Mitochondrial Retinopathies
Abstract
:1. Mitochondrial Bioenergetics
2. Retina Is a Preferential Target of Mitochondrial Dysfunction and Diseases
3. Functional Anatomy of the Retina
4. Physiology of the Photoreceptors
- The photoreceptors—rods and cones—which transmit signals to the outer plexiform layer, where they synapse with bipolar cells and horizontal cells.
- The horizontal cells, which transmit signals horizontally in the outer plexiform layer from the rods and cones to bipolar cells.
- The bipolar cells, which transmit signals vertically from the rods, cones, and horizontal cells to the inner plexiform layer, where they synapse with ganglion cells and amacrine cells.
- The amacrine cells, which transmit signals in two directions, either directly from bipolar cells to RGCs or horizontally within the inner plexiform layer from axons of the bipolar cells to dendrites of the ganglion cells or to other amacrine cells.
- The ganglion cells, or RGCs, which transmit output signals from the retina through the optic nerve into the brain serving both the visual and non-visual pathways (mRGCs). The first leads to formed vision, whereas the second is instrumental to photoentrain circadian rhythms.
5. The Retinal Pigmented Epithelium (RPE)
6. Bioenergetics of Photoreceptors
7. Physiology of RGCs and Related Axons Comprising the Visual Pathway
8. Retinopathy in Mitochondrial Disease
9. Retinal Dystrophy in Mitochondrial Disease
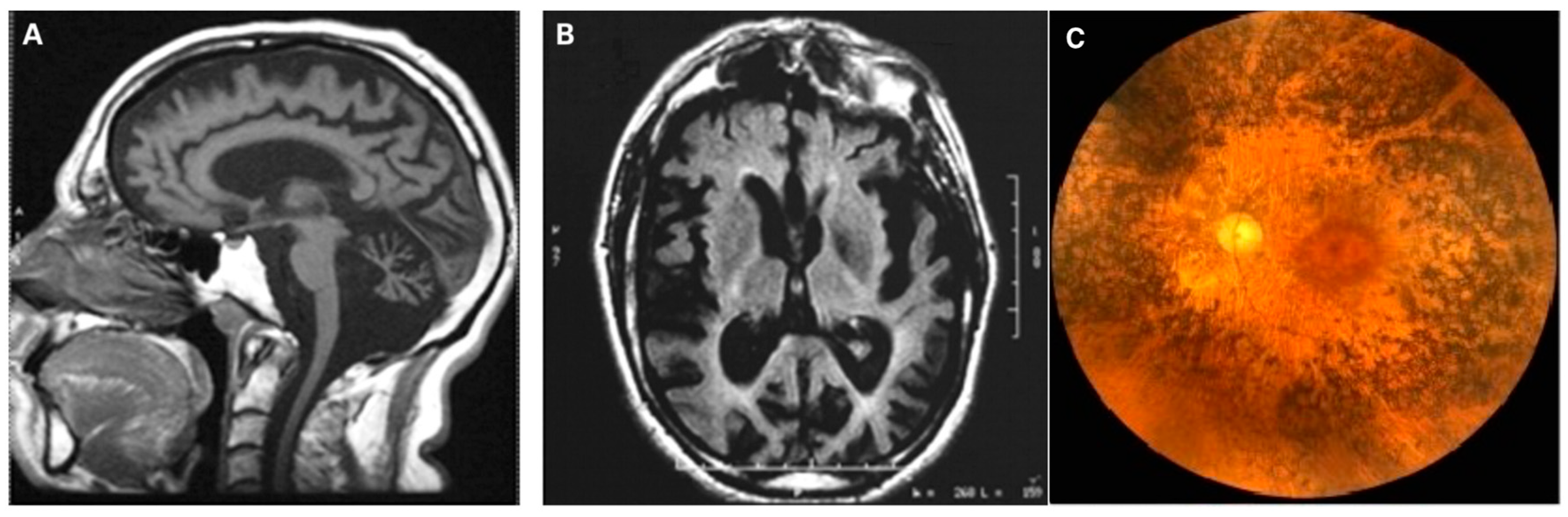
10. Neurogenic Muscle Weakness, Ataxia and Retinitis Pigmentosa (NARP)
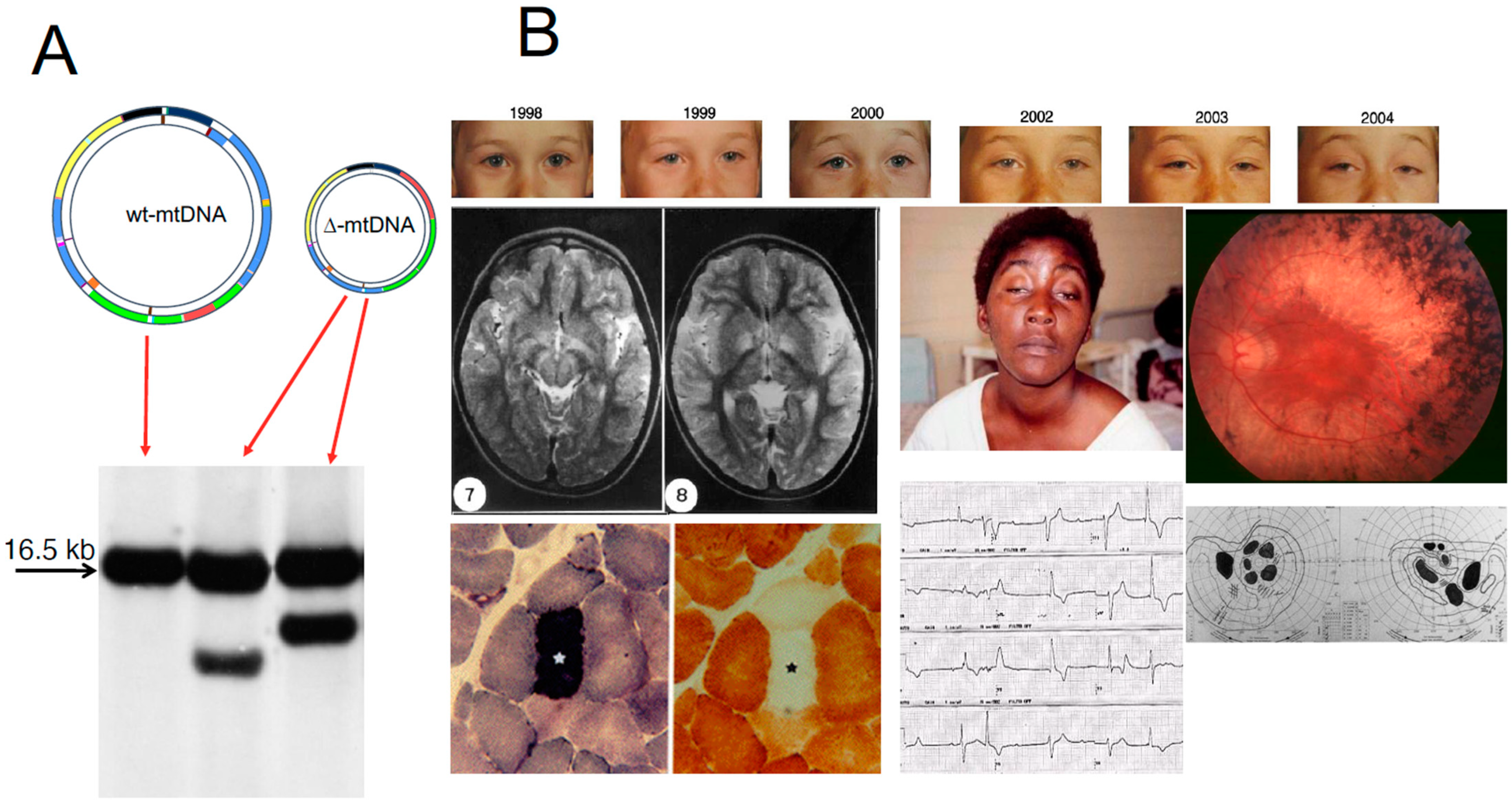
11. Large-Scale Rearrangements of mtDNA
12. Genetic Heterogeneity in Mitochondrial RP
13. Age-Related Macular Degeneration (AMD)
14. Mitochondrial Optic Atrophy
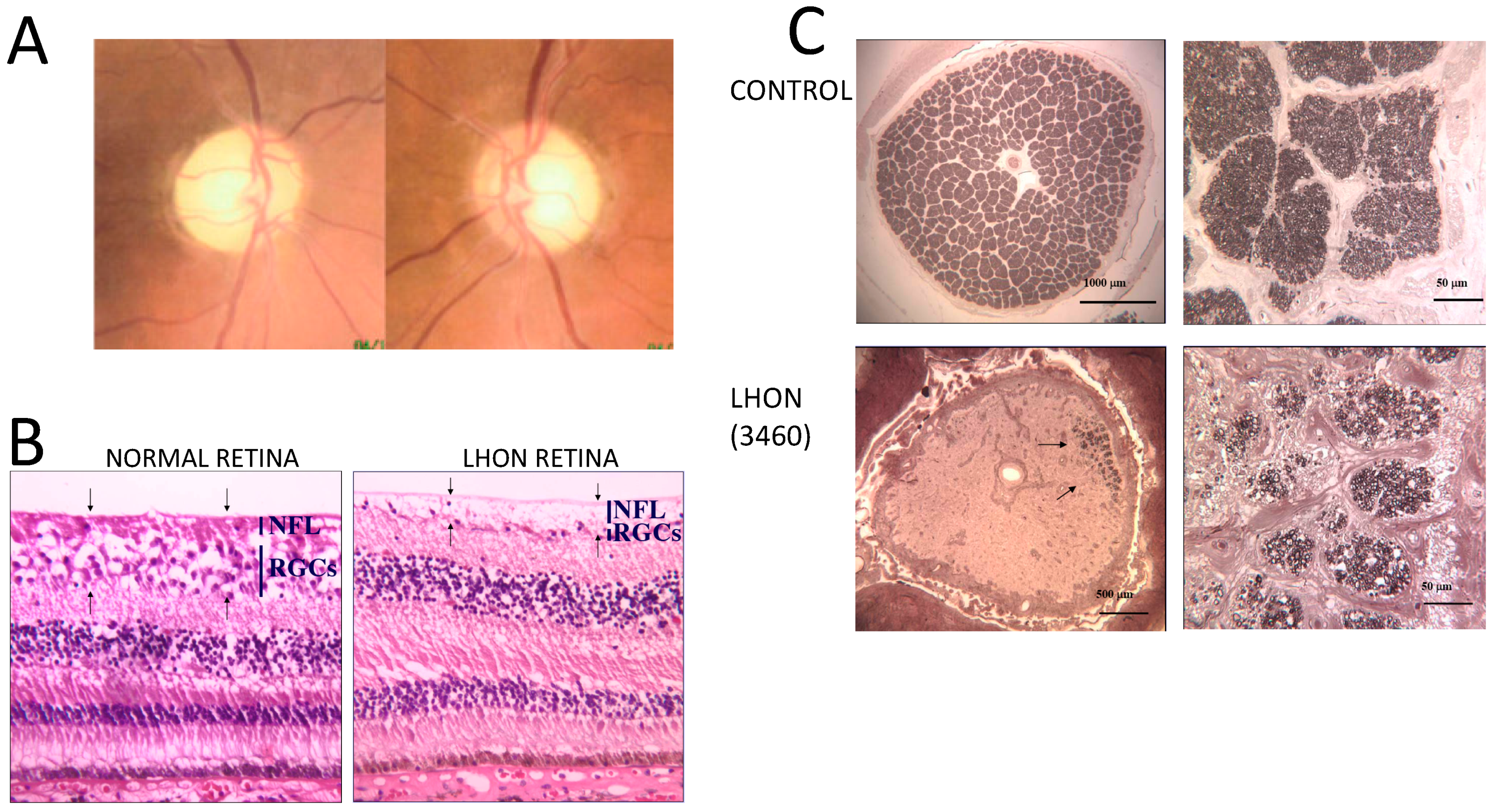
15. Non-Syndromic Optic Atrophy: Leber Hereditary Optic Neuropathy (LHON)
16. Non-Syndromic Optic Atrophy: Autosomal Dominant Optic Atrophy (ADOA)
17. Syndromic Optic Atrophy
“Plus” Forms of LHON and ADOA
18. Rare Mitochondrial Causes of Optic Atrophy
18.1. Optic Atrophy and Charcot–Marie–Tooth (CMT) Peripheral Neuropathy
18.2. Syndromic Optic Atrophy Implicating cI and Altered Interorganellar Cross-Talk—TMEM126 and RTN4P1 Genes
19. Partial Depletion of mtDNA Introduces a New Paradigm—The Cases of SSBP1 and LIG3 Genes
20. Therapeutic Strategies
21. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wallace, D.C. Mitochondria, bioenergetics, and the epigenome in eukaryotic and human evolution. Cold Spring Harb. Symp. Quant. Biol. 2009, 74, 383–393. [Google Scholar] [CrossRef] [PubMed]
- Wallace, D.C. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: A dawn for evolutionary medicine. Annu. Rev. Genet. 2005, 39, 359–407. [Google Scholar] [CrossRef] [Green Version]
- Dzbek, J.; Korzeniewski, B. Control over the contribution of the mitochondrial membrane potential (ΔΨ) and proton gradient (ΔpH) to the protonmotive force (Δp). J. Biol. Chem. 2008, 283, 33232–33239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitchell, P. Chemiosmotic coupling in oxidative and photosynthetic phosphorylation (1966). Biochim. Biophys. Acta Bioenerg. 2011, 1807, 1507–1538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ježek, P.; Holendová, B.; Garlid, K.D.; Jabůrek, M. Mitochondrial Uncoupling Proteins: Subtle Regulators of Cellular Redox Signaling. Antioxid. Redox Signal. 2018, 29, 667–714. [Google Scholar]
- Montava-Garriga, L.; Ganley, I.G. Outstanding Questions in Mitophagy: What We Do and Do Not Know. J. Mol. Biol. 2020, 432, 206–230. [Google Scholar] [CrossRef] [PubMed]
- Calvo, S.E.; Mootha, V.K. The mitochondrial proteome and human disease. Annu. Rev. Genom. Hum. Genet. 2010, 11, 25–44. [Google Scholar] [CrossRef] [Green Version]
- Wallace, D.C. Why do we still have a maternally inherited mitochondrial DNA? Insights from evolutionary medicine. Annu. Rev. Biochem. 2007, 76, 781–821. [Google Scholar] [CrossRef] [Green Version]
- Gorman, G.S.; Chinnery, P.F.; DiMauro, S.; Hirano, M.; Koga, Y.; McFarland, R.; Suomalainen, A.; Thorburn, D.R.; Zeviani, M.; Turnbull, D.M. Mitochondrial diseases. Nat. Rev. Dis. Primers 2016, 2, 16080. [Google Scholar] [CrossRef]
- Zeviani, M.; Di Donato, S. Mitochondrial disorders. Brain 2004, 127, 2153–2172. [Google Scholar] [CrossRef]
- DiMauro, S.; Schon, E.A.; Carelli, V.; Hirano, M. The clinical maze of mitochondrial neurology. Nat. Rev. Neurol. 2013, 9, 429–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DiMauro, S.; Schon, E.A. Mitochondrial respiratory−chain diseases. N. Engl. J. Med. 2003, 348, 2656–2668. [Google Scholar] [CrossRef]
- Schon, E.A.; DiMauro, S. Mitochondrial mutations: Genotype to phenotype. Novartis Found. Symp. 2007, 287, 214–225. [Google Scholar] [PubMed]
- McFarland, R.; Taylor, R.W.; Turnbull, D.M. A neurological perspective on mitochondrial disease. Lancet Neurol. 2010, 9, 829–840. [Google Scholar] [CrossRef]
- Carelli, V.; Barboni, P.; Sadun, A.A. Mitochondrial ophthalmology. In Mitochondrial Medicine; DiMauro, S., Hirano, M., Schon, E., Eds.; Informa Healthcare: Oxford, UK, 2006. [Google Scholar]
- Miller, N.R.; Newman, N.J.; Biousse, V.; Kerrison, J.B. (Eds.) Walsh & Hoyt’s Clinical Neuro-Ophthalmology, 6th ed.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2005; ISBN 0-7817-4814-3. [Google Scholar]
- La Morgia, C.; Carelli, V.; Sadun, A.A. Retina and melanopsin neurons. Handb. Clin. Neurol. 2021, 179, 315–329. [Google Scholar]
- Strauss, O. The retinal pigment epithelium in visual function. Physiol. Rev. 2005, 85, 845–881. [Google Scholar] [CrossRef] [Green Version]
- Steinberg, R.H.; Linsenmeier, R.A.; Griff, E.R. Three light-evoked responses of the retinal pigment epithelium. Vis. Res. 1983, 23, 1315–1323. [Google Scholar] [CrossRef]
- Baylor, D. How photons start vision. Proc. Natl. Acad. Sci. USA 1996, 93, 560–565. [Google Scholar] [CrossRef] [Green Version]
