Loss of BMP2 and BMP4 Signaling in the Dental Epithelium Causes Defective Enamel Maturation and Aberrant Development of Ameloblasts
Abstract
:1. Introduction
2. Results
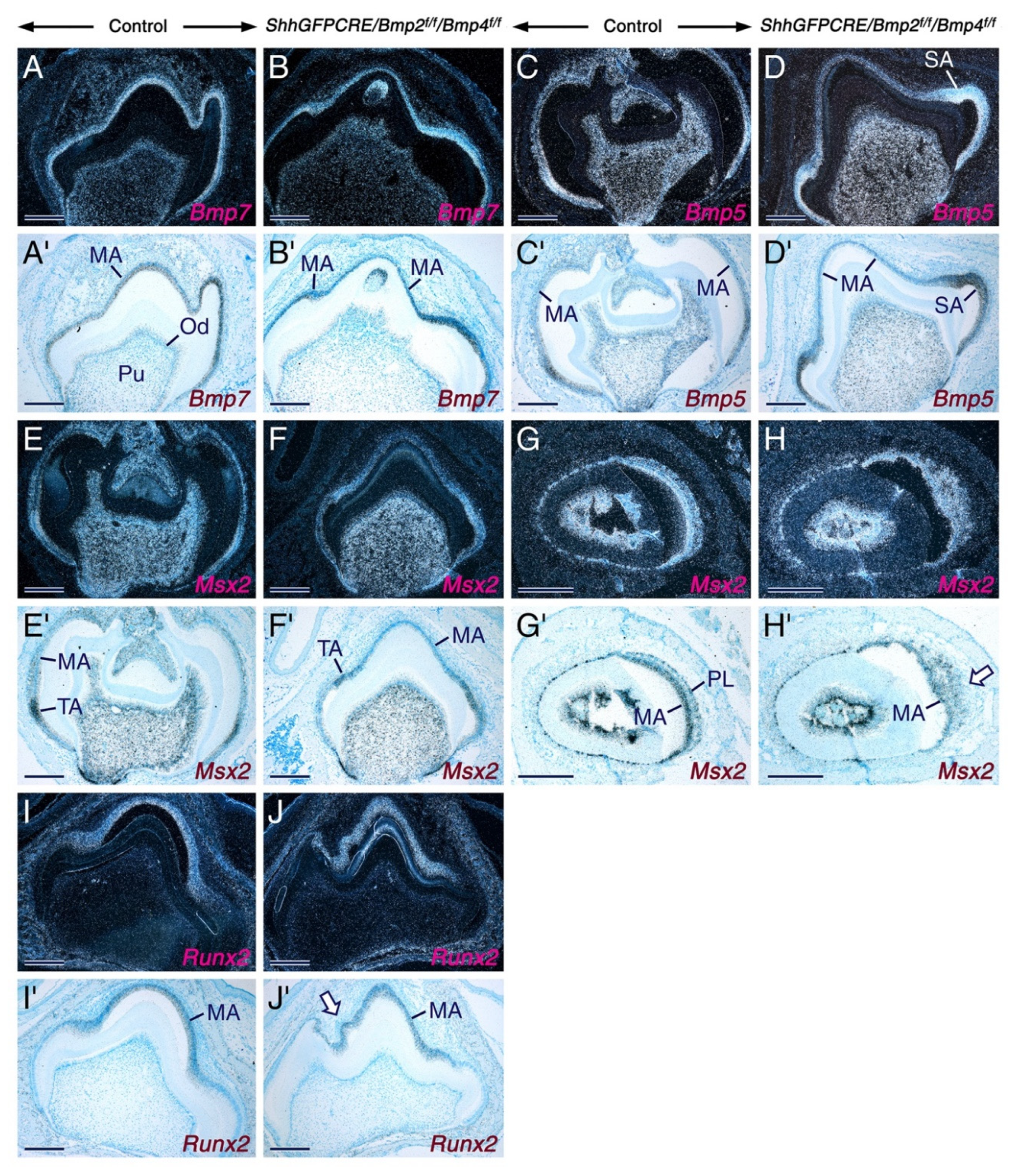
2.1. Normal Tooth Development before the Maturation Stage of Amelogenesis upon Combined Loss of Bmp2 and Bmp4 Gene Function in the Dental Epithelium
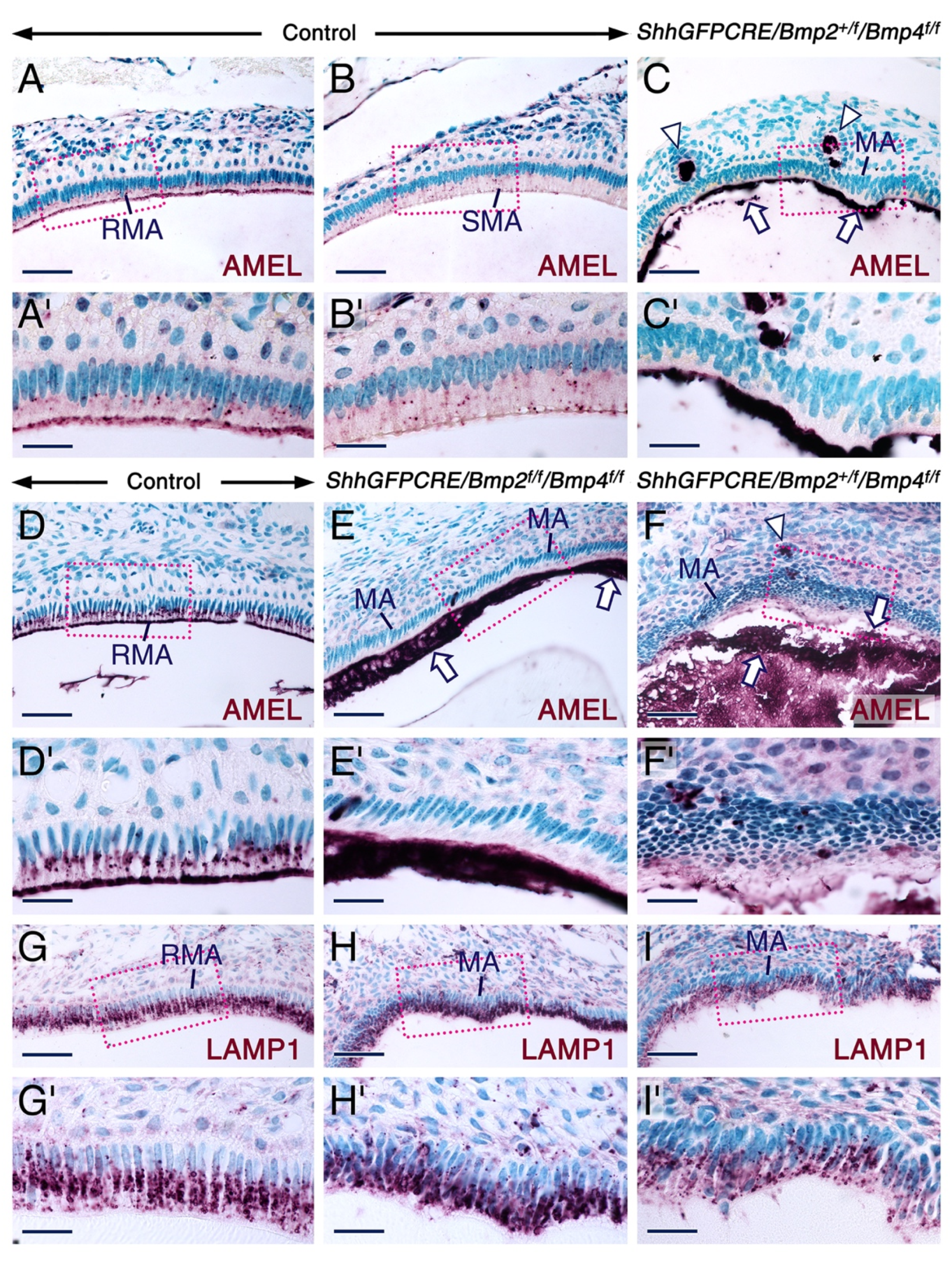
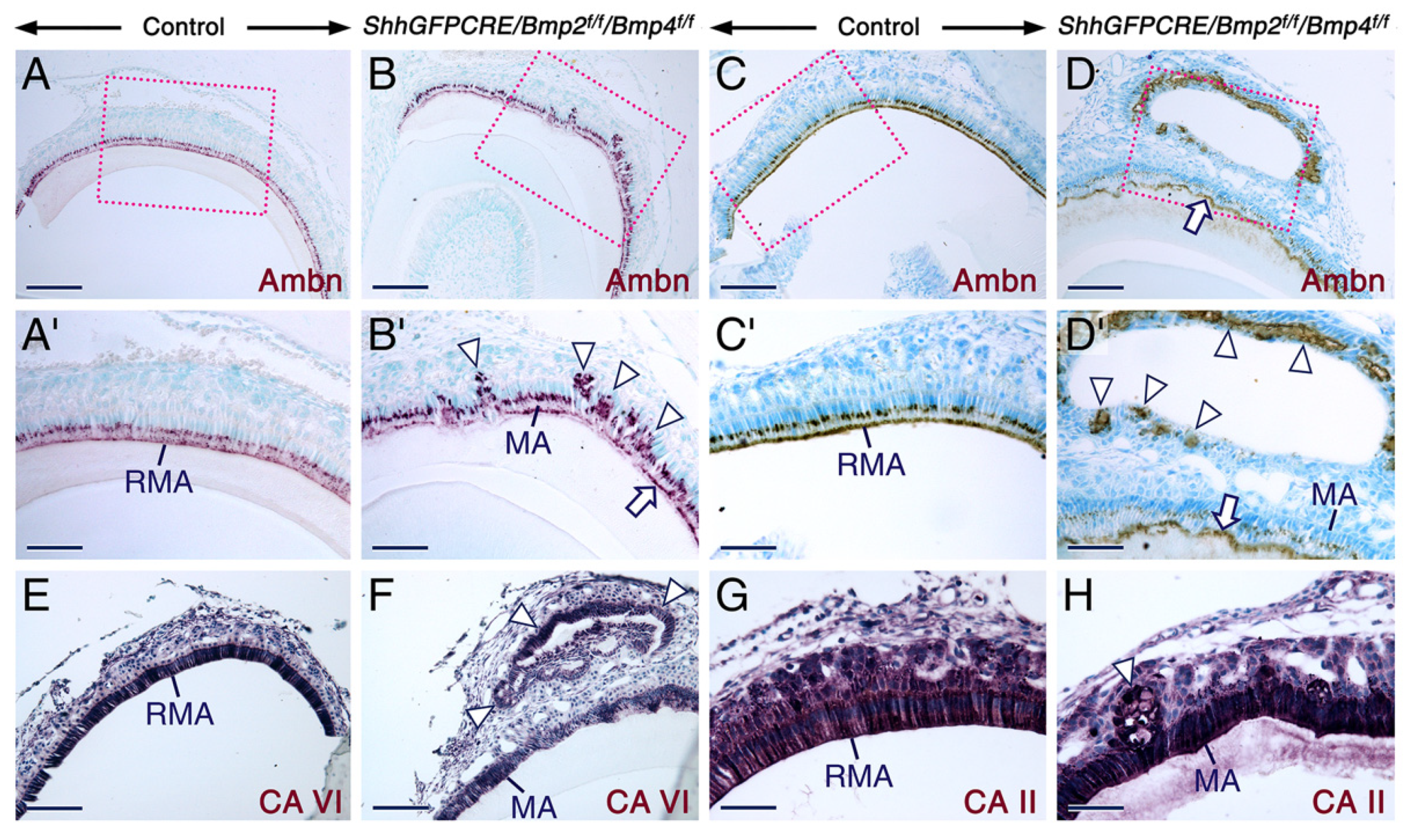
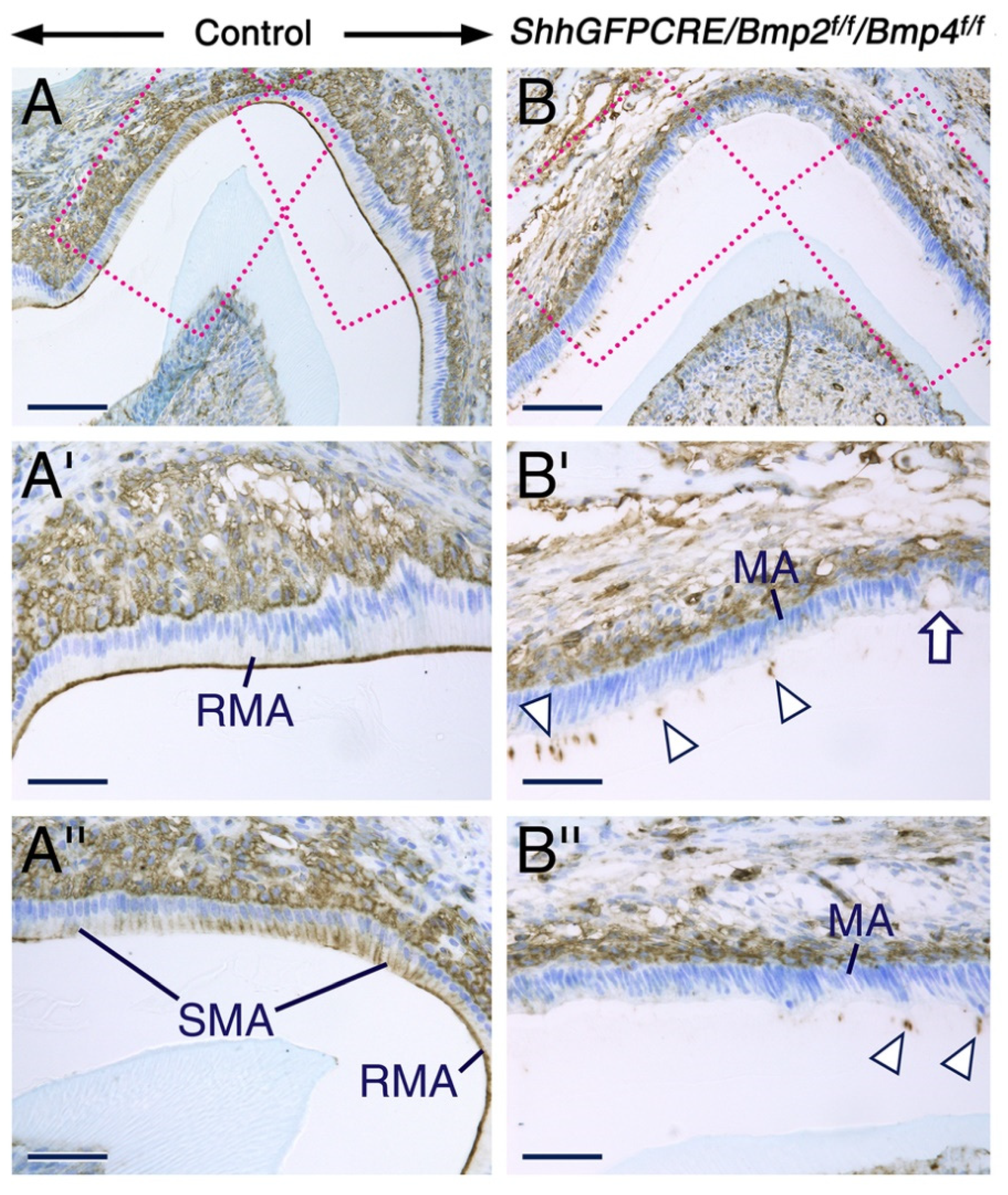
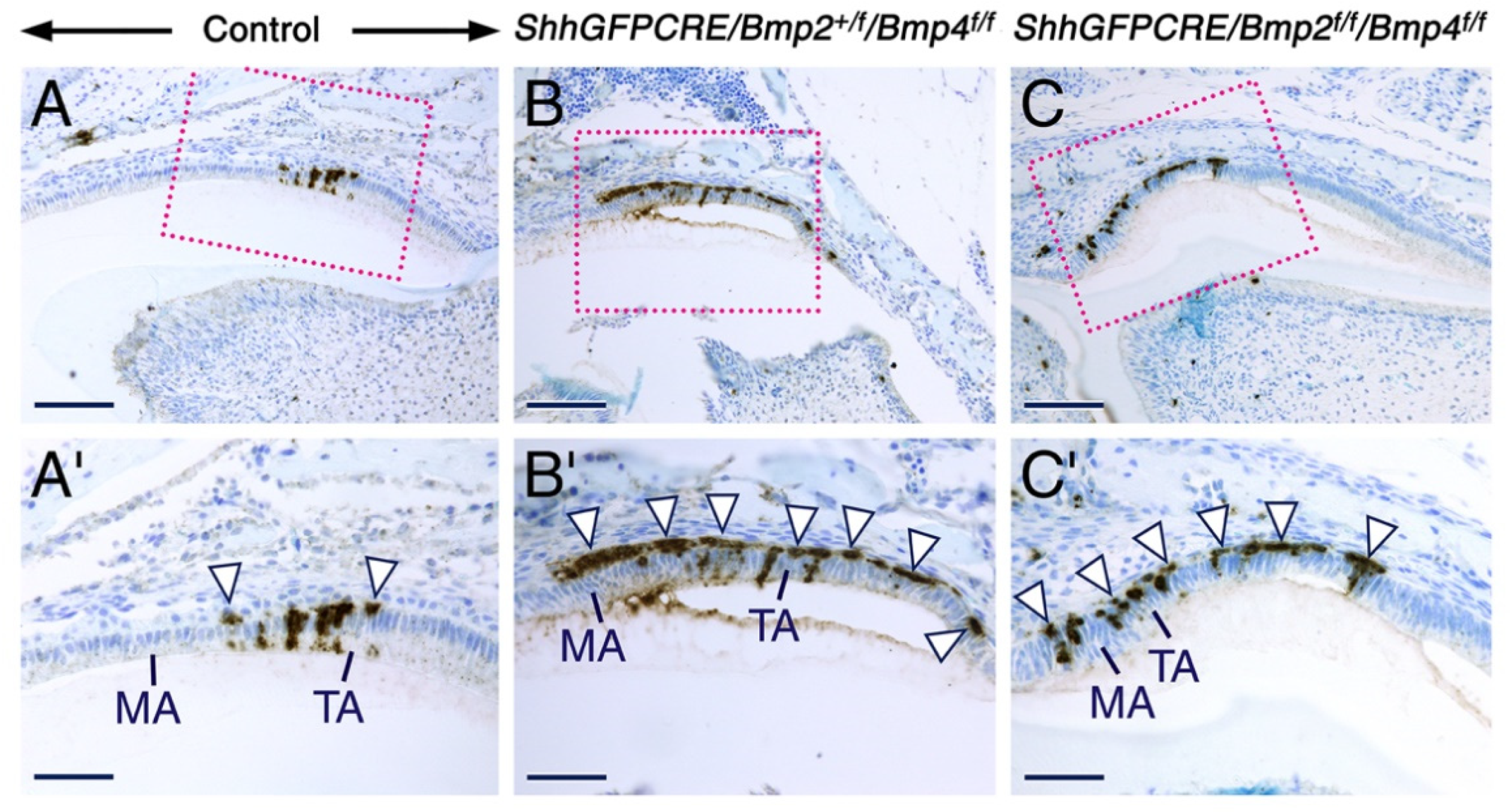
2.2. Combined Loss of BMP2 and BMP4 Signaling in the Dental Epithelium Causes Development of Dysfunctional Maturation-Stage Ameloblasts Lacking a Ruffled Border and Exhibiting Altered Cell-Cell and Cell-Matrix Contacts
2.3. Altered Alkaline Phosphatase Activity in Maturation Stage Ameloblasts upon Combined Loss of Epithelial BMP2 and BMP4 Signaling
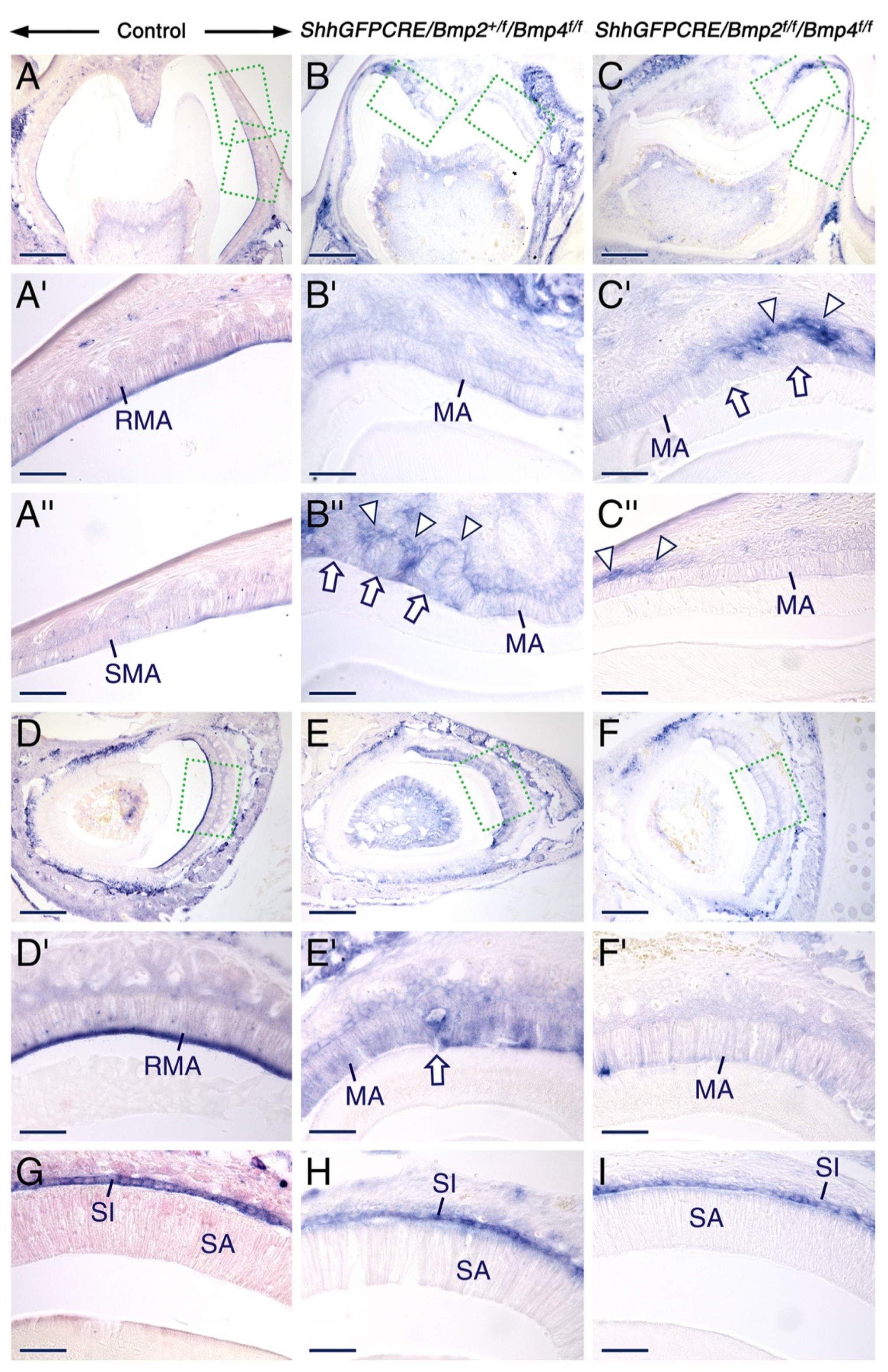
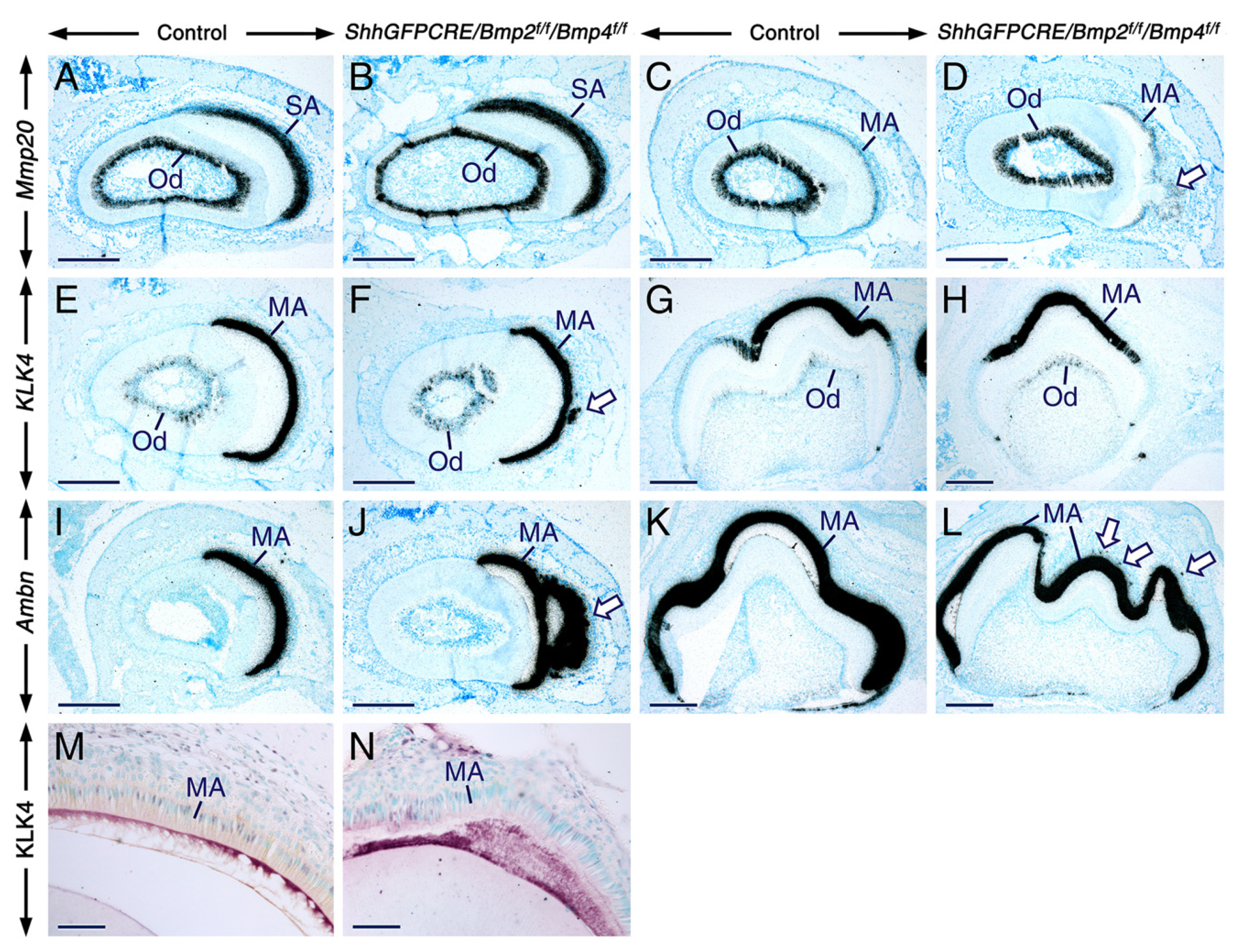
2.4. Normal Gene Expression Patterns in Secretory and Maturation-Stage Ameloblasts and Unaltered KLK4 Secretion upon Combined Loss of Epithelial BMP2 and BMP4 Signaling
2.5. Combined Loss of BMP2 and BMP4 Signaling in the Dental Epithelium Causes Massive Apoptosis in the Stratum Intermedium
