Structure Driven Prediction of Chromatographic Retention Times: Applications to Pharmaceutical Analysis
Abstract
:1. Introduction
2. Results and Discussion
2.1. Development of the Statistical Retention Models
2.1.1. Data Collection, Molecular Descriptor Calculation and Data Preprocessing
2.1.2. Generation of Training and Test Sets
2.1.3. Selection of Molecular Descriptors
2.1.4. Selection of Regression Algorithm
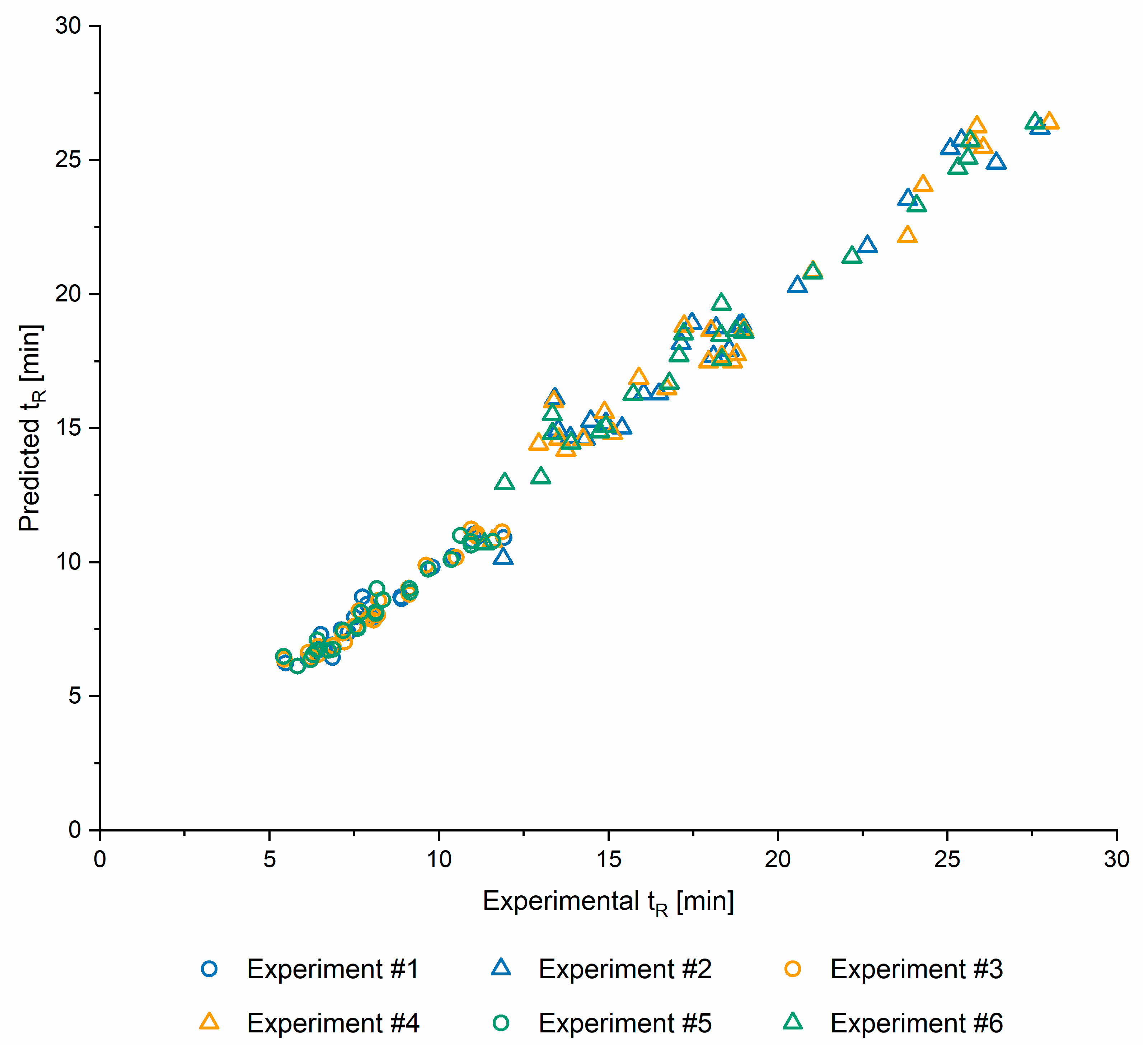
2.1.5. Model Validation
2.2. Application to Method Development
3. Materials and Methods
3.1. Instrumentation
3.2. Chemicals and Reagents
3.3. Software
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| RP-LC | Reversed-Phase Liquid Chromatography |
| API | Active Pharmaceutical Ingredient |
| KPSS | Key Predictive Sample Set |
| QSRR | Quantitative Structure Retention Relationship |
| R | Correlation Coefficient |
| ES | Evolutionary Search |
| MLR | Multiple Linear Regression |
| RMSE | Root Mean Square Error |
| SVM | Support Vector Machine |
| GPR | Gaussian Processes Regression |
| RF | Random Forest |
| PLS | Partial Least Squares |
| RtModelEXP | Retention model built from experimental retention times |
| RtModelQSRR | Retention model built from QSRR predicted retention times |
| RC | Resolution Coefficient |
| Rslimit | Minimal satisfactory resolution between two components |
| Rsi,j | Actual chromatographic resolution between two components in the mixture |
References
- International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use. ICH Harmonised Tripartite Guideline: Specifications: Test Procedures and Acceptance Criteria for New Drug Substances and New Drug Products: Chemical Substances Q6A. Available online: https://database.ich.org/sites/default/files/Q6A%20Guideline.pdf (accessed on 14 November 2020).
- International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use. ICH Harmonised Tripartite Guideline: Impurities in New Drug Substances Q3A(R2). Available online: https://database.ich.org/sites/default/files/Q3A%28R2%29%20Guideline.pdf (accessed on 31 July 2020).
- International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use. ICH Harmonised Tripartite Guideline: Validation of Analytical Procedures: Text and Methodology Q2(R1). Available online: https://database.ich.org/sites/default/files/Q2%28R1%29%20Guideline.pdf (accessed on 31 July 2020).
- Olsen, B.A.; Sreedhara, A.; Baertschi, S.W. Impurity investigations by phases of drug and product development. TrAC, Trends Anal. Chem. 2018, 101, 17–23. [Google Scholar] [CrossRef]
