The ctpF Gene Encoding a Calcium P-Type ATPase of the Plasma Membrane Contributes to Full Virulence of Mycobacterium tuberculosis
Abstract
:1. Introduction
2. Results
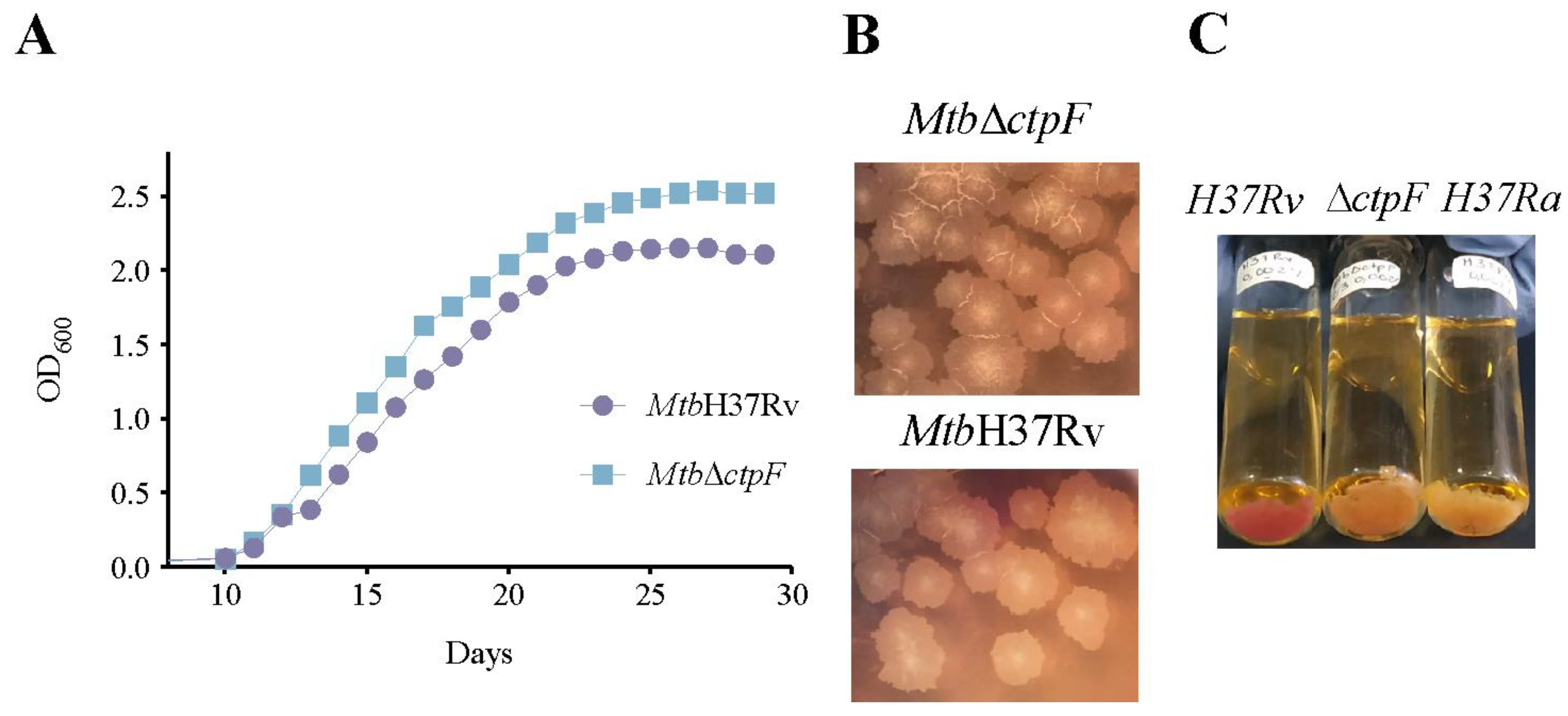
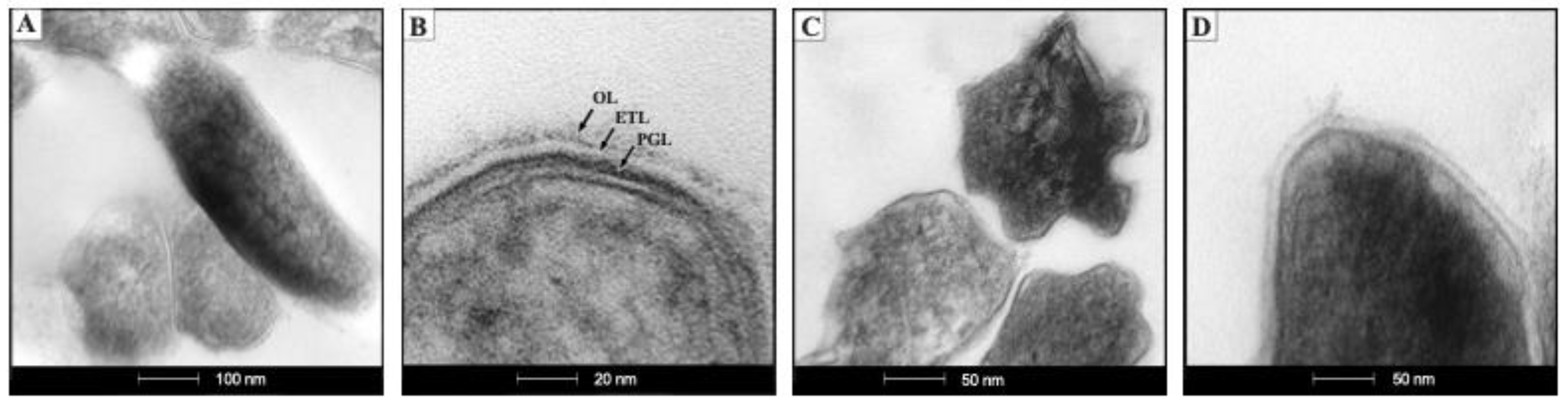
2.1. The ctpF Deletion Does Not Alter the Mtb Growth Kinetics in Standard Culture
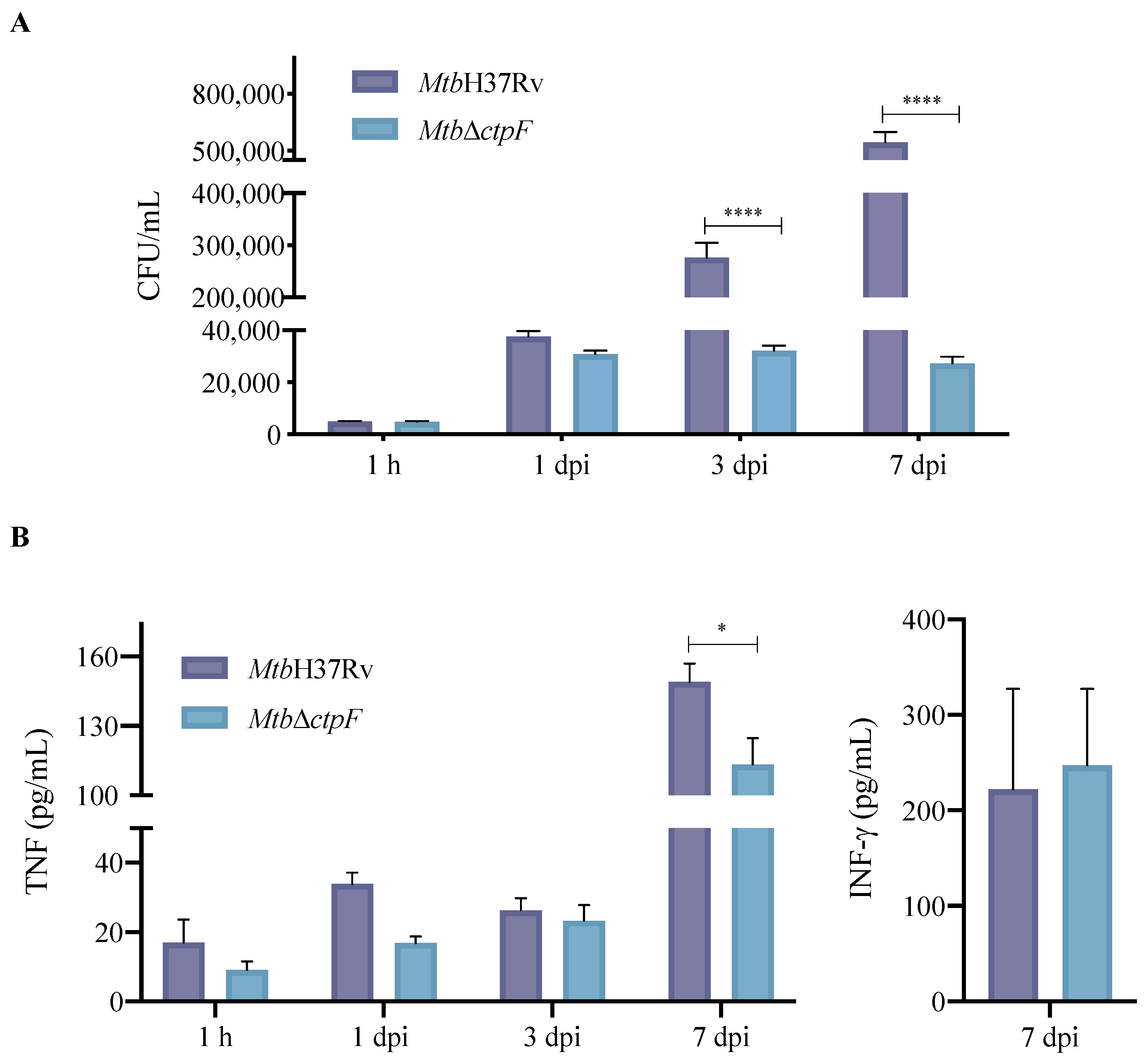
2.2. ctpF Is Required for Mtb Intracellular Proliferation in MH-S Cells
2.3. Deletion of ctpF Does Not Significantly Affect the Production of IFN-γ and TNF in Infected MH-S Cells
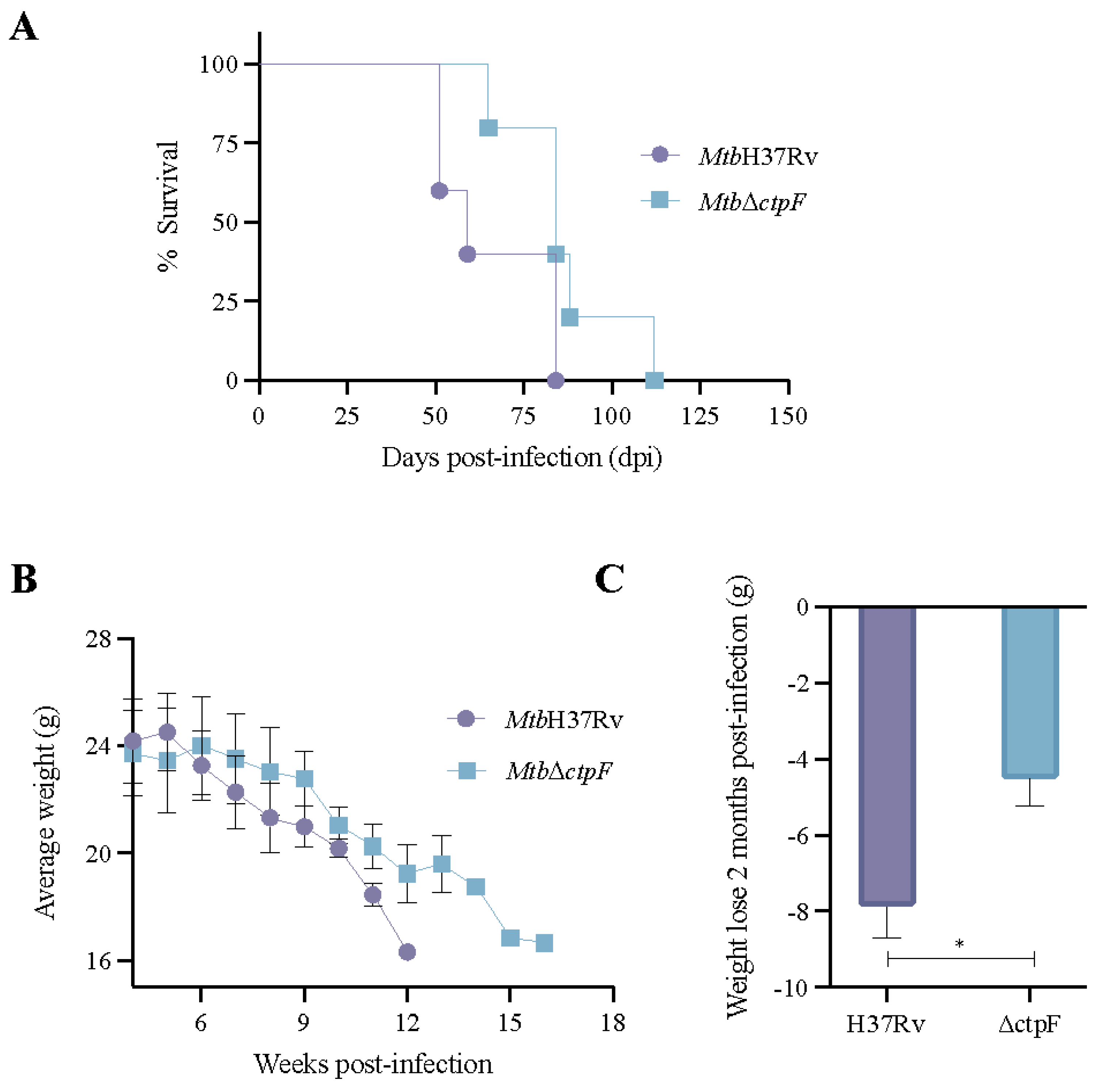
2.4. The MtbΔctpF Strain Shows Attenuated Virulence in Mice
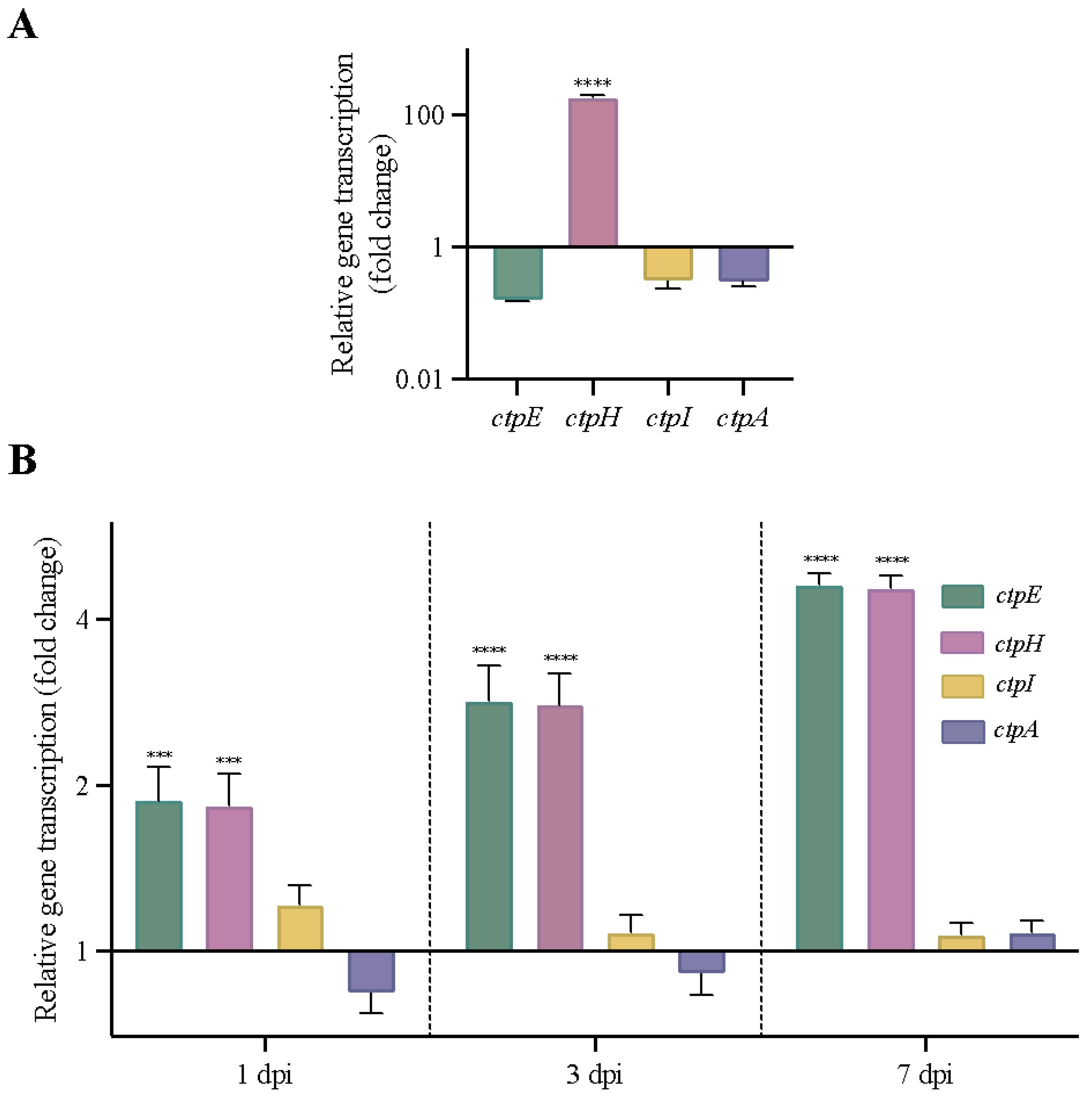
2.5. Other Genes Encoding P2-Type ATPases Are Activated in MtbΔctpF Strain under Stress Conditions
2.6. ctpF Transcription Was Higher during Experimental Latent TB Infection
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
4.2. NR Staining
4.3. MH-S Cells Culture
4.4. Infection of MH-S Cells with Mtb Strains
4.5. Quantification of Cytokines in MH-S Cells Infected with Mtb Strains
4.6. Progressive Pulmonary and Latent TB Infection in BALB/c Mice
4.7. RNA Extraction and cDNA Synthesis
4.8. qRT-PCR Analysis
4.9. TEM
4.10. Ethics Statement
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Global Tuberculosis Report 2020; World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- Martin, C.; Aguilo, N.; Marinova, D.; Gonzalo-Asensio, J. Update on TB vaccine pipeline. Appl. Sci. 2020, 10, 2632. [Google Scholar] [CrossRef] [Green Version]
- Pérez, I.; Uranga, S.; Sayes, F.; Frigui, W.; Samper, S.; Arbués, A.; Aguiló, N.; Brosch, R.; Martín, C.; Gonzalo-Asensio, J. Live attenuated TB vaccines representing the three modern Mycobacterium tuberculosis lineages reveal that the Euro–American genetic background confers optimal vaccine potential. EBioMedicine 2020, 55, 102761. [Google Scholar] [CrossRef]
- Sable, S.B.; Posey, J.E.; Scriba, T.J. Tuberculosis Vaccine Development: Progress in Clinical Evaluation. Clin. Microbiol. Rev. 2019, 33, e00100-19. [Google Scholar] [CrossRef]
- Pando, R.H.; Shin, S.J.; Clark, S.; Casonato, S.; Zambrano, M.B.; Kim, H.; Boldrin, F.; Espinoza, D.M.; Provvedi, R.; Arbues, A.; et al. Construction and characterization of the double unmarked Mycobacterium tuberculosis mutant sigE/fadD26 as a vaccine candidate. Infect. Immun. 2019, 88, e00496-19. [Google Scholar] [CrossRef]
- Andersen, P.; Scriba, T.J. Moving tuberculosis vaccines from theory to practice. Nat. Rev. Immunol. 2019, 19, 550–562. [Google Scholar] [CrossRef]