3. Discussion
3.1. BMP2 and BMP4 Signaling in the Dental Epithelium Is not Required for Early Stages of Tooth Development
3.2. Maturation-Stage Ameloblast Morphological Differentiation and Function Depend on Epithelial BMP2 and BMP4 Inputs to Ensure Proper Enamel Maturation
3.3. Defective Resorptive Activities of Bmp2/Bmp4-Deficient Maturation-Stage Ameloblasts Is Likely a Cause for Failure of Proper Enamel Maturation
3.4. Signaling from BMP2 and BMP4 in the Dental Epithelium Is Required for Survival of the Stratum Intermedium and for Normal Organization of Maturation-Stage Ameloblasts
4. Materials and Methods
4.1. Ethics Statement
4.2. Mouse Lines
4.3. Histology, Immunohistochemistry and In Situ Hybridization
4.4. β-Galactosidase Histochemistry
4.5. Tissue Non-Specific Alkaline Phosphatase Activity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Ambn/Ambn | Ameloblastin protein/gene |
| AMEL/AmelX | Amelogenin protein/gene |
| BMP/Bmp | Bone morphogenetic protein/gene |
| CA | Carbonic anhydrase |
| CRE/CRE | (Cyclization recombination) DNA recombinase protein/gene |
| DPP/dpp | Drosophila decapentaplegic protein/gene |
