metab. Care 2000, 3, 473–478. [Google Scholar] [CrossRef]
- Maes, D.; Collins, D.; Declercq, L.; Foyouzi-Yousseffi, R.; Gan, D.; Mammone, T.; Pelle, E.; Marenus, K.; Gedeon, H. Improving cellular function through modulation of energy metabolism. Int. J. Cosmet. Sci. 2004, 26, 268–269. [Google Scholar] [CrossRef]
- Declerq, L. Cosmetic benefits from modulation of cellular energy metabolism. In Energy Skin Delivery Systems: Transdermals, Dermatologicals, and Cosmetic Actives, 1st ed.; Wiley-Blackwell: Ames, NJ, USA, 2006; pp. 117–124. [Google Scholar]
- Nacarelli, T.; Azar, A.; Sell, C. Inhibition of mTOR prevents ROS production initiated by ethidium bromide-induced mitochondrial DNA depletion. Front. Endocrinol. 2014, 5, 122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Z.; Huo, J.; Ding, X.; Yang, M.; Li, L.; Dai, J.; Hosoe, K.; Kubo, H.; Mori, M.; Higuchi, K. Coenzyme Q10 improves lipid metabolism and ameliorates obesity by regulating CaMKII-mediated PDE4 inhibition. Sci. Rep. 2017, 7, 8253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, B.; Chen, J.; Cheng, A.S.; Ko, B.C. Depletion of sirtuin 1 (SIRT1) leads to epigenetic modifications of telomerase (TERT) gene in hepatocellular carcinoma cells. PLoS ONE 2014, 9, e84931. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Gan, Q.; Han, L.; Li, J.; Zhang, H.; Sun, Y.; Zhang, Z.; Tong, T. SIRT1 overexpression antagonizes cellular senescence with activated ERK/S6k1 signaling in human diploid fibroblasts. PLoS ONE 2008, 3, e1710. [Google Scholar] [CrossRef] [PubMed]
- Ming, M.; Zhao, B.; Shea, C.R.; Shah, P.; Qiang, L.; White, S.R.; Sims, D.M.; He, Y.-Y. Loss of sirtuin 1 (SIRT1) disrupts skin barrier integrity and sensitizes mice to epicutaneous allergen challenge. J. Allergy Clin. Immunol. 2015, 135, 936–945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michan, S.; Sinclair, D. Sirtuins in mammals: Insights into their biological function. Biochem. J. 2007, 404, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salminen, A.; Kaarniranta, K.; Kauppinen, A. Crosstalk between oxidative stress and SIRT1: Impact on the aging process. Int. J. Mol. Sci. 2013, 14, 3834–3859. [Google Scholar] [CrossRef] [Green Version]
- Khan, R.S.; Dine, K.; Sarma, J.D.; Shindler, K.S. SIRT1 activating compounds reduce oxidative stress mediated neuronal loss in viral induced CNS demyelinating disease. Acta Neuropathol. Commun. 2014, 2, 3. [Google Scholar] [CrossRef] [Green Version]
- Zarzuelo, M.J.; López-Sepúlveda, R.; Sánchez, M.; Romero, M.; Gómez-Guzmán, M.; Ungvary, Z.; Pérez-Vizcaíno, F.; Jiménez, R.; Duarte, J. SIRT1 inhibits NADPH oxidase activation and protects endothelial function in the rat aorta: Implications for vascular aging. Biochem. Pharmacol. 2013, 85, 1288–1296. [Google Scholar] [CrossRef]
- Lopez-Lluch, G.; Hunt, N.; Jones, B.; Zhu, M.; Jamieson, H.; Hilmer, S.; Cascajo, M.; Allard, J.; Ingram, D.; Navas, P.; et al. Calorie restriction induces mitochondrial biogenesis and bioenergetic efficiency. Proc. Natl. Acad. Sci. USA 2006, 103, 1768–1773. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.-Y.; Wu, L.-Y.; Zheng, X.-Q.; Lu, J.-L.; Wu, M.-Y.; Liang, Y.-R. Green Tea Polyphenols Attenuating Ultraviolet B–Induced Damage to Human Retinal Pigment Epithelial Cells in Vitro. Investig. Ophthalmol. Vis. Sci. 2010, 51, 6665–6670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, B.C.; Surjana, D.; Halliday, G.M.; Damian, D.L. Nicotinamide enhances repair of ultraviolet radiation-induced DNA damage in primary melanocytes. Exp. Dermatol. 2014, 23, 509–511. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Halliday, G.M.; Surjana, D.; Damian, D.L. Nicotinamide prevents ultraviolet radiation-induced cellular energy loss. Photochem. Photobiol. 2010, 86, 942–948. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, E.L.; Shieh, W.M.; Huang, A.C. Mapping the role of NAD metabolism in prevention and treatment of carcinogenesis. In ADP-Ribosylation Reactions: From Bacterial Pathogenesis to Cancer; Springer: Berlin/Heidelberg, Germany, 1999; pp. 69–74. [Google Scholar]