- Nieuwenhuizen, N.E.; Kaufmann, S.H.E. Next-generation vaccines based on Bacille Calmette-Guérin. Front. Immunol. 2018, 9, 121. [Google Scholar] [CrossRef]
- Gonzalo-Asensio, J.; Marinova, D.; Martin, C.; Aguilo, N. MTBVAC: Attenuating the human pathogen of tuberculosis (TB) toward a promising vaccine against the TB epidemic. Front. Immunol. 2017, 8, 1803. [Google Scholar] [CrossRef] [Green Version]
- Cadena, A.M.; Hopkins, F.F.; Maiello, P.; Carey, A.F.; Wong, E.A.; Martin, C.J.; Gideon, H.P.; DiFazio, R.M.; Andersen, P.; Lin, P.L.; et al. Concurrent infection with Mycobacterium tuberculosis confers robust protection against secondary infection in macaques. PLoS Pathog. 2018, 14, e1007305. [Google Scholar] [CrossRef] [Green Version]
- Gong, W.; Liang, Y.; Wu, X. The current status, challenges, and future developments of new tuberculosis vaccines. Hum. Vaccines Immunother. 2018, 14, 1697–1716. [Google Scholar] [CrossRef] [Green Version]
- Yatime, L.; Buch-Pedersen, M.J.; Musgaard, M.; Morth, J.P.; Winther, A.M.L.; Pedersen, B.P.; Olesen, C.; Andersen, J.P.; Vilsen, B.; Schiøtt, B.; et al. P-type ATPases as drug targets: Tools for medicine and science. Biochim. Biophys. Acta-Bioenerg. 2009, 1787, 207–220. [Google Scholar] [CrossRef] [Green Version]
- Bublitz, M.; Morth, J.P.; Nissen, P. P-type ATPases at a glance. J. Cell Sci. 2011, 124, 3917. [Google Scholar] [CrossRef] [Green Version]
- Morth, J.P.; Pedersen, B.P.; Buch-Pedersen, M.J.; Andersen, J.P.; Vilsen, B.; Palmgren, M.G.; Nissen, P. A structural overview of the plasma membrane Na+,K+-ATPase and H+-ATPase ion pumps. Nat. Rev. Mol. Cell Biol. 2011, 12, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Novoa-Aponte, L. Potencial de las ATPasas Tipo P Como Dianas Terapéuticas o en el diseño de Mutantes Atenuados de Mycobacterium tuberculosis. Ph.D. Thesis, Universidad Nacional de Colombia, Bogotá, Colombia, 2016. [Google Scholar]
- Soldati, T.; Neyrolles, O. Mycobacteria and the Intraphagosomal Environment: Take It with a Pinch of Salt(s)! Traffic 2012, 13, 1042–1052. [Google Scholar] [CrossRef] [PubMed]
- Neyrolles, O.; Wolschendorf, F.; Mitra, A.; Niederweis, M. Mycobacteria, metals, and the macrophage. Immunol. Rev. 2015, 264, 249–263. [Google Scholar] [CrossRef] [PubMed]
- Rathnayake, I.V.N.; Megharaj, M.; Bolan, N.; Naidu, R. Tolerance of Heavy Metals by Gram Positive Soil Bacteria. Int. J. Environ. Eng. 2010, 3, 270–274. [Google Scholar] [CrossRef]