- Kanow, M.A.; Giarmarco, M.M.; Jankowski, C.S.; Tsantilas, K.; Engel, A.L.; Du, J.; Linton, J.D.; Farnsworth, C.C.; Sloat, S.R.; Rountree, A.; et al. Biochemical adaptations of the retina and retinal pigment epithelium support a metabolic ecosystem in the vertebrate eye. eLife 2017, 6, e28899. [Google Scholar] [CrossRef]
- Anderson, B., Jr.; Saltzman, H.A. Retinal oxygen utilization measured by hyperbaric blackout. Arch. Ophthalmol. 1964, 72, 792–795. [Google Scholar] [CrossRef]
- Cringle, S.J.; Yu, D.Y.; Yu, P.K.; Su, E.N. Intraretinal oxygen consumption in the rat in vivo. Investig. Ophthalmol. Vis. Sci. 2002, 43, 1922–1927. [Google Scholar]
- Winkler, B.S. Glycolytic and oxidative metabolism in relation to retinal function. J. Gen. Physiol. 1981, 77, 667–692. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Kondo, M.; Bill, A. Glucose metabolism in cat outer retina. Effects of light and hyperoxia. Investig. Ophthalmol. Vis. Sci. 1997, 38, 48–55. [Google Scholar]
- Chinchore, Y.; Begaj, T.; Wu, D.; Drokhlyansky, E.; Cepko, C.L. Glycolytic reliance promotes anabolism in photoreceptors. eLife 2017, 6, e25946. [Google Scholar] [CrossRef]
- Narayan, D.S.; Chidlow, G.; Wood, J.P.; Casson, R.J. Glucose metabolism in mammalian photoreceptor inner and outer segments. Clin. Exp. Ophthalmol. 2017, 45, 730–741. [Google Scholar] [CrossRef] [Green Version]
- Petit, L.; Ma, S.; Cipi, J.; Cheng, S.Y.; Zieger, M.; Hay, N.; Punzo, C. Aerobic glycolysis is essential for normal rod function and controls secondary cone death in retinitis pigmentosa. Cell Rep. 2018, 23, 2629–2642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carelli, V.; Ross-Cisneros, F.N.; Sadun, A.A. Mitochondrial dysfunction as a cause of optic neuropathies. Prog. Retin. Eye Res. 2004, 23, 53–89. [Google Scholar] [CrossRef]
- Yu-Wai-Man, P.; Griffiths, P.G.; Chinnery, P.F. Mitochondrial optic neuropathies—Disease mechanisms and therapeutic strategies. Prog. Retin. Eye Res. 2011, 30, 81–114. [Google Scholar] [CrossRef] [Green Version]
- Lenaers, G.; Neutzner, A.; Le Dantec, Y.; Jüschke, C.; Xiao, T.; Decembrini, S.; Swirski, S.; Kieninger, S.; Agca, C.; Kim, U.S.; et al. Dominant optic atrophy: Culprit mitochondria in the optic nerve. Prog. Retin. Eye Res. 2021, 83, 100935. [Google Scholar] [CrossRef] [PubMed]
- Pan, B.X.; Ross-Cisneros, F.N.; Carelli, V.; Rue, K.S.; Salomao, S.R.; Moraes-Filho, M.N.; Moraes, M.N.; Berezovsky, A.; Belfort R, J.r.; Sadun, A.A. Mathematically modelling the involvement of axons in Leber’s hereditary optic neuropathy. Investig. Ophthalmol. Vis. Sci. 2012, 53, 7608–7617. [Google Scholar] [CrossRef] [Green Version]
- Ames, A.; Li, Y.Y.; Heher, E.C.; Kimble, C.R. Energy metabolism of rabbit retina as related to function: High cost of Na+ transport. J. Neurosci. 1992, 12, 840–853. [Google Scholar] [CrossRef] [Green Version]
- Niven, J.E.; Anderson, J.C.; Laughlin, S.B. Fly photoreceptors demonstrate energy-information trade-offs in neural coding. PLoS Biol. 2007, 5, e116. [Google Scholar] [CrossRef] [Green Version]
- Okawa, H.; Sampath, A.P.; Laughlin, S.B.; Fain, G.L. ATP consumption by mammalian rod photoreceptors in darkness and in light. Curr. Biol. 2008, 18, 1917–1921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Punzo, C.; Kornacker, K.; Cepko, C.L. Stimulation of the insulin/mTOR pathway delays cone death in a mouse model of retinitis pigmentosa. Nat. Neurosci. 2009, 12, 44–52. [Google Scholar] [CrossRef] [Green Version]
- Aït-Ali, N.; Fridlich, R.; Millet-Puel, G.; Clérin, E.; Delalande, F.; Jaillard, C.; Blond, F.; Perrocheau, L.; Reichman, S.; Byrne, L.C.; et al. Rod-derived cone viability factor promotes cone survival by stimulating aerobic glycolysis. Cell 2015, 161, 817–832. [Google Scholar] [CrossRef] [Green Version]
- Narayan, D.S.; Wood, J.P.; Chidlow, G.; Casson, R.J. A review of the mechanisms of cone degeneration in retinitis pigmentosa. Acta Ophthalmol. 2016, 94, 748–754. [Google Scholar] [CrossRef]
- Wang, W.; Lee, S.J.; Scott, P.A.; Lu, X.; Emery, D.; Liu, Y.; Ezashi, T.; Roberts, M.R.; Ross, J.W.; Kaplan, H.J.; et al. Two-step reactivation of dormant cones in retinitis pigmentosa. Cell Rep. 2016, 15, 372–385. [Google Scholar] [CrossRef] [Green Version]
- Casson, R.J.; Han, G.; Ebneter, A.; Chidlow, G.; Glihotra, J.; Newland, H.; Wood, J.P. Glucose-induced temporary visual recovery in primary open-angle glaucoma: A double-blind, randomized study. Ophthalmology 2014, 121, 1203–1211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shibeeb, O.; Chidlow, G.; Han, G.; Wood, J.P.; Casson, R.J. Effect of subconjunctival glucose on retinal ganglion cell survival in experimental retinal ischaemia and contrast sensitivity in human glaucoma. Clin. Exp. Ophthalmol. 2016, 44, 24–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holt, I.J.; Harding, A.E.; Petty, R.K.; Morgan-Hughes, J.A. A new mitochondrial disease associated with mitochondrial DNA heteroplasmy. Am. J. Hum. Genet. 1990, 46, 428–433. [Google Scholar] [PubMed]
- Uziel, G.; Moroni, I.; Lamantea, E.; Fratta, G.M.; Ciceri, E.; Carrara, F.; Zeviani, M. Mitochondrial disease associated with the T8993G mutation of the mitochondrial ATPase 6 gene: A clinical, biochemical, and molecular study in six families. J. Neurol. Neurosurg. Psychiatry 1997, 63, 16–22. [Google Scholar] [CrossRef] [Green Version]
- Thorburn, D.R.; Rahman, J.; Rahman, S. Mitochondrial DNA-Associated Leigh Syndrome and NARP. 2003 Oct 30 [Updated 2017 Sep 28]. In GeneReviews® [Internet]; Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, E., Bean, L.J.H., Gripp, K.W., Mirzaa, G.M., Amemiya, A., Eds.; University of Wash-ington: Seattle, WA, USA, 1993–2021. Available online: https://www.ncbi.nlm.nih.gov/books/NBK1173/ (accessed on 20 December 2021).