- Baertschi, S.W.; Alsante, K.M.; Reed, R.A. (Eds.) Pharmaceutical Stress Testing: Predicting Drug Degradation, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2011. [Google Scholar] [CrossRef]
- International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use. ICH Harmonised Tripartite Guideline: Stability Testing of New Drug Substances and Products Q1A(R2). Available online: https://database.ich.org/sites/default/files/Q1A%28R2%29%20Guideline.pdf (accessed on 22 February 2021).
- Fekete, S.; Fekete, J.; Molnár, I.; Ganzler, K. Rapid high performance liquid chromatography method development with high prediction accuracy, using 5 cm long narrow bore columns packed with sub-2 μm particles and Design Space computer modeling. J. Chromatogr. A 2009, 1216, 7816–7823. [Google Scholar] [CrossRef]
- Szucs, R.; Brunelli, C.; Lestremau, F.; Hanna-Brown, M. Liquid chromatography in the pharmaceutical industry. In Liquid Chromatography: Applications, 2nd ed.; Fanali, S., Haddad, P.R., Poole, C.F., Riekkola, M.-L., Eds.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 515–537. [Google Scholar] [CrossRef]
- Witting, M.; Böcker, S. Current status of retention time prediction in metabolite identification. J. Sep. Sci. 2020, 43, 1746–1754. [Google Scholar] [CrossRef]
- Taraji, M.; Haddad, P.R.; Amos, R.I.J.; Talebi, M.; Szucs, R.; Dolan, J.W.; Pohl, C.A. Chemometric-assisted method development in hydrophilic interaction liquid chromatography: A review. Anal. Chim. Acta 2018, 1000, 20–40. [Google Scholar] [CrossRef]
- Kaliszan, R. Quantitative structure property (retention) relationships in liquid chromatography. In Liquid Chromatography: Fundamentals and Instrumentation, 2nd ed.; Fanali, S., Haddad, P.R., Poole, C.F., Riekkola, M.-L., Eds.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 553–572. [Google Scholar] [CrossRef]
- Bouwmeester, R.; Martens, L.; Degroeve, S. Comprehensive and Empirical Evaluation of Machine Learning Algorithms for Small Molecule LC Retention Time Prediction. Anal. Chem. 2019, 91, 3694–3703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mauri, A.; Consonni, V.; Pavan, M.; Todeschini, R. DRAGON software: An easy approach to molecular descriptor calculations. MATCH Commun. Math. Comput. Chem. 2006, 56, 237–248. [Google Scholar]
- Cruciani, G.; Crivori, P.; Carrupt, P.A.; Testa, B. Molecular fields in quantitative structure-permeation relationships: The VolSurf approach. J. Mol. Struct. THEOCHEM 2000, 503, 17–30. [Google Scholar] [CrossRef]
- Valdés-Martiní, J.R.; Marrero-Ponce, Y.; García-Jacas, C.R.; Martinez-Mayorga, K.; Barigye, S.J.; Vaz D‘Almeida, Y.S.; Pham-The, H.; Pérez-Giménez, F.; Morell, C.A. QuBiLS-MAS, open source multi-platform software for atom- and bond-based topological (2D) and chiral (2.5D) algebraic molecular descriptors computations. J. Cheminformatics 2017, 9, 35. [Google Scholar] [CrossRef]