| EMPs | Enamel matrix proteins |
| ERM | Ezrin/radixin/moesin |
| f | Floxed allele |
| KLK4/Klk4 | Kallikrein-4 protein/gene |
| LAMP1 | Lysosome-associated membrane protein |
| MMP20/Mmp20 | Matrix metalloproteinase 20 protein/gene |
| MSX2/Msx2 | Muscle segment homeobox 2 protein/gene |
| RMA | Ruffle-ended maturation-stage ameloblasts |
| RUNX2/Runx2 | Runt-related transcription factor 2 protein/gene |
| SHH/Shh | Sonic Hedgehog protein/gene |
| SMA | Smooth-ended maturation-stage ameloblasts |
| TNAP/Alpl | Tissue non-specific alkaline phosphatase protein/gene |
| ZO1 | Zonula occludens1 |
References
- Reith, E.J. The stages of amelogenesis as observed in molar teeth of young rats. J. Ultrastruc. Res. 1970, 30, 111–151. [Google Scholar] [CrossRef]
- Warshawsky, H.; Smith, C.E. Morphological classification of rat incisor ameloblasts. Anat. Rec. 1974, 179, 423–446. [Google Scholar] [CrossRef] [PubMed]
- Karcher-Djuricic, V.; Staubli, A.; Meyer, J.M.; Ruch, J.V. Acellular dental matrices promote functional differentiation of ameloblasts. Differentiation 1985, 29, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Zeichner-David, M.; Diekwisch, T.; Fincham, A.; Lau, E.; MacDougall, M.; Moradian-Oldak, J.; Simmer JSnead, M.; Slavkin, H.C. Control of ameloblast differentiation. Int. J. Dev. Biol. 1995, 39, 69–92. [Google Scholar] [PubMed]