- Spikes, T.E.; Montgomery, M.G.; Walker, J.E. Structure of the dimeric ATP synthase from bovine mitochondria. Proc. Natl. Acad. Sci. USA 2020, 117, 23519–23526. [Google Scholar] [CrossRef] [PubMed]
- Mori, M.; Mytinger, J.R.; Martin, L.C.; Bartholomew, D.; Hickey, S. m.8993T>G-Associated Leigh Syndrome with Hypocitrullinemia on Newborn Screening. JIMD Rep. 2014, 17, 47–51. [Google Scholar] [PubMed] [Green Version]
- Zeviani, M.; Moraes, C.T.; DiMauro, S.; Nakase, H.; Bonilla, E.; Schon, E.A.; Rowland, L.P. Deletions of mitochondrial DNA in Kearns-Sayre syndrome. Neurology 1998, 51, 1525–1533. [Google Scholar] [CrossRef]
- Poulton, J.; Deadman, M.E.; Gardiner, R.M. Duplications of mitochondrial DNA in mitochondrial myopathy. Lancet 1989, 1, 236–240. [Google Scholar] [CrossRef]
- Mita, S.; Schmidt, B.; Schon, E.A.; DiMauro, S.; Bonilla, E. Detection of “deleted” mitochondrial genomes in cytochrome-c oxidase-deficient muscle fibers of a patient with Kearns-Sayre syndrome. Proc. Natl. Acad. Sci. USA 1989, 86, 9509–9513. [Google Scholar] [CrossRef] [Green Version]
- Barkovich, A.J.; Good, W.V.; Koch, T.K.; Berg, B.O. Mitochondrial disorders: Analysis of their clinical and imaging characteristics. AJNR Am. J. Neuroradiol. 1993, 14, 1119–1137. [Google Scholar] [PubMed]
- Shoffner, J.M.; Lott, M.T.; Voljavec, A.S.; Soueidan, S.A.; Costigan, D.A.; Wallace, D.C. Spontaneous Kearns-Sayre/chronic external ophthalmoplegia plus syndrome associated with a mitochondrial DNA deletion: A slip-replication model and metabolic therapy. Proc. Natl. Acad. Sci. USA 1989, 86, 7952–7956. [Google Scholar] [CrossRef] [Green Version]
- Rötig, A.; Cormier, V.; Blanche, S.; Bonnefont, J.P.; Ledeist, F.; Romero, N.; Schmitz, J.; Rustin, P.; Fischer, A.; Saudubray, J.M.; et al. Pearson’s marrow-pancreas syndrome. A multisystem mitochondrial disorder in infancy. J. Clin. Investig. 1990, 86, 1601–1608. [Google Scholar] [CrossRef]
- Shanske, S.; Tang, Y.; Hirano, M.; Nishigaki, Y.; Tanji, K.; Bonilla, E.; Sue, C.; Krishna, S.; Carlo, J.R.; Willner, J.; et al. Identical mitochondrial DNA deletion in a woman with ocular myopathy and in her son with pearson syndrome. Am. J. Hum. Genet. 2002, 71, 679–683. [Google Scholar] [CrossRef] [Green Version]
- Schon, E.A.; Rizzuto, R.; Moraes, C.T.; Nakase, H.; Zeviani, M.; DiMauro, S. A direct repeat is a hotspot for large-scale deletion of human mitochondrial DNA. Science 1989, 244, 346–349. [Google Scholar] [CrossRef] [PubMed]
- Chinnery, P.F.; Turnbull, D.M. Mitochondrial DNA mutations in the pathogenesis of human disease. Mol. Med. Today 2000, 6, 425–3432. [Google Scholar] [CrossRef]
- Hayashi, J.; Ohta, S.; Kikuchi, A.; Takemitsu, M.; Goto, Y.; Nonaka, I. Introduction of disease-related mitochondrial DNA deletions into HeLa cells lacking mitochondrial DNA results in mitochondrial dysfunction. Proc. Natl. Acad. Sci. USA 1991, 88, 10614–10618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mariotti, C.; Tiranti, V.; Carrara, F.; Dallapiccola, B.; DiDonato, S.; Zeviani, M. Defective respiratory capacity and mitochondrial protein synthesis in transformant cybrids harboring the tRNA(Leu(UUR)) mutation associated with maternally inherited myopathy and cardiomyopathy. J. Clin. Investig. 1994, 93, 1102–1107. [Google Scholar] [CrossRef] [Green Version]
- Birtel, J.; von Landenberg, C.; Gliem, M.; Gliem, C.; Reimann, J.; Kunz, W.S.; Herrmann, P.; Betz, C.; Caswell, R.; Nesbitt, V.; et al. Mitochondrial Retinopathy. Ophthalmol. Retin. 2021. [Google Scholar] [CrossRef] [PubMed]
- Gehrs, K.M.; Anderson, D.H.; Johnson, L.V.; Hageman, G.S. Age-related macular degeneration—Emerging pathogenetic and therapeutic concepts. Ann. Med. 2006, 38, 450–471. [Google Scholar] [CrossRef]
- Terluk, M.R.; Kapphahn, R.J.; Soukup, L.M.; Gong, H.; Gallardo, C.; Montezuma, S.R.; Ferrington, D.A. Investigating mitochondria as a target for treating age-related macular degeneration. J. Neurosci. 2015, 35, 7304–7311. [Google Scholar] [CrossRef] [Green Version]
- Brown, E.E.; DeWeerd, A.J.; Ildefonso, C.J.; Lewin, A.S.; Ash, J.D. Mitochondrial oxidative stress in the retinal pigment epithelium (RPE) led to metabolic dysfunction in both the RPE and retinal photoreceptors. Redox Biol. 2019, 24, 101201. [Google Scholar] [CrossRef]
- Fisher, C.R.; Ferrington, D.A. Perspective on AMD pathobiology: A bioenergetic crisis in the RPE. Investig. Ophthalmol. Vis. Sci. 2018, 59, AMD41–AMD47. [Google Scholar] [CrossRef] [Green Version]
- Liang, F.Q.; Godley, B.F. Oxidative stress-induced mitochondrial DNA damage in human retinal pigment epithelial cells: A possible mechanism for RPE aging and age-related macular degeneration. Exp. Eye Res. 2003, 76, 397–403. [Google Scholar] [CrossRef]
- Feher, J.; Kovacs, I.; Artico, M.; Cavallotti, C.; Papale, A.; Balacco Gabrieli, C. Mitochondrial alterations of retinal pigment epithelium in age-related macular degeneration. Neurobiol. Aging 2006, 27, 983–993. [Google Scholar] [CrossRef]
- Karunadharma, P.P.; Nordgaard, C.L.; Olsen, T.W.; Ferrington, D.A. Mitochondrial DNA damage as a potential mechanism for age-related macular degeneration. Investig. Ophthalmol. Vis. Sci. 2010, 51, 5470–5479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, H.; Xu, H.; Liang, F.Q.; Liang, H.; Gupta, P.; Havey, A.N.; Boulton, M.E.; Godley, B.F. Mitochondrial DNA damage and repair in RPE associated with aging and age-related macular degeneration. Investig. Ophthalmol. Vis. Sci. 2011, 52, 3521–3529. [Google Scholar] [CrossRef]
- Blasiak, J.; Hoser, G.; Bialkowska-Warzecha, J.; Pawlowska, E.; Skorski, T. Reactive oxygen species and mitochondrial DNA damage and repair in BCR-ABL1 cells resistant to Imatinib. Biores. Open Access. 2015, 4, 334–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shokolenko, I.; Venediktova, N.; Bochkareva, A.; Wilson, G.L.; Alexeyev, M.F. Oxidative stress induces degradation of mitochondrial DNA. Nucleic Acids Res. 2009, 37, 2539–2548. [Google Scholar] [CrossRef] [Green Version]
- Nissanka, N.; Moraes, C.T. Mitochondrial DNA damage and reactive oxygen species in neurodegenerative disease. FEBS Lett. 2018, 592, 728–742. [Google Scholar] [CrossRef]
- Kaarniranta, K.; Pawlowska, E.; Szczepanska, J.; Jablkowska, A.; Blasiak, J. Role of mitochondrial DNA damage in ROS-mediated pathogenesis of age-related macular degeneration (AMD). Int. J. Mol. Sci. 2019, 20, 2374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carelli, V.; La Morgia, C.; Ross-Cisneros, F.N.; Sadun, A.A. Optic neuropathies: The tip of the neurodegeneration iceberg. Hum. Mol. Genet. 2017, 26, R139–R150. [Google Scholar] [CrossRef] [Green Version]
- Wallace, D.C.; Singh, G.; Lott, M.T.; Hodge, J.A.; Schurr, T.G.; Lezza, A.M.; Elsas, L.J.; Nikoskelainen, E.K. Mitochondrial DNA mutation associated with Leber’s hereditary optic neuropathy. Science 1988, 242, 1427–1430. [Google Scholar] [CrossRef]
- Erickson, R.P. Leber’s optic atrophy, a possible example of maternal inheritance. Am. J. Hum. Genet. 1972, 24, 348–349. [Google Scholar]
- Achilli, A.; Iommarini, L.; Olivieri, A.; Pala, M.; Hooshiar Kashani, B.; Reynier, P.; La Morgia, C.; Valentino, M.L.; Liguori, R.; Pizza, F.; et al. Rare primary mitochondrial DNA mutations and probable synergistic variants in Leber’s hereditary optic neuropathy. PLoS ONE 2012, 7, e42242. [Google Scholar] [CrossRef]
- Gerber, S.; Ding, M.G.; Gérard, X.; Zwicker, K.; Zanlonghi, X.; Rio, M.; Serre, V.; Hanein, S.; Munnich, A.; Rotig, A.; et al. Compound heterozygosity for severe and hypomorphic NDUFS2 mutations cause non-syndromic LHON-like optic neuropathy. J. Med. Genet. 2017, 54, 346–356. [Google Scholar] [CrossRef]
- Stenton, S.L.; Sheremet, N.L.; Catarino, C.B.; Andreeva, N.A.; Assouline, Z.; Barboni, P.; Barel, O.; Berutti, R.; Bychkov, I.; Caporali, L.; et al. Impaired complex I repair causes recessive Leber’s hereditary optic neuropathy. J. Clin. Investig. 2021, 131, e138267. [Google Scholar] [CrossRef]
- Leber, T. Uber hereditare und congenital-angelegte Sehnervenleiden. Arch. Ophthalmol. 1871, 17, 249–291. [Google Scholar]