- Campbell, A.K. Intracellular Calcium, 1st ed.; John Wiley & Sons: Hoboken, NJ, USA, 2015; ISBN 9780470695111. [Google Scholar]
- Naseem, R.; Wann, K.T.; Holland, I.B.; Campbell, A.K. ATP Regulates Calcium Efflux and Growth in E. coli. J. Mol. Biol. 2009, 391, 42–56. [Google Scholar] [CrossRef]
- Tisa, L.S.; Adler, J. Calcium ions are involved in Escherichia coli chemotaxis. Proc. Natl. Acad. Sci. USA 1992, 89, 11804–11808. [Google Scholar] [CrossRef] [Green Version]
- Werthén, M.; Lundgren, T. Intracellular Ca2+ Mobilization and Kinase Activity during Acylated Homoserine Lactone-dependent Quorum Sensing in Serratia liquefaciens. J. Biol. Chem. 2001, 276, 6468–6472. [Google Scholar] [CrossRef] [Green Version]
- Burns, D.A.; Minton, N.P. Sporulation studies in Clostridium difficile. J. Microbiol. Methods 2011, 87, 133–138. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Zhang, X.; Shi, Y.; Zhou, Y.; Zhang, W.; Su, X.D.; Xia, B.; Zhao, J.; Jin, C. Structures of anabaena calcium-binding protein CcbP: Insights into Ca2+ signaling during heterocyst differentiation. J. Biol. Chem. 2011, 286, 12381–12388. [Google Scholar] [CrossRef] [Green Version]
- Jones, H.E.; Holland, I.B.; Campbell, A.K. Direct measurement of free Ca2+ shows different regulation of Ca2+ between the periplasm and the cytosol of Escherichia coli. Cell Calcium 2002, 32, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Barrán-Berdón, A.L.; Rodea-Palomares, I.; Leganés, F.; Fernández-Piñas, F. Free Ca2+ as an early intracellular biomarker of exposure of cyanobacteria to environmental pollution. Anal. Bioanal. Chem. 2011, 400, 1015–1029. [Google Scholar] [CrossRef] [PubMed]
- Domínguez, D.C. Calcium Signaling in Prokaryotes. Calcium Signal Transduct. 2018, 5, 89–106. [Google Scholar] [CrossRef] [Green Version]
- Domínguez, D.C.; Guragain, M.; Patrauchan, M. Calcium binding proteins and calcium signaling in prokaryotes. Cell Calcium 2015, 57, 151–165. [Google Scholar] [CrossRef]
- Waditee, R.; Hossain, G.S.; Tanaka, Y.; Nakamura, T.; Shikata, M.; Takano, J.; Takabe, T.; Takabe, T. Isolation and functional characterization of Ca2+/H+ antiporters from cyanobacteria. J. Biol. Chem. 2004, 279, 4330–4338. [Google Scholar] [CrossRef] [Green Version]
- Malik, Z.A.; Iyer, S.S.; Kusner, D.J. Mycobacterium tuberculosis phagosomes exhibit altered calmodulin-dependent signal transduction: Contribution to inhibition of phagosome-lysosome fusion and intracellular survival in human macrophages. J. Immunol. 2001, 166, 3392–3401. [Google Scholar] [CrossRef] [Green Version]