- Fang, E.F.; Scheibye-Knudsen, M.; Brace, L.E.; Kassahun, H.; SenGupta, T.; Nilsen, H.; Mitchell, J.R.; Croteau, D.L.; Bohr, V.A. Defective mitophagy in XPA via PARP-1 hyperactivation and NAD+/SIRT1 reduction. Cell 2014, 157, 882–896. [Google Scholar] [CrossRef] [Green Version]
- Teixeira, J.; Deus, C.M.; Borges, F.; Oliveira, P.J. Mitochondria: Targeting mitochondrial reactive oxygen species with mitochondriotropic polyphenolic-based antioxidants. Int. J. Biochem. Cell Biol. 2018, 97, 98–103. [Google Scholar] [CrossRef]
- Pinnell, S.R. Cutaneous photodamage, oxidative stress, and topical antioxidant protection. J. Am. Acad. Dermatol. 2003, 48, 1–22. [Google Scholar] [CrossRef]
- Bachelor, M.A.; Bowden, G.T. UVA-mediated activation of signaling pathways involved in skin tumor promotion and progression. Semin. Cancer Biol. 2004, 14, 131–138. [Google Scholar] [CrossRef]
- Fuchs, J.; Packer, L. Photooxidative stress in the skin. Oxidative Stress Oxid. Antioxid. 1991, 26, 559–583. [Google Scholar]
- Heck, D.E.; Gerecke, D.R.; Vetrano, A.M.; Laskin, J.D. Solar ultraviolet radiation as a trigger of cell signal transduction. Toxicol. Appl. Pharmacol. 2004, 195, 288–297. [Google Scholar] [CrossRef]
- Xu, P.; Zhang, M.; Wang, X.; Yan, Y.; Chen, Y.; Wu, W.; Zhang, L.; Zhang, L. Antioxidative effect of quetiapine on acute ultraviolet-B-induced skin and HaCaT cell damage. Int. J. Mol. Sci. 2018, 19, 953. [Google Scholar] [CrossRef] [Green Version]
- Syed, D.N.; Malik, A.; Hadi, N.; Sarfaraz, S.; Afaq, F.; Mukhtar, H. Photochemopreventive effect of pomegranate fruit extract on UVA-mediated activation of cellular pathways in normal human epidermal keratinocytes. Photochem. Photobiol. 2006, 82, 398–405. [Google Scholar] [CrossRef] [PubMed]
- Davinelli, S.; Bertoglio, J.C.; Polimeni, A.; Scapagnini, G. Cytoprotective polyphenols against chronological skin aging and cutaneous photodamage. Curr. Pharm. Des. 2018, 24, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Mirshafiey, A.; Hosseini, S.; Afraei, S.; Rastkari, N.T.; Zavareh, F.; Azizi, G. Anti-aging property of G2013 molecule as a novel immunosuppressive agent on enzymatic and non-enzymatic oxidative stress determinants in rat model. Curr. Drug Discov. Technol. 2016, 13, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, F.; Mahdian-Shakib, A.; Jadidi-Niaragh, F.; Enderami, S.E.; Mohammadi, H.; Hemmatzadeh, M.; Mohammed, H.A.; Anissian, A.; Kokhaei, P.; Mirshafiey, A. Anti-inflammatory and anti-tumor effects of α-l-guluronic acid (G2013) on cancer-related inflammation in a murine breast cancer model. Biomed. Pharmacother. 2018, 98, 793–800. [Google Scholar] [CrossRef]
- Schniertshauer, D.; Gebhard, D.; Bergemann, J. Age-dependent loss of mitochondrial function in epithelial tissue can be reversed by coenzyme Q10. J. Aging Res. 2018, 2018, 2090–2212. [Google Scholar] [CrossRef] [Green Version]
- Krutmann, J.; Schroeder, P. Role of mitochondria in photoaging of human skin: The defective powerhouse model. J. Investig. Dermatol. Symp. Proc. 2009, 14, 44–49. [Google Scholar] [CrossRef] [Green Version]
- Brunk, U.T.; Terman, A. The mitochondrial-lysosomal axis theory of aging: Accumulation of damaged mitochondria as a result of imperfect autophagocytosis. Eur. J. Biochem. 2002, 269, 1996–2002. [Google Scholar] [CrossRef]
- Clayton, D.A.; Doda, J.N.; Friedberg, E.C. The absence of a pyrimidine dimer repair mechanism in mammalian mitochondria. Proc. Natl. Acad. Sci. USA 1974, 71, 2777–2781. [Google Scholar] [CrossRef] [Green Version]