- Yu-Wai-Man, P.; Griffiths, P.G.; Howell, N.; Turnbull, D.M.; Chinnery, P.F. The epidemiology of Leber hereditary optic neuropathy in the North East of England. Am. J. Hum. Genet. 2003, 72, 333–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mascialino, B.; Leinonen, M.; Meier, T. Meta-analysis of the prevalence of Leber hereditary optic neuropathy mtDNA mutations in Europe. Eur. J. Ophthalmol. 2012, 22, 461–465. [Google Scholar] [CrossRef] [PubMed]
- Carelli, V.; Carbonelli, M.; de Coo, I.F.; Kawasaki, A.; Klopstock, T.; Lagrèze, W.A.; La Morgia, C.; Newman, N.J.; Orssaud, C.; Pott, J.W.R.; et al. International Consensus Statement on the Clinical and Therapeutic Management of Leber Hereditary Optic Neuropathy. J. Neuroophthalmol. 2017, 37, 371–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nikoskelainen, E.; Hoyt, W.F.; Nummelin, K. Ophthalmoscopic findings in Leber’s hereditary optic neuropathy. I. Fundus findings in asymptomatic family members. Arch. Ophthalmol. 1982, 100, 1597–1602. [Google Scholar] [CrossRef]
- Savini, G.; Barboni, P.; Valentino, M.L.; Montagna, P.; Cortelli, P.; De Negri, A.M.; Sadun, F.; Bianchi, S.; Longanesi, L.; Zanini, M.; et al. Retinal nerve fiber layer evaluation by optical coherence tomography in unaffected carriers with Leber’s hereditary optic neuropathy mutations. Ophthalmology 2005, 112, 127–131. [Google Scholar] [CrossRef]
- Balducci, N.; Cascavilla, M.L.; Ciardella, A.; La Morgia, C.; Triolo, G.; Parisi, V.; Bandello, F.; Sadun, A.A.; Carelli, V.; Barboni, P. Peripapillary vessel density changes in Leber’s hereditary optic neuropathy: A new biomarker. Clin. Exp. Ophthalmol. 2018, 46, 1055–1062. [Google Scholar] [CrossRef] [Green Version]
- Nikoskelainen, E.; Hoyt, W.F.; Nummelin, K. Ophthalmoscopic findings in Leber’s hereditary optic neuropathy. II. The fundus findings in the affected family members. Arch. Ophthalmol. 1983, 101, 1059–1068. [Google Scholar] [CrossRef] [PubMed]
- Barboni, P.; Carbonelli, M.; Savini, G.; Ramos Cdo, V.; Carta, A.; Berezovsky, A.; Salomao, S.R.; Carelli, V.; Sadun, A.A. Natural history of Leber’s hereditary optic neuropathy: Longitudinal analysis of the retinal nerve fiber layer by optical coherence tomography. Ophthalmology 2010, 117, 623–627. [Google Scholar] [CrossRef]
- Hwang, T.J.; Karanjia, R.; Moraes-Filho, M.N.; Gale, J.; Tran, J.S.; Chu, E.R.; Salomao, S.R.; Berezovsky, A.; Belfort, R.J.; Moraes, M.N.; et al. Natural History of Conversion of Leber’s Hereditary Optic Neuropathy: A Prospective Case Series. Ophthalmology 2017, 124, 843–850. [Google Scholar] [CrossRef] [PubMed]
- Newman, N.J.; Carelli, V.; Taiel, M.; Yu-Wai-Man, P. Visual Outcomes in Leber Hereditary Optic Neuropathy Patients With the m.11778G>A (MTND4) Mitochondrial DNA Mutation. J. Neuroophthalmol. 2020, 40, 547–557. [Google Scholar] [CrossRef]
- Barboni, P.; Savini, G.; Valentino, M.L.; La Morgia, C.; Bellusci, C.; De Negri, A.M.; Sadun, F.; Carta, A.; Carbonelli, M.; Sadun, A.A.; et al. Leber’s hereditary optic neuropathy with childhood onset. Investig. Ophthalmol. Vis. Sci. 2006, 47, 5303–5309. [Google Scholar] [CrossRef] [Green Version]
- Majander, A.; Bowman, R.; Poulton, J.; Antcliff, R.J.; Reddy, M.A.; Michaelides, M.; Webster, A.R.; Chinnery, P.F.; Votruba, M.; Moore, A.T.; et al. Childhood-onset Leber hereditary optic neuropathy. Br. J. Ophthalmol. 2017, 101, 1505–1509. [Google Scholar] [CrossRef] [Green Version]
- Dimitriadis, K.; Leonhardt, M.; Yu-Wai-Man, P.; Kirkman, M.A.; Korsten, A.; De Coo, I.F.; Chinnery, P.F.; Klopstock, T. Leber’s hereditary optic neuropathy with late disease onset: Clinical and molecular characteristics of 20 patients. Orphanet J. Rare Dis. 2014, 9, 158. [Google Scholar] [CrossRef] [Green Version]
- Yu-Wai-Man, P.; Bateman, D.E.; Hudson, G.; Griffiths, P.G.; Chinnery, P.F. Leber hereditary optic neuropathy presenting in a 75-year-old man. J. Neuroophthalmol. 2008, 28, 155. [Google Scholar] [CrossRef] [PubMed]
- Caporali, L.; Maresca, A.; Capristo, M.; Del Dotto, V.; Tagliavini, F.; Valentino, M.L.; La Morgia, C.; Carelli, V. Incomplete penetrance in mitochondrial optic neuropathies. Mitochondrion 2017, 36, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Carelli, V.; Achilli, A.; Valentino, M.L.; Rengo, C.; Semino, O.; Pala, M.; Olivieri, A.; Mattiazzi, M.; Pallotti, F.; Carrara, F.; et al. Haplogroup effects and recombination of mitochondrial DNA: Novel clues from the analysis of Leber hereditary optic neuropathy pedigrees. Am. J. Hum. Genet. 2006, 78, 564–574. [Google Scholar] [CrossRef] [Green Version]
- Hudson, G.; Carelli, V.; Spruijt, L.; Gerards, M.; Mowbray, C.; Achilli, A.; Pyle, A.; Elson, J.; Howell, N.; La Morgia, C.; et al. Clinical expression of Leber hereditary optic neuropathy is affected by the mitochondrial DNA-haplogroup background. Am. J. Hum. Genet. 2007, 81, 228–233. [Google Scholar] [CrossRef] [Green Version]
- Jiang, P.; Jin, X.; Peng, Y.; Wang, M.; Liu, H.; Liu, X.; Zhang, Z.; Ji, Y.; Zhang, J.; Liang, M.; et al. The exome sequencing identified the mutation in YARS2 encoding the mitochondrial tyrosyl-tRNA synthetase as a nuclear modifier for the phenotypic manifestation of Leber’s hereditary optic neuropathy-associated mitochondrial DNA mutation. Hum. Mol. Genet. 2016, 25, 584–596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.; Liang, X.; Ji, Y.; Ai, C.; Liu, J.; Zhu, L.; Nie, Z.; Jin, X.; Wang, C.; Zhang, J.; et al. PRICKLE3 linked to ATPase biogenesis manifested Leber’s hereditary optic neuropathy. J. Clin. Investig. 2020, 130, 4935–4946. [Google Scholar] [CrossRef] [PubMed]
- Hudson, G.; Keers, S.; Yu-Wai-Man, P.; Griffiths, P.; Huoponen, K.; Savontaus, M.L.; Nikoskelainen, E.; Zeviani, M.; Carrara, F.; Horvath, R.; et al. Identification of an X-chromosomal locus and haplotype modulating the phenotype of a mitochondrial DNA disorder. Am. J. Hum. Genet. 2005, 77, 1086–1091. [Google Scholar] [CrossRef] [Green Version]
- Shankar, S.P.; Fingert, J.H.; Carelli, V.; Valentino, M.L.; King, T.M.; Daiger, S.P.; Salomao, S.R.; Berezovsky, A.; Belfort, R.J.; Braun, T.A.; et al. Evidence for a novel x-linked modifier locus for leber hereditary optic neuropathy. Ophthalmic Genet. 2008, 29, 17–24. [Google Scholar] [CrossRef]
- Sadun, F.; De Negri, A.M.; Carelli, V.; Salomao, S.R.; Berezovsky, A.; Andrade, R.; Moraes, M.; Passos, A.; Belfort, R.; da Rosa, A.B.; et al. Ophthalmologic findings in a large pedigree of 11778/Haplogroup J Leber hereditary optic neuropathy. Am. J. Ophthalmol. 2004, 137, 271–277. [Google Scholar] [CrossRef]
- Kirkman, M.A.; Yu-Wai-Man, P.; Korsten, A.; Leonhardt, M.; Dimitriadis, K.; De Coo, I.F.; Klopstock, T.; Chinnery, P.F. Gene-environment interactions in Leber hereditary optic neuropathy. Brain 2009, 132 Pt 9, 2317–2326. [Google Scholar] [CrossRef] [PubMed]
- Carelli, V.; d’Adamo, P.; Valentino, M.L.; La Morgia, C.; Ross-Cisneros, F.N.; Caporali, L.; Maresca, A.; Loguercio Polosa, P.; Barboni, P.; De Negri, A.; et al. Parsing the differences in affected with LHON: Genetic versus environmental triggers of disease conversion. Brain 2016, 139 Pt 3, e17. [Google Scholar] [CrossRef] [Green Version]
- Baracca, A.; Solaini, G.; Sgarbi, G.; Lenaz, G.; Baruzzi, A.; Schapira, A.H.; Martinuzzi, A.; Carelli, V. Severe impairment of complex I-driven adenosine triphosphate synthesis in leber hereditary optic neuropathy cybrids. Arch. Neurol. 2005, 62, 730–736. [Google Scholar] [CrossRef]
- Floreani, M.; Napoli, E.; Martinuzzi, A.; Pantano, G.; De Riva, V.; Trevisan, R.; Bisetto, E.; Valente, L.; Carelli, V.; Dabbeni-Sala, F. Antioxidant defences in cybrids harboring mtDNA mutations associated with Leber’s hereditary optic neuropathy. FEBS J. 2005, 272, 1124–1135. [Google Scholar] [CrossRef]
- Danielson, S.R.; Wong, A.; Carelli, V.; Martinuzzi, A.; Schapira, A.H.; Cortopassi, G.A. Cells bearing mutations causing Leber’s hereditary optic neuropathy are sensitized to Fas-Induced apoptosis. J. Biol. Chem. 2002, 277, 5810–5815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zanna, C.; Ghelli, A.; Porcelli, A.M.; Martinuzzi, A.; Carelli, V.; Rugolo, M. Caspase-independent death of Leber’s hereditary optic neuropathy cybrids is driven by energetic failure and mediated by AIF and Endonuclease G. Apoptosis 2005, 10, 997–1007. [Google Scholar] [CrossRef]
- Giordano, C.; Montopoli, M.; Perli, E.; Orlandi, M.; Fantin, M.; Ross-Cisneros, F.N.; Caparrotta, L.; Martinuzzi, A.; Ragazzi, E.; Ghelli, A.; et al. Oestrogens ameliorate mitochondrial dysfunction in Leber’s hereditary optic neuropathy. Brain 2011, 134 Pt 1, 220–234. [Google Scholar] [CrossRef] [PubMed]