- Sharma, S.; Meena, L.S. Potential of Ca2+ in Mycobacterium tuberculosis H37Rv Pathogenesis and Survival. Appl. Biochem. Biotechnol. 2017, 181, 762–771. [Google Scholar] [CrossRef]
- Ward, S.K.; Abomoelak, B.; Hoye, E.A.; Steinberg, H.; Talaat, A.M. CtpV: A putative copper exporter required for full virulence of Mycobacterium tuberculosis. Mol. Microbiol. 2010, 77, 1096–1110. [Google Scholar] [CrossRef] [Green Version]
- Botella, H.; Peyron, P.; Levillain, F.; Poincloux, R.; Poquet, Y.; Brandli, I.; Wang, C.; Tailleux, L.; Tilleul, S.; Charrire, G.M.; et al. Mycobacterial P 1-Type ATPases mediate resistance to Zinc poisoning in human macrophages. Cell Host Microbe 2011, 10, 248–259. [Google Scholar] [CrossRef]
- Padilla-Benavides, T.; Long, J.E.; Raimunda, D.; Sassetti, C.M.; Argüello, J.M. A novel P1B-type Mn2+-transporting ATPase is required for secreted protein metallation in mycobacteria. J. Biol. Chem. 2013, 288, 11334–11347. [Google Scholar] [CrossRef] [Green Version]
- Palmgren, M.G.; Nissen, P. P-type ATPases. Annu. Rev. Biophys. 2011, 40, 243–266. [Google Scholar] [CrossRef] [PubMed]
- Agranoff, D. Metal ion transport and regulation in mycobacterium tuberculosis. Front. Biosci. 2004, 9, 2996–3006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Novoa-Aponte, L.; Leon-Torres, A.; Patino-Ruiz, M.; Cuesta-Bernal, J.; Salazar, L.M.; Landsman, D.; Marino-Ramirez, L.; Soto, C.Y. In silico identification and characterization of the ion transport specificity for P-type ATPases in the Mycobacterium tuberculosis complex. BMC Struct. Biol. 2012, 12, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, H.K.; Shrivastava, S.; Sharma, R. A Novel Calcium Uptake Transporter of Uncharacterized P-Type ATPase Family Supplies Calcium for Cell Surface Integrity in Mycobacterium smegmatis. MBio 2017, 8, e01388-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maya-Hoyos, M. ATPasas Tipo P2 Como Blancos Para la Atenuación de Mycobacterium tuberculosis. Ph.D. Thesis, Universidad Nacional de Colombia, Bogotá, Colombia, 2021. [Google Scholar]
- Novoa-Aponte, L.; Soto Ospina, C.Y. Mycobacterium tuberculosis P-type ATPases: Possible targets for drug or vaccine development. Biomed Res. Int. 2014, 2014, 296986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maya-Hoyos, M.; Rosales, C.; Novoa-Aponte, L.; Castillo, E.; Soto, C.Y. The P-type ATPase CtpF is a plasma membrane transporter mediating calcium efflux in Mycobacterium tuberculosis cells. Heliyon 2019, 5, e02852. [Google Scholar] [CrossRef]