- Yap, C.W. PaDEL-descriptor: An open source software to calculate molecular descriptors and fingerprints. J. Comput. Chem. 2011, 32, 1466–1474. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.-S.; Xu, Q.-S.; Hu, Q.-N.; Liang, Y.-Z. ChemoPy: Freely available python package for computational biology and chemoinformatics. Bioinformatics 2013, 29, 1092–1094. [Google Scholar] [CrossRef] [PubMed]
- Steinbeck, C.; Hoppe, C.; Kuhn, S.; Floris, M.; Guha, R.; Willighagen, E.L. Recent Developments of the Chemistry Development Kit (CDK) - An Open-Source Java Library for Chemo- and Bioinformatics. Curr. Pharm. Des. 2006, 12, 2111–2120. [Google Scholar] [CrossRef] [Green Version]
- Witten, I.H.; Frank, E.; Hall, M.A.; Pal, C.J. Data Mining: Practical Machine Learning Tools and Techniques, 4th ed.; Morgan Kaufmann: Cambridge, MA, USA, 2016. [Google Scholar]
- Haddad, P.R.; Taraji, M.; Szücs, R. Prediction of Analyte Retention Time in Liquid Chromatography. Anal. Chem. 2021, 93, 228–256. [Google Scholar] [CrossRef]
- Henneman, A.; Palmblad, M. Retention Time Prediction and Protein Identification. In Mass Spectrometry Data Analysis in Proteomics; Matthiesen, R., Ed.; Humana: New York, NY, USA, 2020; pp. 115–132. [Google Scholar] [CrossRef]
- Moruz, L.; Käll, L. Peptide retention time prediction. Mass Spectrom. Rev. 2017, 36, 615–623. [Google Scholar] [CrossRef]
- Krokhin, O.V.; Spicer, V. Predicting Peptide Retention Times for Proteomics. Curr. Protoc. Bioinformatics 2010, 13.14.11–13.14.15. [Google Scholar] [CrossRef]
- Tarasova, I.A.; Masselon, C.D.; Gorshkov, A.V.; Gorshkov, M.V. Predictive chromatography of peptides and proteins as a complementary tool for proteomics. Analyst 2016, 141, 4816–4832. [Google Scholar] [CrossRef]
- Krokhin, O. Peptide retention prediction in reversed-phase chromatography: Proteomic applications. Expert Rev. Proteomics 2012, 9, 1–4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, Y.; Talebi, M.; Amos, R.I.J.; Szucs, R.; Dolan, J.W.; Pohl, C.A.; Haddad, P.R. Retention prediction in reversed phase high performance liquid chromatography using quantitative structure-retention relationships applied to the Hydrophobic Subtraction Model. J. Chromatogr. A 2018, 1541, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Amos, R.I.J.; Talebi, M.; Szucs, R.; Dolan, J.W.; Pohl, C.A.; Haddad, P.R. Retention Index Prediction Using Quantitative Structure-Retention Relationships for Improving Structure Identification in Nontargeted Metabolomics. Anal. Chem. 2018, 90, 9434–9440. [Google Scholar] [CrossRef]