- Ruch, J.V. Determinism of odontogenesis. Cell Biol. Rev. 1987, 14, 1–112. [Google Scholar]
- Ruch, J.V. Patterned distribution of differentiating dental cells: Facts and hypotheses. J. Biol. Buccale 1990, 18, 91–98. [Google Scholar]
- Smith, C.E. Cellular and chemical events during enamel maturation. Crit. Rev. Oral Biol. Med. 1998, 9, 128–161. [Google Scholar] [CrossRef]
- Lu, Y.; Papagerakis, P.; Yamakoshi, Y.; Hu, J.C.-C.; Bartlett, J.D.; Simmer, J.P. Functions of KLK4 and MMP-20 in dental enamel formation. Biol. Chem. 2008, 389, 695–700. [Google Scholar] [CrossRef] [Green Version]
- Smith, C.E.L.; Poulter, J.A.; Antanaviciute, A.; Kirkham, J.; Brookes, S.J.; Inglehearn, C.F.; Mighell, A.J. Amelogenesis imperfecta; genes, proteins, and pathways. Front. Physiol. 2017, 8, 835. [Google Scholar] [CrossRef] [Green Version]
- Bègue-Kirn, C.; Krebsbach, P.H.; Bartlett, J.D.; Butler, W.T. Dentin sialoprotein, dentin phosphoprotein, enamelysin and ameloblastin: Tooth-specific molecules that are distinctively expressed during murine dental differentiation. Eur. J. Oral Sci. 1998, 106, 963–970. [Google Scholar] [CrossRef]