- Peddireddy, V.; Doddam, S.N.; Ahmed, N. Mycobacterial dormancy systems and host responses in tuberculosis. Front. Immunol. 2017, 8, 84. [Google Scholar] [CrossRef] [Green Version]
- Zheng, H.; Abramovitch, R.B. Inhibiting DosRST as a new approach to tuberculosis therapy. Future Med. Chem. 2020, 12, 457–467. [Google Scholar] [CrossRef]
- Guirado, E.; Mbawuike, U.; Keiser, T.L.; Arcos, J.; Azad, A.K.; Wang, S.H.; Schlesinger, L.S. Characterization of host and microbial determinants in individuals with latent tuberculosis infection using a human granuloma model. MBio 2015, 6, e02537-14. [Google Scholar] [CrossRef] [Green Version]
- Park, H.D.; Guinn, K.M.; Harrell, M.I.; Liao, R.; Voskuil, M.I.; Tompa, M.; Schoolnik, G.K.; Sherman, D.R. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol. Microbiol. 2003, 48, 833–843. [Google Scholar] [CrossRef] [Green Version]
- Leistikow, R.L.; Morton, R.A.; Bartek, I.L.; Frimpong, I.; Wagner, K.; Voskuil, M.I. The Mycobacterium tuberculosis DosR regulon assists in metabolic homeostasis and enables rapid recovery from nonrespiring dormancy. J. Bacteriol. 2010, 192, 1662–1670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garg, R.; Borbora, S.M.; Bansia, H.; Rao, S.; Singh, P.; Verma, R.; Balaji, K.N.; Nagaraja, V. Mycobacterium tuberculosis Calcium Pump CtpF Modulates the Autophagosome in an mTOR-Dependent Manner. Front. Cell. Infect. Microbiol. 2020, 10, 461. [Google Scholar] [CrossRef] [PubMed]
- Sassetti, C.M.; Boyd, D.H.; Rubin, E.J. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 2003, 48, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Cardona, P.J.; Soto, C.Y.; Martín, C.; Giquel, B.; Agustí, G.; Guirado, E.; Sirakova, T.; Kolattukudy, P.; Julián, E.; Luquin, M. Neutral-red reaction is related to virulence and cell wall methyl-branched lipids in Mycobacterium tuberculosis. Microbes Infect. 2006, 8, 183–190. [Google Scholar] [CrossRef]
- Bonilla, D.L.; Bhattacharya, A.; Sha, Y.; Xu, Y.; Xiang, Q.; Kan, A.; Jagannath, C.; Komatsu, M.; Eissa, N.T. Autophagy regulates phagocytosis by modulating the expression of scavenger receptors. Immunity 2013, 39, 537–547. [Google Scholar] [CrossRef] [Green Version]
- Cooper, A.M.; Mayer-Barber, K.D.; Sher, A. Role of innate cytokines in mycobacterial infection. Mucosal Immunol. 2011, 4, 252–260. [Google Scholar] [CrossRef]