- Giordano, C.; Iommarini, L.; Giordano, L.; Maresca, A.; Pisano, A.; Valentino, M.L.; Caporali, L.; Liguori, R.; Deceglie, S.; Roberti, M.; et al. Efficient mitochondrial biogenesis drives incomplete penetrance in Leber’s hereditary optic neuropathy. Brain 2014, 137 Pt 2, 335–353. [Google Scholar] [CrossRef] [Green Version]
- Giordano, L.; Deceglie, S.; d’Adamo, P.; Valentino, M.L.; La Morgia, C.; Fracasso, F.; Roberti, M.; Cappellari, M.; Petrosillo, G.; Ciaravolo, S.; et al. Cigarette toxicity triggers Leber’s hereditary optic neuropathy by affecting mtDNA copy number, oxidative phosphorylation and ROS detoxification pathways. Cell Death Dis. 2015, 6, e2021. [Google Scholar] [CrossRef] [Green Version]
- Alexander, C.; Votruba, M.; Pesch, U.E.; Thiselton, D.L.; Mayer, S.; Moore, A.; Rodriguez, M.; Kellner, U.; Leo-Kottler, B.; Auburger, G.; et al. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat. Genet. 2000, 26, 211–215. [Google Scholar] [CrossRef] [PubMed]
- Delettre, C.; Lenaers, G.; Griffoin, J.M.; Gigarel, N.; Lorenzo, C.; Belenguer, P.; Pelloquin, L.; Grosgeorge, J.; Turc-Carel, C.; Perret, E.; et al. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat. Genet. 2000, 26, 207–210. [Google Scholar] [CrossRef]
- Caporali, L.; Magri, S.; Legati, A.; Del Dotto, V.; Tagliavini, F.; Balistreri, F.; Nasca, A.; La Morgia, C.; Carbonelli, M.; Valentino, M.L.; et al. ATPase Domain AFG3L2 Mutations Alter OPA1 Processing and Cause Optic Neuropathy. Ann. Neurol. 2020, 88, 18–32. [Google Scholar] [CrossRef] [Green Version]
- Charif, M.; Chevrollier, A.; Gueguen, N.; Bris, C.; Goudenège, D.; Desquiret-Dumas, V.; Leruez, S.; Colin, E.; Meunier, A.; Vignal, C.; et al. Mutations in the m-AAA proteases AFG3L2 and SPG7 are causing isolated dominant optic atrophy. Neurol. Genet. 2020, 6, e428. [Google Scholar] [CrossRef]
- Charif, M.; Gueguen, N.; Ferré, M.; Elkarhat, Z.; Khiati, S.; LeMao, M.; Chevrollier, A.; Desquiret-Dumas, V.; Goudenège, D.; Bris, C.; et al. Dominant ACO2 mutations are a frequent cause of isolated optic atrophy. Brain Commun. 2021, 3, fcab063. [Google Scholar] [CrossRef]
- Reynier, P.; Amati-Bonneau, P.; Verny, C.; Olichon, A.; Simard, G.; Guichet, A.; Bonnemains, C.; Malecaze, F.; Malinge, M.C.; Pelletier, J.B.; et al. OPA3 gene mutations responsible for autosomal dominant optic atrophy and cataract. J. Med. Genet. 2004, 41, e110. [Google Scholar] [CrossRef] [Green Version]
- Reynier, P.; Amati-Bonneau, P.; Verny, C.; Olichon, A.; Simard, G.; Guichet, A.; Bonnemains, C.; Malecaze, F.; Malinge, M.C.; Pelletier, J.B.; et al. Autosomal dominant optic atrophy associated with hearing impairment and impaired glucose regulation caused by a missense mutation in the WFS1 gene. J. Med. Genet. 2006, 43, 435–440. [Google Scholar]
- Gerber, S.; Charif, M.; Chevrollier, A.; Chaumette, T.; Angebault, C.; Kane, M.S.; Paris, A.; Alban, J.; Quiles, M.; Delettre, C.; et al. Mutations in DNM1L, as in OPA1, result in dominant optic atrophy despite opposite effects on mitochondrial fusion and fission. Brain 2017, 140, 2586–2596. [Google Scholar] [CrossRef]
- Jurkute, N.; Leu, C.; Pogoda, H.M.; Arno, G.; Robson, A.G.; Nürnberg, G.; Altmüller, J.; Thiele, H.; Motameny, S.; Toliat, M.R.; et al. SSBP1 mutations in dominant optic atrophy with variable retinal degeneration. Ann. Neurol. 2019, 86, 368–383. [Google Scholar] [CrossRef]
- Del Dotto, V.; Ullah, F.; Di Meo, I.; Magini, P.; Gusic, M.; Maresca, A.; Caporali, L.; Palombo, F.; Tagliavini, F.; Baugh, E.H.; et al. SSBP1 mutations cause mtDNA depletion underlying a complex optic atrophy disorder. J. Clin. Investig. 2020, 130, 108–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piro-Mégy, C.; Sarzi, E.; Tarrés-Solé, A.; Péquignot, M.; Hensen, F.; Quilès, M.; Manes, G.; Chakraborty, A.; Sénéchal, A.; Bocquet, B.; et al. Dominant mutations in mtDNA maintenance gene SSBP1 cause optic atrophy and foveopathy. J. Clin. Investig. 2020, 130, 143–156. [Google Scholar] [CrossRef] [PubMed]
- Gerber, S.; Charif, M.; Chevrollier, A.; Chaumette, T.; Angebault, C.; Kane, M.S.; Paris, A.; Alban, J.; Quiles, M.; Delettre, C.; et al. OPA1 mutations associated with dominant optic atrophy impair oxidative phosphorylation and mitochondrial fusion. Brain 2008, 131 Pt 2, 352–367. [Google Scholar]
- Kjier, P. Infantile optic atrophy with dominant mode of inheritance: A clinical and genetic study of 19 Danish families. Acta Ophthalmol. Suppl. 1959, 164 (Suppl. S54), 1–147. [Google Scholar]
- Lee, J.; Jung, S.C.; Hong, Y.B.; Yoo, J.H.; Koo, H.; Lee, J.H.; Hong, H.D.; Kim, S.B.; Chung, K.W.; Choi, B.O. Recessive optic atrophy, sensorimotor neuropathy and cataract associated with novel compound heterozygous mutations in OPA1. Mol. Med. Rep. 2016, 14, 33–40. [Google Scholar] [CrossRef] [Green Version]
- Nikoskelainen, E.K.; Marttila, R.J.; Huoponen, K.; Juvonen, V.; Lamminen, T.; Sonninen, P.; Savontaus, M.L. Leber’s “plus”: Neurological abnormalities in patients with Leber’s hereditary optic neuropathy. J. Neurol. Neurosurg. Psychiatry 1995, 59, 160–164. [Google Scholar] [CrossRef] [Green Version]
- Yu-Wai-Man, P.; Griffiths, P.G.; Gorman, G.S.; Lourenco, C.M.; Wright, A.F.; Auer-Grumbach, M.; Toscano, A.; Musumeci, O.; Valentino, M.L.; Caporali, L.; et al. Multi-system neurological disease is common in patients with OPA1 mutations. Brain 2010, 133, 771–786. [Google Scholar] [CrossRef]
- Murakami, T.; Mita, S.; Tokunaga, M.; Maeda, H.; Ueyama, H.; Kumamoto, T.; Uchino, M.; Ando, M. Hereditary cerebellar ataxia with Leber’s hereditary optic neuropathy mitochondrial DNA 11778 mutation. J. Neurol. Sci. 1996, 142, 111–113. [Google Scholar] [CrossRef]
- La Morgia, C.; Achilli, A.; Iommarini, L.; Barboni, P.; Pala, M.; Olivieri, A.; Zanna, C.; Vidoni, S.; Tonon, C.; Lodi, R.; et al. Rare mtDNA variants in Leber hereditary optic neuropathy families with recurrence of myoclonus. Neurology 2008, 70, 762–770. [Google Scholar] [CrossRef]
- Bursle, C.; Riney, K.; Stringer, J.; Moore, D.; Gole, G.; Kearns, L.S.; Mackey, D.A.; Coman, D. Leber Hereditary Optic Neuropathy and Longitudinally Extensive Transverse Myelitis. JIMD Rep. 2018, 42, 53–60. [Google Scholar] [PubMed]
- Berardo, A.; Emmanuele, V.; Vargas, W.; Tanji, K.; Naini, A.; Hirano, M. Leber hereditary optic neuropathy plus dystonia, and transverse myelitis due to double mutations in MT-ND4 and MT-ND6. J. Neurol. 2020, 267, 823–829. [Google Scholar] [CrossRef] [PubMed]
- Mnatsakanyan, L.; Ross-Cisneros, F.N.; Carelli, V.; Wang, M.Y.; Sadun, A.A. Axonal degeneration in peripheral nerves in a case of Leber hereditary optic neuropathy. J. Neuroophthalmol. 2011, 31, 6–11. [Google Scholar] [CrossRef] [PubMed]
- Carelli, V.; Bellan, M. Myelin, mitochondria, and autoimmunity: What’s the connection? Neurology 2008, 70 Pt 2, 1075–1076. [Google Scholar] [CrossRef]
- Bargiela, D.; Chinnery, P.F. Mitochondria in neuroinflammation—Multiple sclerosis (MS), leber hereditary optic neuropathy (LHON) and LHON-MS. Neurosci Lett. 2019, 710, 132932. [Google Scholar] [CrossRef]
- Pfeffer, G.; Burke, A.; Yu-Wai-Man, P.; Compston, D.A.; Chinnery, P.F. Clinical features of MS associated with Leber hereditary optic neuropathy mtDNA mutations. Neurology 2013, 81, 2073–2081. [Google Scholar] [CrossRef] [Green Version]
- Harding, A.E.; Sweeney, M.G.; Miller, D.H.; Mumford, C.J.; Kellar-Wood, H.; Menard, D.; McDonald, W.I.; Compston, D.A. Occurrence of a multiple sclerosis-like illness in women who have a Leber’s hereditary optic neuropathy mitochondrial DNA mutation. Brain 1992, 115 Pt 4, 979–989. [Google Scholar] [CrossRef]
- Amati-Bonneau, P.; Guichet, A.; Olichon, A.; Chevrollier, A.; Viala, F.; Miot, S.; Ayuso, C.; Odent, S.; Arrouet, C.; Verny, C.; et al. OPA1 R445H mutation in optic atrophy associated with sensorineural deafness. Ann. Neurol. 2005, 58, 958–963. [Google Scholar] [CrossRef]
- Amati-Bonneau, P.; Valentino, M.L.; Reynier, P.; Gallardo, M.E.; Bornstein, B.; Boissière, A.; Campos, Y.; Rivera, H.; de la Aleja, J.G.; Carroccia, R.; et al. OPA1 mutations induce mitochondrial DNA instability and optic atrophy ‘plus’ phenotypes. Brain 2008, 131 Pt 2, 338–351. [Google Scholar] [CrossRef] [Green Version]