- Hu, J.C.; Sun, X.; Zhang, C.; Liu, S.; Bartlett, J.D.; Simmer, J.P. Enamelysin and Kallikrein-4 mRNA expression in developing mouse molars. Eur. J. Oral Sci. 2002, 110, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Simmer, J.P.; Hu, J.C. Expression, structure and function of enamel proteinases. Connect. Tissue Res. 2002, 43, 441–443. [Google Scholar] [CrossRef] [PubMed]
- Fincham, A.G.; Moradian-Oldak, J.; Simmer, J.P. The structural biology of the developing dental enamel matrix. J. Struct. Biol. 1999, 126, 270–299. [Google Scholar] [CrossRef] [PubMed]
- Kallenbach, E. Fine structure of rat incisor ameloblasts in transition between enamel secretion and maturation stages. Tissue Cell 1974, 6, 173–190. [Google Scholar] [CrossRef]
- Smith, C.E.; Nanci, A. Overview of morphological changes in enamel organ cells associated with major events in amelogenesis. Int. J. Dev. Biol. 1995, 39, 153–161. [Google Scholar]
- Josephsen, K.; Fejerskov, O. Ameloblast modulation in the mature zone of the rat incisor enamel organ, a light and electron microscopy study. J. Anat. 1977, 124, 45–70. [Google Scholar]