- Bogdan, C.; Schleicher, U. Production of interferon-γ by myeloid cells—Fact or fancy? Trends Immunol. 2006, 27, 282–290. [Google Scholar] [CrossRef]
- Jordao, L.; Bleck, C.K.E.; Mayorga, L.; Griffiths, G.; Anes, E. On the killing of mycobacteria by macrophages. Cell. Microbiol. 2007, 10, 529–548. [Google Scholar] [CrossRef]
- Barber-Mayer, K.D.; Barber, D.L. Innate and adaptive cellular immune responses to Mycobacterium tuberculosis infection. Cold Spring Harb. Perspect. Med. 2015, 5, a018424. [Google Scholar] [CrossRef] [Green Version]
- O’Garra, A.; Redford, P.S.; McNab, F.W.; Bloom, C.I.; Wilkinson, R.J.; Berry, M.P.R. The immune response in tuberculosis. Annu. Rev. Immunol. 2013, 31, 475–527. [Google Scholar] [CrossRef]
- León-Torres, A.; Novoa-Aponte, L.; Soto, C.Y. CtpA, a putative Mycobacterium tuberculosis P-type ATPase, is stimulated by copper (I) in the mycobacterial plasma membrane. BioMetals 2015, 28, 713–724. [Google Scholar] [CrossRef] [PubMed]
- Doddam, S.N.; Peddireddy, V.; Ahmed, N. Mycobacterium tuberculosis DosR Regulon Gene Rv2004c Encodes a Novel Antigen with Pro-inflammatory Functions and Potential Diagnostic Application for Detection of Latent Tuberculosis. Front. Immunol. 2017, 8, 712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagner, D.; Maser, J.; Lai, B.; Cai, Z.; Barry, C.E.; Honer zu Bentrup, K.; Russell, D.G.; Bermudez, L.E. Elemental Analysis of Mycobacterium avium-, Mycobacterium tuberculosis-, and Mycobacterium smegmatis-Containing Phagosomes Indicates Pathogen-Induced Microenvironments within the Host Cell’s Endosomal System. J. Immunol. 2005, 174, 1491–1500. [Google Scholar] [CrossRef] [Green Version]
- Nava, A.R.; Mauricio, N.; Sanca, A.J.; Domínguez, D.C. Evidence of Calcium Signaling and Modulation of the LmrS Multidrug Resistant Efflux Pump Activity by Ca2+ Ions in S. aureus. Front. Microbiol. 2020, 11, 573388. [Google Scholar] [CrossRef] [PubMed]
- King, M.M.; Kayastha, B.B.; Franklin, M.J.; Patrauchan, M.A. Calcium Regulation of Bacterial Virulence. Adv. Exp. Med. Biol. 2020, 1131, 827–855. [Google Scholar] [CrossRef]
- Brennan, P.J. The Envelope of Mycobacteria. Annu. Rev. Biochem. 1995, 64, 29–63. [Google Scholar] [CrossRef]
- Jarlier, V.; Nikaido, H. Mycobacterial cell wall: Structure and role in natural resistance to antibiotics. FEMS Microbiol. Lett. 1994, 123, 11–18. [Google Scholar] [CrossRef]
- Torrelles, J.B.; Schlesinger, L.S. Diversity in Mycobacterium tuberculosis mannosylated cell wall determinants impacts adaptation to the host. Tuberculosis 2010, 90, 84–93. [Google Scholar] [CrossRef] [Green Version]