- Hudson, G.; Amati-Bonneau, P.; Blakely, E.L.; Stewart, J.D.; He, L.; Schaefer, A.M.; Griffiths, P.G.; Ahlqvist, K.; Suomalainen, A.; Reynier, P.; et al. Mutation of OPA1 causes dominant optic atrophy with external ophthalmoplegia, ataxia, deafness and multiple mitochondrial DNA deletions: A novel disorder of mtDNA maintenance. Brain 2008, 131, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Carelli, V.; Musumeci, O.; Caporali, L.; Zanna, C.; La Morgia, C.; Del Dotto, V.; Porcelli, A.M.; Rugolo, M.; Valentino, M.L.; Iommarini, L.; et al. Syndromic parkinsonism and dementia associated with OPA1 missense mutations. Ann. Neurol. 2015, 78, 21–38. [Google Scholar] [CrossRef] [Green Version]
- Rouzier, C.; Bannwarth, S.; Chaussenot, A.; Chevrollier, A.; Verschueren, A.; Bonello-Palot, N.; Fragaki, K.; Cano, A.; Pouget, J.; Pellissier, J.F.; et al. The MFN2 gene is responsible for mitochondrial DNA instability and optic atrophy ‘plus’ phenotype. Brain 2012, 135 Pt 1, 23–34. [Google Scholar] [CrossRef] [Green Version]
- Gorman, G.S.; Pfeffer, G.; Griffin, H.; Blakely, E.L.; Kurzawa-Akanbi, M.; Gabriel, J.; Sitarz, K.; Roberts, M.; Schoser, B.; Pyle, A.; et al. Clonal expansion of secondary mitochondrial DNA deletions associated with spinocerebellar ataxia type 28. JAMA Neurol. 2015, 72, 106–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfeffer, G.; Gorman, G.S.; Griffin, H.; Kurzawa-Akanbi, M.; Blakely, E.L.; Wilson, I.; Sitarz, K.; Moore, D.; Murphy, J.L.; Alston, C.L.; et al. Mutations in the SPG7 gene cause chronic progressive external ophthalmoplegia through disordered mitochondrial DNA maintenance. Brain 2014, 137 Pt 5, 1323–1336. [Google Scholar] [CrossRef] [Green Version]
- Verny, C.; Loiseau, D.; Scherer, C.; Lejeune, P.; Chevrollier, A.; Gueguen, N.; Guillet, V.; Dubas, F.; Reynier, P.; Amati-Bonneau, P.; et al. Multiple sclerosis-like disorder in OPA1-related autosomal dominant optic atrophy. Neurology 2008, 70 Pt 2, 1152–1153. [Google Scholar] [CrossRef] [Green Version]
- Yu-Wai-Man, P.; Spyropoulos, A.; Duncan, H.J.; Guadagno, J.V.; Chinnery, P.F. A multiple sclerosis-like disorder in patients with OPA1 mutations. Ann. Clin. Transl. Neurol. 2016, 3, 723–729. [Google Scholar] [CrossRef] [PubMed]
- Behr, C. Die komplizierte, hereditar-familiare Optikusatrophie des Kindesalters – ein bisher nicht beschriebener Symptomkomplex. Klin Mbl. Augenheilkd. 1909, 47, 138–160. [Google Scholar]
- Carelli, V.; Sabatelli, M.; Carrozzo, R.; Rizza, T.; Schimpf, S.; Wissinger, B.; Zanna, C.; Rugolo, M.; La Morgia, C.; Caporali, L.; et al. ‘Behr syndrome’ with OPA1 compound heterozygote mutations. Brain 2015, 138, e321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kleffner, I.; Wessling, C.; Gess, B.; Korsukewitz, C.; Allkemper, T.; Schirmacher, A.; Young, P.; Senderek, J.; Husstedt, I.W. Behr syndrome with homozygous C19ORF12 mutation. J. Neurol Sci. 2015, 357, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Kleffner, I.; Wessling, C.; Gess, B.; Korsukewitz, C.; Allkemper, T.; Schirmacher, A.; Young, P.; Senderek, J.; Husstedt, I.W. Disturbed mitochondrial dynamics and neurodegenerative disorders. Nat. Rev. Neurol. 2015, 11, 11–24. [Google Scholar]
- De Brito, O.M.; Scorrano, L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature 2008, 456, 605–610. [Google Scholar] [CrossRef]
- Giacomello, M.; Pyakurel, A.; Glytsou, C.; Scorrano, L. The cell biology of mitochondrial membrane dynamics. Nat. Rev. Mol. Cell Biol. 2020, 21, 204–224. [Google Scholar] [CrossRef] [PubMed]
- Züchner, S.; Mersiyanova, I.V.; Muglia, M.; Bissar-Tadmouri, N.; Rochelle, J.; Dadali, E.L.; Zappia, M.; Nelis, E.; Patitucci, A.; Senderek, J.; et al. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat. Genet. 2004, 36, 449–451. [Google Scholar] [CrossRef]
- Nicholson, G.A.; Magdelaine, C.; Zhu, D.; Grew, S.; Ryan, M.M.; Sturtz, F.; Vallat, J.M.; Ouvrier, R.A. Severe early-onset axonal neuropathy with homozygous and compound heterozygous MFN2 mutations. Neurology. 2008, 70, 1678–1681. [Google Scholar] [CrossRef]
- Züchner, S.; De Jonghe, P.; Jordanova, A.; Claeys, K.G.; Guergueltcheva, V.; Cherninkova, S.; Hamilton, S.R.; Van Stavern, G.; Krajewski, K.M.; Stajich, J.; et al. Axonal neuropathy with optic atrophy is caused by mutations in mitofusin 2. Ann Neurol. 2006, 59, 276–281. [Google Scholar] [CrossRef] [PubMed]
- Pareyson, D.; Saveri, P.; Sagnelli, A.; Piscosquito, G. Mitochondrial dynamics and inherited peripheral nerve diseases. Neurosci Lett. 2015, 596, 66–77. [Google Scholar] [CrossRef]
- Abrams, A.J.; Fontanesi, F.; Tan, N.B.L.; Buglo, E.; Campeanu, I.J.; Rebelo, A.P.; Kornberg, A.J.; Phelan, D.G.; Stark, Z.; Zuchner, S. Insights into the genotype-phenotype correlation and molecular function of SLC25A46. Hum. Mutat. 2018, 39, 1995–2007. [Google Scholar] [CrossRef] [PubMed]
- Janer, A.; Prudent, J.; Paupe, V.; Fahiminiya, S.; Majewski, J.; Sgarioto, N.; Des Rosiers, C.; Forest, A.; Lin, Z.Y.; Gingras, A.C.; et al. SLC 25A46 is required for mitochondrial lipid homeostasis and cristae maintenance and is responsible for Leigh syndrome. EMBO Mol. Med. 2016, 8, 1019–1038. [Google Scholar] [CrossRef]
- Charlesworth, G.; Balint, B.; Mencacci, N.E.; Carr, L.; Wood, N.W.; Bhatia, K.P. SLC25A46 mutations underlie progressive myoclonic ataxia with optic atrophy and neuropathy. Mov Disord. 2016, 31, 1249–1251. [Google Scholar] [CrossRef]
- Wan, J.; Steffen, J.; Yourshaw, M.; Mamsa, H.; Andersen, E.; Rudnik-Schöneborn, S.; Pope, K.; Howell, K.B.; McLean, C.A.; Kornberg, A.J.; et al. Loss of function of SLC25A46 causes lethal congenital pontocerebellar hypoplasia. Brain 2016, 139, 2877–2890. [Google Scholar] [CrossRef] [Green Version]
- Formosa, L.E.; Reljic, B.; Sharpe, A.J.; Hock, D.H.; Muellner-Wong, L.; Stroud, D.A.; Ryan, M.T. Optic atrophy-associated TMEM126A is an assembly factor for the ND4-module of mitochondrial complex I. Proc. Natl. Acad. Sci. USA 2021, 118, e2019665118. [Google Scholar] [CrossRef] [PubMed]
- D’Angelo, L.; Astro, E.; De Luise, M.; Kurelac, I.; Umesh-Ganesh, N.; Ding, S.; Fearnley, I.M.; Gasparre, G.; Zeviani, M.; Porcelli, A.M.; et al. NDUFS3 depletion permits complex I maturation and reveals TMEM126A/OPA7 as an assembly factor binding the ND4-module intermediate. Cell Rep. 2021, 35, 109002. [Google Scholar] [CrossRef] [PubMed]
- Hanein, S.; Perrault, I.; Roche, O.; Gerber, S.; Khadom, N.; Rio, M.; Boddaert, N.; Jean-Pierre, M.; Brahimi, N.; Serre, V.; et al. TMEM126A, encoding a mitochondrial protein, is mutated in autosomal-recessive nonsyndromic optic atrophy. Am. J. Hum. Genet. 2009, 84, 493–498. [Google Scholar] [CrossRef] [Green Version]
- La Morgia, C.; Caporali, L.; Tagliavini, F.; Palombo, F.; Carbonelli, M.; Liguori, R.; Barboni, P.; Carelli, V. First TMEM126A missense mutation in an Italian proband with optic atrophy and deafness. Neurol. Genet. 2019, 5, e329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Angebault, C.; Guichet, P.O.; Talmat-Amar, Y.; Charif, M.; Gerber, S.; Fares-Taie, L.; Gueguen, N.; Halloy, F.; Moore, D.; Amati-Bonneau, P.; et al. Recessive Mutations in RTN4IP1 Cause Isolated and Syndromic Optic Neuropathies. Am. J. Hum. Genet. 2015, 97, 754–760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charif, M.; Nasca, A.; Thompson, K.; Gerber, S.; Makowski, C.; Mazaheri, N.; Bris, C.; Goudenège, D.; Legati, A.; Maroofian, R.; et al. Neurologic phenotypes associated with mutations in RTN4IP1 (OPA10) in children and young adults. JAMA Neurol. 2018, 75, 105–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajabian, F.; Manitto, M.P.; Palombo, F.; Caporali, L.; Grazioli, A.; Starace, V.; Arrigo, A.; Cascavilla, M.L.; La Morgia, C.; Barboni, P.; et al. Combined Optic Atrophy and Rod-Cone Dystrophy Expands the RTN4IP1 (Optic Atrophy 10) Phenotype. J. Neuroophthalmol. 2021, 41, e290–e292. [Google Scholar] [CrossRef]
- Meunier, I.; Bocquet, B.; Charif, M.; Dhaenens, C.M.; Manes, G.; Amati-Bonneau, P.; Roubertie, A.; Zanlonghi, X.; Lenaers, G. A rod-cone dystrophy is systematically associated to the RTNPIPI recessive optic atrophy. Retina 2021, 41, 1771–1779. [Google Scholar] [CrossRef] [PubMed]
- Maresca, A.; Carelli, V. Molecular Mechanisms behind Inherited Neurodegeneration of the Optic Nerve. Biomolecules 2021, 11, 496. [Google Scholar] [CrossRef] [PubMed]