- Garant, P.R.; Nalbandian, J. The fine structure of the papillary region of the mouse enamel organ. Arch. Oral Biol. 1968, 13, 1167–1185. [Google Scholar] [CrossRef]
- Yamamoto, K.; Nishimoto, T.; Matsuo, S.; Wakisaka, S.; Nakata, T.; Akai, M. Scanning electron microscopy of the papillary layer of the rat incisor enamel organ. Arch. Oral Biol. 1983, 28, 283–285. [Google Scholar] [CrossRef]
- Joseph, B.K.; Gobé, G.C.; Savage, N.W.; Young, W.G. Expression and localization of sulphated glycoprotein-2 mRNA in the rat incisor tooth ameloblasts: Relationships with apoptosis. Int. J. Pathol. 1994, 75, 313–320. [Google Scholar]
- Smith, C.E.; Warshawsky, H. Quantitative analysis of cell turnover in the enamel organ of the rat incisor. Evidence of ameloblast death immediately after enamel matrix secretion. Anat. Rec. 1977, 187, 63–98. [Google Scholar]
- Nanci, A.; Slavkin, H.C.; Smith, C.E. Immunocytochemical and radiographic evidence for secretion and intracellular degradation of enamel proteins by ameloblasts during the maturation stage of amelogenesis in rat incisors. Anat. Rec. 1987, 217, 107–123. [Google Scholar] [CrossRef] [PubMed]