- Flynn, J.L.; Chan, J. Immunology of tuberculosis. Annu. Rev. Immunol. 2001, 19, 93–129. [Google Scholar] [CrossRef]
- Jayachandran, R.; Sundaramurthy, V.; Combaluzier, B.; Mueller, P.; Korf, H.; Huygen, K.; Miyazaki, T.; Albrecht, I.; Massner, J.; Pieters, J. Survival of Mycobacteria in Macrophages Is Mediated by Coronin 1-Dependent Activation of Calcineurin. Cell 2007, 130, 37–50. [Google Scholar] [CrossRef] [Green Version]
- Trimble, W.S.; Grinstein, S. TB or not TB: Calcium Regulation in Mycobacterial Survival. Cell 2007, 130, 12–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forrellad, M.A.; Klepp, L.I.; Gioffré, A.; García, J.S.; Morbidoni, H.R.; de la Paz Santangelo, M.; Cataldi, A.A.; Bigi, F. Virulence factors of the mycobacterium tuberculosis complex. Virulence 2013, 4, 3–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosch, J.W.; Sublett, J.; Gao, G.; Wang, Y.D.; Tuomanen, E.I. Calcium efflux is essential for bacterial survival in the eukaryotic host. Mol. Microbiol. 2008, 70, 435–444. [Google Scholar] [CrossRef] [Green Version]
- Hernández-Pando, R.; Orozcoe, H.; Sampieri, A.; Pavón, L.; Velasquillo, C.; Larriva-Sahd, J.; Alcocer, J.M.; Madrid, M.V. Correlation between the kinetics of Th1, Th2 cells and pathology in a murine model of experimental pulmonary tuberculosis. Immunology 1996, 89, 26–33. [Google Scholar] [PubMed]
- Hernandez-Pando, R.; Orozco, H.; Honour, J.; Silva, P.; Leyva, R.; Rook, G.A.W. Adrenal changes in murine pulmonary tuberculosis; a clue to pathogenesis? FEMS Immunol. Med. Microbiol. 1995, 12, 63–72. [Google Scholar] [CrossRef]
- Santos, P.; Lopez-Vallejo, F.; Ramírez, D.; Caballero, J.; Mata Espinosa, D.; Hernández-Pando, R.; Soto, C.Y. Identification of Mycobacterium tuberculosis CtpF as a target for designing new antituberculous compounds. Bioorg. Med. Chem. 2019, 28, 115256. [Google Scholar] [CrossRef]
- Rodríguez, J.E.; Ramírez, A.S.; Salas, L.P.; Helguera-Repetto, C.; Gonzalez-y-Merchand, J.; Soto, C.Y.; Hernández-Pando, R. Transcription of Genes Involved in Sulfolipid and Polyacyltrehalose Biosynthesis of Mycobacterium tuberculosis in Experimental Latent Tuberculosis Infection. PLoS ONE 2013, 8, e58378. [Google Scholar] [CrossRef] [PubMed]
- Rustad, T.R.; Roberts, D.M.; Liao, R.P.; Sherman, D.R. Isolation of mycobacterial RNA. Methods Mol. Biol. 2009, 465, 13–22. [Google Scholar] [CrossRef]