- Demple, B.; Harrison, L. Repair of oxidative damage to DNA: Enzymology and biology. Annu. Rev. Biochem. 1994, 63, 915–948. [Google Scholar] [CrossRef]
- Wallace, D.C. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: A dawn for evolutionary medicine. Annu. Rev. Genet. 2005, 39, 359–407. [Google Scholar] [CrossRef] [Green Version]
- Finley, L.W.; Haigis, M.C. The coordination of nuclear and mitochondrial communication during aging and calorie restriction. Ageing Res. Rev. 2009, 8, 173–188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butow, R.A.; Avadhani, N.G. Mitochondrial signaling: The retrograde response. Mol. Cell 2004, 14, 1–15. [Google Scholar] [CrossRef]
- Chae, S.; Ahn, B.Y.; Byun, K.; Cho, Y.M.; Yu, M.-H.; Lee, B.; Hwang, D.; Park, K.S. A systems approach for decoding mitochondrial retrograde signaling pathways. Sci. Signal. 2013, 6, rs4. [Google Scholar] [CrossRef]
- Jazwinski, S.M.; Kriete, A. The yeast retrograde response as a model of intracellular signaling of mitochondrial dysfunction. Front. Physiol. 2012, 3, 139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, R.; Nigam, L.; Singh, A.P.; Singh, K.; Subbarao, N.; Dey, S. Design, synthesis of allosteric peptide activator for human SIRT1 and its biological evaluation in cellular model of Alzheimer’s disease. Eur. J. Med. Chem. 2017, 127, 909–916. [Google Scholar] [CrossRef] [PubMed]
- Beher, D.; Wu, J.; Cumine, S.; Kim, K.W.; Lu, S.C.; Atangan, L.; Wang, M. Resveratrol is not a direct activator of SIRT1 enzyme activity. Chem. Biol. Drug Des. 2009, 74, 619–624. [Google Scholar] [CrossRef] [PubMed]
- Higashida, K.; Kim, S.H.; Jung, S.R.; Asaka, M.; Holloszy, J.O.; Han, D.-H. Effects of resveratrol and SIRT1 on PGC-1α activity and mitochondrial biogenesis: A reevaluation. PLoS Biol. 2013, 11, e1001603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guarente, L. Calorie restriction and sirtuins revisited. Genes Dev. 2013, 27, 2072–2085. [Google Scholar] [CrossRef] [Green Version]
- Satoh, A.; Imai, S.-I. Hypothalamic Sirt1 in aging. Aging Albany NY 2014, 6, 1. [Google Scholar] [CrossRef] [Green Version]
- Sweeney, G.; Song, J. The association between PGC-1α and Alzheimer’s disease. Anat. Cell Biol. 2016, 49, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Schaefer, M.; Werner, S. Nrf2—A regulator of keratinocyte redox signaling. Free Radic. Biol. Med. 2015, 88, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Janjetovic, Z.; Jarrett, S.G.; Lee, E.F.; Duprey, C.; Reiter, R.J.; Slominski, A.T. Melatonin and its metabolites protect human melanocytes against UVB-induced damage: Involvement of NRF2-mediated pathways. Sci. Rep. 2017, 7, 1274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langie, S.A.; Kowalczyk, P.; Tudek, B.; Zabielski, R.; Dziaman, T.; Oliński, R.; van Schooten, F.J.; Godschalk, R.W. The effect of oxidative stress on nucleotide-excision repair in colon tissue of newborn piglets. Mutat. Res. Genet. Toxicol. Environ. Mutagenesis 2010, 695, 75–80. [Google Scholar] [CrossRef]
- Han, W.; Ming, M.; Zhao, R.; Pi, J.; Wu, C.; He, Y.-Y. Nrf1 CNC-bZIP protein promotes cell survival and nucleotide excision repair through maintaining glutathione homeostasis. J. Biol. Chem. 2012, 287, 18788–18795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodgers, J.T.; Lerin, C.; Haas, W.; Gygi, S.P.; Spiegelman, B.M.; Puigserver, P. Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature 2005, 434, 113. [Google Scholar] [CrossRef]
- Lehman, J.J.; Barger, P.M.; Kovacs, A.; Saffitz, J.E.; Medeiros, D.M.; Kelly, D.P. Peroxisome proliferator–activated receptor γ coactivator-1 promotes cardiac mitochondrial biogenesis. J. Clin. Investig. 2000, 106, 847–856. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.; Puigserver, P.; Andersson, U.; Zhang, C.; Adelmant, G.; Mootha, V.; Troy, A.; Cinti, S.; Lowell, B.; Scarpulla, R.C. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 1999, 98, 115–124. [Google Scholar] [CrossRef] [Green Version]