- Fong, C.D.; Hammarström, L.; Lundmark, C.; Wurtz, T.; Slaby, I. Expression patterns of RNAS for amelin and amelogenin in developing rat molars and incisors. Adv. Dent. Res. 1996, 10, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Inage, T.; Shimokawa, H.; Teranishi, Y.; Iwase, T.; Toda, Y.; Moro, I. Immunocytochemical demonstration of amelogenins and enamelins secreted by ameloblasts during the secretory and maturation stages. Arch. Histol. Cytol. 1989, 52, 213–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inage, T.; Shimokawa, H.; Wakao, K.; Sasaki, S. Gene expression and localization of amelogenin in the rat incisor. Adv. Dent. Res. 1996, 10, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Wurtz, T.; Lundmark, C.; Christersson, C.; Bawden, J.W.; Slaby, I.; Hammartröm, L. Expression of amelogenin mRNA sequences during development of rat molars. J. Bone Miner. Res. 1996, 11, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Barron, M.J.; Brookes, S.J.; Kirkham, J.; Shore, R.C.; Hunt, C.; Mironiv, A.; Kingswell, N.J.; Maycock, J.; Shuttleworth, C.A.; Dixon, M.J. A mutation in the mouse Amelx tri-tirosyl domain results in impaired secretion of amelogenin and phenocopies amelogenesis imperfecta. Hum. Mol. Genet. 2010, 19, 1230–1247. [Google Scholar] [CrossRef]