- Taraji, M.; Haddad, P.R.; Amos, R.I.J.; Talebi, M.; Szucs, R.; Dolan, J.W.; Pohl, C.A. Rapid Method Development in Hydrophilic Interaction Liquid Chromatography for Pharmaceutical Analysis Using a Combination of Quantitative Structure-Retention Relationships and Design of Experiments. Anal. Chem. 2017, 89, 1870–1878. [Google Scholar] [CrossRef] [PubMed]
- Mauri, A.; Consonni, V.; Todeschini, R. Molecular descriptors. In Handbook of Computational Chemistry, 2nd ed.; Leszczynski, J., Kaczmarek-Kedziera, A., Puzyn, T., Papadopoulos, M.G., Reis, H., Shukla, M.K., Eds.; Springer: Cham, Switzerland, 2017; pp. 2065–2093. [Google Scholar] [CrossRef]
- Leardi, R. Genetic algorithms in chemistry. J. Chromatogr. A 2007, 1158, 226–233. [Google Scholar] [CrossRef] [PubMed]
- Hall, M.; Frank, E.; Holmes, G.; Pfahringer, B.; Reutemann, P.; Witten, I.H. The WEKA Data Mining Software: An Update. SIGKDD Explor. 2009, 11, 10–18. [Google Scholar] [CrossRef]
- Kotthoff, L.; Thornton, C.; Hoos, H.H.; Hutter, F.; Leyton-Brown, K. Auto-WEKA 2.0: Automatic model selection and hyperparameter optimization in WEKA. J. Mach. Learn. Res. 2017, 18, 826–830. [Google Scholar]
- Willett, P.; Barnard, J.M.; Downs, G.M. Chemical Similarity Searching. J. Chem. Inf. Comput. Sci. 1998, 38, 983–996. [Google Scholar] [CrossRef] [Green Version]
- Aalizadeh, R.; Thomaidis, N.S.; Bletsou, A.A.; Gago-Ferrero, P. Quantitative Structure-Retention Relationship Models to Support Nontarget High-Resolution Mass Spectrometric Screening of Emerging Contaminants in Environmental Samples. J. Chem. Inf. Model. 2016, 56, 1384–1398. [Google Scholar] [CrossRef] [PubMed]
- Passarin, P.B.S.; Lourenço, F.R. Modeling an in silico platform to predict chromatographic profiles of UV filters using ChromSimulator. Microchem. J. 2020, 157, 105002. [Google Scholar] [CrossRef]
- Shevade, S.K.; Keerthi, S.S.; Bhattacharyya, C.; Murthy, K.R.K. Improvements to the SMO Algorithm for SVM Regression. IEEE Trans. Neural Netw. 2000, 11, 1188–1193. [Google Scholar] [CrossRef] [Green Version]
- Smola, A.J.; Schölkopf, B. A tutorial on support vector regression. Stat. Comput. 2004, 14, 199–222. [Google Scholar] [CrossRef] [Green Version]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Frank, E.; Hall, M.A.; Witten, I.H. The WEKA Workbench. Online Appendix for “Data Mining: Practical Machine Learning Tools and Techniques”, 4th ed.; Morgan Kaufmann, 2016; Available online: https://www.cs.waikato.ac.nz/ml/weka/Witten_et_al_2016_appendix.pdf (accessed on 14 November 2020).