- Chiang, H.-M.; Chen, C.-W.; Lin, T.-Y.; Kuo, Y.-H. N-phenethyl caffeamide and photodamage: Protecting skin by inhibiting type I procollagen degradation and stimulating collagen synthesis. Food Chem. Toxicol. 2014, 72, 154–161. [Google Scholar] [CrossRef]
- Wu, P.-Y.; Huang, C.-C.; Chu, Y.; Huang, Y.-H.; Lin, P.; Liu, Y.-H.; Wen, K.-C.; Lin, C.-Y.; Hsu, M.-C.; Chiang, H.-M. Alleviation of ultraviolet B-induced photodamage by Coffea arabica extract in human skin fibroblasts and hairless mouse skin. Int. J. Mol. Sci. 2017, 18, 782. [Google Scholar] [CrossRef] [Green Version]
- Tirosh, O.; Sen, C.; Roy, S.; Packer, L. Cellular and mitochondrial changes in glutamate-induced HT4 neuronal cell death. Neuroscience 2000, 97, 531–541. [Google Scholar] [CrossRef]
- Wölfle, U.; Esser, P.R.; Simon-Haarhaus, B.; Martin, S.F.; Lademann, J.; Schempp, C.M. UVB-induced DNA damage, generation of reactive oxygen species, and inflammation are effectively attenuated by the flavonoid luteolin in vitro and in vivo. Free Radic. Biol. Med. 2011, 50, 1081–1093. [Google Scholar] [CrossRef] [PubMed]
- Chiang, H.-M.; Chan, S.-Y.; Chu, Y.; Wen, K.-C. Fisetin ameliorated photodamage by suppressing the mitogen-activated protein kinase/matrix metalloproteinase pathway and nuclear factor-κB pathways. J. Agric. Food Chem. 2015, 63, 4551–4560. [Google Scholar] [CrossRef] [PubMed]
- Patanè, G.; Anello, M.; Piro, S.; Vigneri, R.; Purrello, F.; Rabuazzo, A.M. Role of ATP production and uncoupling protein-2 in the insulin secretory defect induced by chronic exposure to high glucose or free fatty acids and effects of peroxisome proliferator-activated receptor-γ inhibition. Diabetes 2002, 51, 2749–2756. [Google Scholar] [CrossRef] [Green Version]
- Kumar, M.J.; Nicholls, D.G.; Andersen, J.K. METABOLISM AND BIOENERGETICS-Oxidative a-Ketoglutarate Dehydrogenase Inhibition via Subtle Elevations in Monoamine Oxidase B Levels Results in Loss of Spare Respiratory Capacity. IMPLICATIONS FOR. J. Biol. Chem. 2003, 278, 46432–46439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Humphries, K.M.; Szweda, L.I. Selective inactivation of α-ketoglutarate dehydrogenase and pyruvate dehydrogenase: Reaction of lipoic acid with 4-hydroxy-2-nonenal. Biochemistry 1998, 37, 15835–15841. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- De Belder, A.; Granath, K. Preparation and properties of fluorescein-labelled dextrans. Carbohydr. Res. 1973, 30, 375–378. [Google Scholar] [CrossRef]









| Formula | Mw | Source | Purity | Storage | Properties |
|---|---|---|---|---|---|
| C36H44O37Na6 | 1206.65 Da | Marine brown algae | ≥96% | 0~−20 °C | Off-white to light yellow powdery or flocculent lyophilisate; odorless, non-irritating odor; hygroscopic |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Q.; Bai, D.; Qin, L.; Shao, M.; Liu, X.; Zhang, S.; Yan, C.; Yu, G.; Hao, J. Protective Effect of l-Hexaguluroic Acid Hexasodium Salt on UVA-Induced Photo-Aging in HaCaT Cells. Int. J. Mol. Sci. 2020, 21, 1201. https://doi.org/10.3390/ijms21041201
Li Q, Bai D, Qin L, Shao M, Liu X, Zhang S, Yan C, Yu G, Hao J. Protective Effect of l-Hexaguluroic Acid Hexasodium Salt on UVA-Induced Photo-Aging in HaCaT Cells. International Journal of Molecular Sciences. 2020; 21(4):1201. https://doi.org/10.3390/ijms21041201
Chicago/Turabian StyleLi, Qiong, Donghui Bai, Ling Qin, Meng Shao, Xi Liu, Shuai Zhang, Chengxiu Yan, Guangli Yu, and Jiejie Hao. 2020. "Protective Effect of l-Hexaguluroic Acid Hexasodium Salt on UVA-Induced Photo-Aging in HaCaT Cells" International Journal of Molecular Sciences 21, no. 4: 1201. https://doi.org/10.3390/ijms21041201