- Moraes, C.T.; Shanske, S.; Tritschler, H.J.; Aprille, J.R.; Andreetta, F.; Bonilla, E.; Schon, E.A.; DiMauro, S. mtDNA depletion with variable tissue expression: A novel genetic abnormality in mitochondrial diseases. Am. J. Hum. Genet. 1991, 48, 492–501. [Google Scholar] [PubMed]
- Jurkute, N.; D’Esposito, F.; Robson, A.G.; Pitceathly, R.D.S.; Cordeiro, F.; Raymond, F.L.; Moore, A.T.; Michaelides, M.; Yu-Wai-Man, P.; Webster, A.R.; et al. SSBP1-Disease Update: Expanding the Genetic and Clinical Spectrum, Reporting Variable Penetrance and Confirming Recessive Inheritance. Invest Ophthalmol Vis Sci. 2021, 62, 12. [Google Scholar] [CrossRef]
- Bonora, E.; Chakrabarty, S.; Kellaris, G.; Tsutsumi, M.; Bianco, F.; Bergamini, C.; Ullah, F.; Isidori, F.; Liparulo, I.; Diquigiovanni, C.; et al. Biallelic variants in LIG3 cause a novel mito-chondrial neurogastrointestinal encephalomyopathy. Brain 2021, 144, 1451–1466. [Google Scholar] [CrossRef]
- Invernizzi, F.; Legati, A.; Nasca, A.; Lamantea, E.; Garavaglia, B.; Gusic, M.; Kopajtich, R.; Prokisch, H.; Zeviani, M.; Lamperti, C.; et al. Myopathic mitochondrial DNA depletion syndrome associated with biallelic variants in LIG3. Brain 2021, 144, e74. [Google Scholar] [CrossRef]
- Srivastava, S.; Moraes, C.T. Manipulating mitochondrial DNA heteroplasmy by a mitochondrially targeted restriction endonuclease. Hum. Mol. Genet. 2001, 10, 3093–3099. [Google Scholar] [CrossRef] [Green Version]
- Zekonyte, U.; Bacman, S.R.; Smith, J.; Shoop, W.; Pereira, C.V.; Tomberlin, G.; Stewart, J.; Jantz, D.; Moraes, C.T. Mitochondrial targeted meganuclease as a platform to eliminate mutant mtDNA in vivo. Nat. Commun. 2021, 12, 3210. [Google Scholar] [CrossRef]
- Bacman, S.R.; Gammage, P.A.; Minczuk, M.; Moraes, C.T. Manipulation of mitochondrial genes and mtDNA heteroplasmy. Methods Cell Biol. 2020, 155, 441–487. [Google Scholar]
- Gueven, N.; Ravishankar, P.; Eri, R.; Rybalka, E. Idebenone: When an antioxidant is not an antioxidant. Redox Biol. 2021, 38, 101812. [Google Scholar] [CrossRef]
- Klopstock, T.; Yu-Wai-Man, P.; Dimitriadis, K.; Rouleau, J.; Heck, S.; Bailie, M.; Atawan, A.; Chattopadhyay, S.; Schubert, M.; Garip, A.; et al. A randomized placebo-controlled trial of idebenone in Leber’s hereditary optic neuropathy. Brain 2011, 134, 2677–2686. [Google Scholar] [CrossRef]
- Carelli, V.; La Morgia, C.; Valentino, M.L.; Rizzo, G.; Carbonelli, M.; De Negri, A.M.; Sadun, F.; Carta, A.; Guerriero, S.; Simonelli, F.; et al. Idebenone treatment in Leber’s hereditary optic neuropathy. Brain 2011, 134, e188. [Google Scholar] [CrossRef]
- Catarino, C.B.; von Livonius, B.; Priglinger, C.; Landau, K.; Newman, N.J.; Votruba, M.; Rudolph, G.; Klopstock, T. Real-World Clinical Experience with Idebenone in the Treatment of Leber Hereditary Optic Neuropathy. J. Neuroophthalmol. 2020, 40, 558–565. [Google Scholar] [CrossRef] [PubMed]
- Yu-Wai-Man, P.; Newman, N.J.; Carelli, V.; La Morgia, C.; Biousse, V.; Bandello, F.M.; Clermont, C.V.; Campillo, L.C.; Leruez, S.; Moster, M.L.; et al. Natural history of patients with Leber hereditary optic neuropathy-results from the REALITY study. Eye (Lond) 2021. [Google Scholar] [CrossRef]
- Mendell, J.R.; Al-Zaidy, S.; Shell, R.; Arnold, W.D.; Rodino-Klapac, L.R.; Prior, T.W.; Lowes, L.; Alfano, L.; Berry, K.; Church, K.; et al. Single-dose gene-replacement therapy for spinal muscular atrophy. N. Engl. J. Med. 2017, 377, 1713–1722. [Google Scholar] [CrossRef]
- Viscomi, C.; Zeviani, M. Strategies for fighting mitochondrial diseases. J. Internal Med. 2020, 287, 665–684. [Google Scholar] [CrossRef] [PubMed]
- Gammage, P.A.; Moraes, C.T.; Minczuk, M. Mitochondrial Genome Engineering: The Revolution May Not Be CRISPR-Ized. Trends Genet. 2018, 34, 101–110. [Google Scholar] [CrossRef] [Green Version]
- Manfredi, G.; Fu, J.; Ojaimi, J.; Sadlock, J.E.; Kwong, J.Q.; Guy, J.; Schon, E.A. Rescue of a deficiency in ATP synthesis by transfer of MTATP6, a mitochondrial DNA-encoded gene, to the nucleus. Nat. Genet. 2002, 30, 394–399. [Google Scholar] [CrossRef] [PubMed]
- Guy, J.; Qi, X.; Pallotti, F.; Schon, E.A.; Manfredi, G.; Carelli, V.; Martinuzzi, A.; Hauswirth, W.W.; Lewin, A.S. Rescue of a mitochondrial deficiency causing Leber Hereditary Optic Neuropathy. Ann. Neurol. 2002, 52, 534–542. [Google Scholar] [CrossRef]
- Oca-Cossio, J.; Kenyon, L.; Hao, H.; Moraes, C.T. Limitations of allotopic expression of mitochondrial genes in mammalian cells. Genetics 2003, 165, 707–720. [Google Scholar] [CrossRef]
- Perales-Clemente, E.; Fernández-Silva, P.; Acín-Pérez, R.; Pérez-Martos, A.; Enríquez, J.A. Allotopic expression of mitochondrial-encoded genes in mammals: Achieved goal, undemonstrated mechanism or impossible task? Nucleic Acids Res. 2011, 39, 225–234. [Google Scholar] [CrossRef] [Green Version]
- Yu-Wai-Man, P.; Newman, N.J.; Carelli, V.; Moster, M.L.; Biousse, V.; Sadun, A.A.; Klopstock, T.; Vignal-Clermont, C.; Sergott, R.C.; Rudolph, G.; et al. Bilateral visual improvement with unilateral gene therapy injection for Leber hereditary optic neuropathy. Sci. Transl. Med. 2020, 12, eaaz7423. [Google Scholar] [CrossRef] [PubMed]
- Calkins, D.J.; Yu-Wai-Man, P.; Newman, N.J.; Taiel, M.; Singh, P.; Chalmey, C.; Rogue, A.; Carelli, V.; Ancian, P.; Sahel, J.A. Biodistribution of intravitreal lenadogene nolparvovec gene therapy in nonhuman primates. Mol. Ther Methods Clin. Dev. 2021, 23, 307–318. [Google Scholar] [CrossRef]
- Newman, N.J.; Yu-Wai-Man, P.; Carelli, V.; Moster, M.L.; Biousse, V.; Vignal-Clermont, C.; Sergott, R.C.; Klopstock, T.; Sadun, A.A.; Barboni, P.; et al. Efficacy and Safety of Intravitreal Gene Therapy for Leber Hereditary Optic Neuropathy Treated within 6 Months of Disease Onset. Ophthalmology 2021, 128, 649–660. [Google Scholar] [CrossRef] [PubMed]
- Newman, N.J.; Yu-Wai-Man, P.; Carelli, V.; Biousse, V.; Moster, M.L.; Vignal-Clermont, C.; Sergott, R.C.; Klopstock, T.; Sadun, A.A.; Girmens, J.F.; et al. Intravitreal Gene Therapy vs. Natural History in Patients With Leber Hereditary Optic Neuropathy Carrying the m.11778G>A ND4 Mutation: Systematic Review and Indirect Comparison. Front Neurol. 2021, 12, 662838. [Google Scholar] [CrossRef]
- Wan, X.; Pei, H.; Zhao, M.J.; Yang, S.; Hu, W.K.; He, H.; Ma, S.Q.; Zhang, G.; Dong, X.Y.; Chen, C.; et al. Efficacy and safety of rAAV2-ND4 treatment for Leber’s hereditary optic neuropathy. Sci. Rep. 2016, 6, 21587. [Google Scholar] [CrossRef] [PubMed]
- Guy, J.; Feuer, W.J.; Davis, J.L.; Porciatti, V.; Gonzalez, P.J.; Koilkonda, R.D.; Yuan, H.; Hauswirth, W.W.; Lam, B.L. Gene therapy for Leber hereditary optic neuropathy: Low- and medium-dose visual results. Ophthalmology 2017, 124, 1621–1634. [Google Scholar] [CrossRef] [PubMed]
- Del Dotto, V.; Fogazza, M.; Lenaers, G.; Rugolo, M.; Carelli, V.; Zanna, C. OPA1: How much do we know to approach therapy? Pharmacol Res. 2018, 131, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Romagnoli, M.; La Morgia, C.; Carbonelli, M.; Di Vito, L.; Amore, G.; Zenesini, C.; Cascavilla, M.L.; Barboni, P.; Carelli, V. Idebenone increases chance of stabilization/recovery of visual acuity in OPA1-dominant optic atrophy. Ann. Clin. Transl Neurol. 2020, 7, 590–594. [Google Scholar] [CrossRef]
- Aleo, S.J.; Del Dotto, V.; Fogazza, M.; Maresca, A.; Lodi, T.; Goffrini, P.; Ghelli, A.; Rugolo, M.; Carelli, V.; Baruffini, E.; et al. Drug repositioning as a therapeutic strategy for neurodegenerations associated with OPA1 mutations. Hum Mol. Genet. 2021, 29, 3631–3645. [Google Scholar] [CrossRef]
- Zaninello, M.; Palikaras, K.; Naon, D.; Iwata, K.; Herkenne, S.; Quintana-Cabrera, R.; Semenzato, M.; Grespi, F.; Ross-Cisneros, F.N.; Carelli, V.; et al. Inhibition of autophagy curtails visual loss in a model of autosomal dominant optic atrophy. Nat. Commun. 2020, 11, 4029. [Google Scholar] [CrossRef] [PubMed]
- Cretin, E.; Lopes, P.; Vimont, E.; Tatsuta, T.; Langer, T.; Gazi, A.; Sachse, M.; Yu-Wai-Man, P.; Reynier, P.; Wai, T. High-throughput screening identifies suppressors of mitochondrial fragmentation in OPA1 fibroblasts. EMBO. Mol. Med. 2021, 13, e13579. [Google Scholar] [CrossRef] [PubMed]





| Retinal Degeneration | ||||
|---|---|---|---|---|
| Gene mutation | Protein change | Main retinal lesions | Syndrome | OMIM |