| Descriptor | Description | Descriptor | Description |
|---|---|---|---|
| CATS2D_03_DL | CATS2D Donor-Lipophilic at lag 03 | LOGP_N-oct | Log Octanol/water |
| CATS2D_09_DA | CATS2D Donor-Acceptor at lag 09 | CATS2D_09_NL | CATS2D Negative-Lipophilic at lag 09 |
| F03[C-O] | Frequency of C-O at topological distance 3 | GATS5s | Geary autocorrelation of lag 5 weighted by I-state |
| GATS6e | Geary autocorrelation of lag 6 weighted by Sanderson electronegativity | GATS6m | Geary autocorrelation of lag 6 weighted by mass |
| GATS7m | Geary autocorrelation of lag 7 weighted by mass | HATS4e | leverage-weighted autocorrelation of lag 4/weighted by Sanderson electronegativity |
| HATS5s | leverage-weighted autocorrelation of lag 5/weighted byI-state | Mor10p | signal 10/weighted by polarizability |
| AMW | average molecular weight | BLTA96 | Verhaar Algae base-line toxicity from MLOGP (mmol/L) |
| Mor24p | signal 24/weighted by polarizability | N-075 | R--N--R/R--N--X |
| nArCOOR | number of esters (aromatic) | NNRS | normalized number of ring systems |
| TDB07m | 3D Topological distance-based descriptors—lag 7 weighted by mass | TDB08s | 3D Topological distance-based descriptors—lag 8 weighted byI-state |
| a_acc | Number of hydrogen bond acceptor atoms | logS | Log of the aqueous solubility |
| PEOE_VSA_NEG | Total negative van der Waals surface area | PEOE_VSA+0 | Sum of vi where qi is in the range of 0.00–0.05 |
| SMR_VSA7 | Sum of vi such that Ri > 0.56 | ACACDO | H-bond acceptor and donor |
| L0LgS | Solubility profiling coefficient | L2LgS | Solubility profiling coefficient |
| pctFU4 | Percent unionized species at pH 4 | pctFU6 | Percent unionized species at pH 6 |
| Algorithm | Settings |
|---|---|
| Support Vector Machine [36,37] | Normalized training data Polynomial Kernel |
| Gaussian Processes | Without hyperparameter tuning Normalized Polynomial Kernel |
| Multiple Linear Regression | M5 attribute selection method |
| Random Forest [38] | WEKA default Setting |
| Partial Least Squares (PLS) | Optimal Number of PLS factors determined using Leave One Out cross validation |
| Experiment #1 | Experiment #2 | Experiment #3 | Experiment #4 | Experiment #5 | Experiment #6 | |
|---|---|---|---|---|---|---|
| RMSE | 0.4262 | 0.9981 | 0.3472 | 1.0133 | 0.4091 | 0.8401 |
| R | 0.9769 | 0.9763 | 0.9851 | 0.9792 | 0.9799 | 0.9874 |
| Experiment | Column Temperature (°C) | Gradient Profile a |
|---|---|---|
| 1 | 20 | Time = 0 min, %B = 5%; Time = 15 min, %B = 95% |
| 2 | 20 | Time = 0 min, %B = 5%; Time = 45 min, %B = 95% |
| 3 | 40 | Time = 0 min, %B = 5%; Time = 15 min, %B = 95% |
| 4 | 40 | Time = 0 min, %B = 5%; Time = 45 min, %B = 95% |
| 5 | 60 | Time = 0 min, %B = 5%; Time = 15 min, %B = 95% |
| 6 | 60 | Time = 0 min, %B = 5%; Time = 45 min, %B = 95% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szucs, R.; Brown, R.; Brunelli, C.; Heaton, J.C.; Hradski, J. Structure Driven Prediction of Chromatographic Retention Times: Applications to Pharmaceutical Analysis. Int. J. Mol. Sci. 2021, 22, 3848. https://doi.org/10.3390/ijms22083848
Szucs R, Brown R, Brunelli C, Heaton JC, Hradski J. Structure Driven Prediction of Chromatographic Retention Times: Applications to Pharmaceutical Analysis. International Journal of Molecular Sciences. 2021; 22(8):3848. https://doi.org/10.3390/ijms22083848
Chicago/Turabian StyleSzucs, Roman, Roland Brown, Claudio Brunelli, James C. Heaton, and Jasna Hradski. 2021. "Structure Driven Prediction of Chromatographic Retention Times: Applications to Pharmaceutical Analysis" International Journal of Molecular Sciences 22, no. 8: 3848. https://doi.org/10.3390/ijms22083848