- López-Torres, M.O.; Marquina-Castillo, B.; Ramos-Espinosa, O.; Mata-Espinosa, D.; Barrios-Payan, J.A.; Baay-Guzman, G.; Yepez, S.H.; Bini, E.; Torre-Villalvazo, I.; Torres, N.; et al. 16α-Bromoepiandrosterone as a new candidate for experimental diabetes–tuberculosis co-morbidity treatment. Clin. Exp. Immunol. 2021, 205, 232–245. [Google Scholar] [CrossRef]
- Cornejo-Granados, F.; López-Leal, G.; Mata-Espinosa, D.A.; Barrios-Payán, J.; Marquina-Castillo, B.; Equihua-Medina, E.; Zatarain-Barrón, Z.L.; Molina-Romero, C.; Hernández-Pando, R.; Ochoa-Leyva, A. Targeted rna-seq reveals the m. Tuberculosis transcriptome from an in vivo infection model. Biology 2021, 10, 848. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2019, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.; Wang, R.; Deng, D.; Chen, Y.; Liu, H.; Wang, T.; Wang, J.; Zhu, X.; Zhu, X.; Zhu, Y.; et al. Comparative genomics of a bovine Mycobacterium tuberculosis isolate and other strains reveals its potential mechanism of bovine adaptation. Front. Microbiol. 2017, 8, 2500. [Google Scholar] [CrossRef] [PubMed]
- De Coste, N.J.; Gadkar, V.J.; Filion, M. Relative and absolute quantitative real-time PCR-based quantifications of hcnC and phlD gene transcripts in natural soil spiked with Pseudomonas sp. strain LBUM300. Appl. Environ. Microbiol. 2011, 77, 41–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maya-Hoyos, M.; Mata-Espinosa, D.; López-Torres, M.O.; Tovar-Vázquez, B.; Barrios-Payán, J.; León-Contreras, J.C.; Ocampo, M.; Hernández-Pando, R.; Soto, C.Y. The ctpF Gene Encoding a Calcium P-Type ATPase of the Plasma Membrane Contributes to Full Virulence of Mycobacterium tuberculosis. Int. J. Mol. Sci. 2022, 23, 6015. https://doi.org/10.3390/ijms23116015
Maya-Hoyos M, Mata-Espinosa D, López-Torres MO, Tovar-Vázquez B, Barrios-Payán J, León-Contreras JC, Ocampo M, Hernández-Pando R, Soto CY. The ctpF Gene Encoding a Calcium P-Type ATPase of the Plasma Membrane Contributes to Full Virulence of Mycobacterium tuberculosis. International Journal of Molecular Sciences. 2022; 23(11):6015. https://doi.org/10.3390/ijms23116015
Chicago/Turabian StyleMaya-Hoyos, Milena, Dulce Mata-Espinosa, Manuel O. López-Torres, Blanca Tovar-Vázquez, Jorge Barrios-Payán, Juan C. León-Contreras, Marisol Ocampo, Rogelio Hernández-Pando, and Carlos Y. Soto. 2022. "The ctpF Gene Encoding a Calcium P-Type ATPase of the Plasma Membrane Contributes to Full Virulence of Mycobacterium tuberculosis" International Journal of Molecular Sciences 23, no. 11: 6015. https://doi.org/10.3390/ijms23116015