| Heteroplasmic 8993T>G in mt-ATPase6 | Leu156Arg in protein A of H+-ATP synthase | Retinal degeneration (retinitis pigmentosa) | NARP/MILS Ataxia and muscle weakness in NARP, maternally inherited Leigh syndrome if heteroplasmy is ≥70% | OMIM #551500. |
| Heteroplasmic 8993T>C in mt-ATPase6 | Leu156Pro in protein A of H+-ATP synthase | Retinal degeneration (retinitis pigmentosa) | NARP/MILS Ataxia and muscle weakness in NARP, maternally inherited Leigh syndrome if heteroplasmy is ≥90% | OMIM #551500. |
| Heteroplasmic single, sporadic large-scale mtDNA deletions | Ablation or disruption of mtDNA genes including at least one tRNA gene | Retinal degeneration (retinitis pigmentosa) | Juvenile KSS or neonatal onset Pearson’ syndrome if children that overcome the haematological failure | OMIM #530000 OMIM #557000 |
| Optic Atrophy (non-syndromic) | ||||
| MTND6*LDYT14459A MTND4*LHON11778A MTND1*LHON3460A MTND6*LHON14484C MTCYB*LHON15257A MTCO3*LHON9438A MTCO3*LHON9804A MTND5*LHON13730A MTND1*LHON4160C MTND2*LHON5244A MTCOI*LHON7444A MTND1*LHON3394C MTND5*LHON13708A MTCYB*LHON15812A MTND2*LHON4917G MTND1*LHON4216C MTND1*LHON4136G MTATP6*LHON9101C MTND4L*LHON10663C | A72V R340H A52T M64V D171N | Maternally inherited LHON Primary ascertained mutations: MTND5*LDYT14459A MTND4*LHON11778A, MTND1*LHON3460A, MTND6*LHON14484C MTCYB*LHON15257A Three additional putative mutations are: MTND5*LHON13730A; MTCO3*LHON9438A MTCO3*LHON9804A. Nine other mutations are ‘secondary’ mutations which may interact with the primary mutation to clinical risk. Among the more important of these mutations are: MTND5*LHON13708A; MTND1*LHON3394C; MTCO1*LHON7444A MTND1*LHON4160C MTND2*LHON5244A | Four mutations are considered ‘primary’ mutations, the presence of which greatly increases the probability of blindness. One mutation is associated with LHON plus dystonia (LDYT). | OMIM #535000 |
| DNAJC30 c.152A>G c.232C>T c.302T>A | DNAJC30 Y51C P78S L101Q | Autosomal recessive LHON. The protein is a chaperone involved in turnover and repair of cI. | All hallmarks of mtLHON are recapitulated, including incomplete penetrance, male predominance, and significant idebenone responsivity. | OMIM #619382 |
| Heterozygous OPA1 Mutations | Truncation, aberration or single aminoacid replacements in OPA1 due to mutations in the corresponding gene | Autosomal dominant optic atrophy (ADOA), probably caused by haploinsufficiency. | Missense mutations in the GTPase domain may often determine an autosomal dominant syndrome with multiple mtDNA deletions leading to muscle weakness with or without peripheral neuropathy, bilateral eyelid ptosis, progressive external ophthalmoplegia and occasionally parkinsonism. | OMIM #165500 |
| Recessive OPA1 mutations, usually in compound heterozygosis | S256R + Q285R c.2708delTTAG + I382M E487K + I383M Etc. | Behr’ syndrome | Besides early-onset optic atrophy, there is ataxia, pyramidal signs, spasticity, and mental retardation | OMIM #210000 |
| C19ORF12 | C19ORF12 | Behr’ syndrome (mitochondrial membrane protein-associated neurodegeneration, MPAN) | Besides optic atrophy in the context of Behr’ syndrome, mutations of this gene cause neurodegeneration with brain iron accumulation NBIA4 | OMIM #614298 |
| Locus Xp11.4-p11.21 | unknown | OPA2 | Besides very slow progression of optic atrophy, affected males may have mental retardation and minor neurological abnormalities | OMIM #311050 |
| OPA3 | OPA3 Possibly an outer mitochondrial membrane lipid metabolism regulator | Autosomal dominant optic atrophy | Cataract, extra-pyramidal signs, 3-methyl-glutaconic aciduria type III | OMIM #165300 |
| Locus 18q12.2-q12.3 | unknown | OPA4 | Mutation found in one German family with autosomal dominant optic atrophy associated with Kidd blood group | OMIM #605293 |
| DRP1 | DNM1L | OPA5 | Autosomal dominant encephalopathy due to defective mitochondrial and peroxisomal fission 1 | OMIM #603850 |
| Locus 8q21-q22 | unknown | OPA6 Found in a consanguineous family of French origin with 4 sibs affected by early-onset, slowly progressive isolated optic atrophy. Disease progression was very slow, with moderate photophobia and dyschromatopsia. | No other clinical sign. | OMIM #258500 |
| TMEM126A | TMEM126A assembly of cI | OPA7 Autosomal recessive juvenile-onset optic atrophy characterized by severe bilateral deficiency in visual acuity, optic disc pallor, and central scotoma. | Occasional sensory-motor axonal neuropathy with focal demyelinating abnormalities. | OMIM #612989 |
| Locus 16q21-q22 | unknown | OPA8 | Occasionally, bilateral sensorineural hearing loss for high frequencies, with abnormal BAEP and SEP. Mitral valve prolapse or insufficiency. Subsarcolemmal accumulations of mitochondria with impaired growth of fibroblasts in galactose, and an abnormally high rate of fusion activity, suggestive of mitochondrial dysfunction. | OMIM #616648 |
| ACO2 | ACO2 Encodes the mitochondrial aconitase, part of the TCA cycle. In yeast ACO2 is essential for mitochondrial DNA maintenance independent of its catalytic activity. | OPA9 Recessive, early onset optic atrophy. | Onset between ages 2 and 6 months with truncal hypotonia, athetosis, seizures, in addition to the ophthalmologic abnormalities, profound psychomotor retardation, with only some achieving rolling, sitting, or recognition of family. Brain MRI showed progressive cerebral and cerebellar degeneration. | OMIM #100850 |
| RTN4IP1 | RTN4IP1 Reticulon, or NOGO-interacting mitochondrial protein. | OPA10 Autosomal recessive optic atrophy. | Ataxia, mental retardation, and seizures, | OMIM #610502 |
| YME1L1 | YME1L1 A mitochondrial metalloprotease involved in the quality control of mitochondrial proteins. | OPA11 Autosomal recessive optic atrophy in a single Saudi Arabian family. | In vitro functional expression studies and studies of patient cells showed that the mutation resulted in degradation of the mutant protein, abnormal processing of YME1L1 substrates PRELID1 and OPA1, increased mitochondrial fragmentation, and impaired cell proliferation. | OMIM #607472 |
| AFG3L2 | AFG3L2 A mitochondrial AAA-protease involved in quality control of mitochondrial proteins. | OPA12 Autosomal dominant optic atrophy. | Autosomal dominant spino-cerebellar ataxia (SCA28), and autosomal recessive spastic ataxia. | OMIM #604581 |
| SSBP1 | SSBP1 Single-stranded mtDNA binding protein, essential for mtDNA replication. | OPA13 Autosomal dominant optic atrophy with retinal and foveal degeneration. | OMIM #600439 | |
| SPG7 | Paraplegin An AAA mitochondrial metalloprotease involved in the quality control of mitochondrial proteins. | Autosomal recessive optic atrophy | Most commonly associated with autosomal recessive HSP7 | OMIM #607259 |
| MFN2 | MFN2 | Dominant optic atrophy | CMT2A | OMIM #609260 |
| SLC25A46 | SLC25A46 (ortholog of yeast UGO1) | Dominant optic atrophy | CMT6B, Leigh syndrome, pontocerebellar hypoplasia (PCH1E) | OMIM #601152 |
| LIGIII | LIG III An essential mitochondrial ligase involved in termination of mtDNA replication, it is present also in the nucleus. | Recessive optic atrophy. | Gut dysmotility and MNGIE-like neurological abnormalities including leukoencephalopathy, epilepsy, migraine, stroke-like episodes, and neurogenic bladder. In one family recessive mutations in LIG3 caused neonatal fatal myopathy with profound mtDNA depletion. | OMIM *600940. |
| Miscellaneous mutations in mtDNA and mitochondrion-related nuclear genes | Subunits of the MRC, as well as accessory proteins involved in the formation, turnover and quality control of MRCcomplexes and OXPHOS. The most frequent protein involved is SURF1, a putative chaperone of cIV. | Non-syndromic optic atrophy. | Necrotizing Encephalomyelopathy or Leigh Syndrome | OMIM #256000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeviani, M.; Carelli, V. Mitochondrial Retinopathies. Int. J. Mol. Sci. 2022, 23, 210. https://doi.org/10.3390/ijms23010210
Zeviani M, Carelli V. Mitochondrial Retinopathies. International Journal of Molecular Sciences. 2022; 23(1):210. https://doi.org/10.3390/ijms23010210
Chicago/Turabian StyleZeviani, Massimo, and Valerio Carelli. 2022. "Mitochondrial Retinopathies" International Journal of Molecular Sciences 23, no. 1: 210. https://doi.org/10.3390/ijms23010210