- Lee, S.K.; Krebsbach, P.H.; Matsuki, Y.; Nanci, A.; Yamada, K.M.; Yamada, Y. Ameloblastin expression in rat incisors and human tooth germs. Int. J. Dev. Biol. 1996, 40, 1141–1150. [Google Scholar]
- Iwasaki, K.; Bajenova, E.; Somogyi-Ganss, E.; Miller, M.; Nguyen, V.; Nourkeyhani, H.; Gao, Y.; Wendell, M.; Ganss, B. Amelotin-a novel secreted, ameloblast-specific protein. J. Dent. Res. 2005, 84, 1127–1132. [Google Scholar] [CrossRef]
- Moffatt, P.; Smith, C.E.; St-Arnaud, R.; Simmons, D.J.; Wright, T.; Nanci, A. Cloning of rat amelotin and localization of the protein to the basal lamina of maturation-stage ameloblasts. Biochem. J. 2006, 399, 37–46. [Google Scholar] [CrossRef] [Green Version]
- Moffatt, P.; Smith, C.E.; St-Arnaud, R.; Nanci, A. Characterization of Apin, a secreted protein highly expressed in tooth-associated epithelia. J. Cell Biochem. 2008, 103, 941–956. [Google Scholar] [CrossRef]
- Park, J.-C.; Park, J.-T.; Son, H.-H.; Jeong, M.-J.; Lee, C.-S.; Cho, M., II; Dey, R. The amyloid protein APin is highly expressed during enamel mineralization and maturation in rat incisors. Eur. J. Oral Sci. 2007, 115, 153–260. [Google Scholar] [CrossRef] [PubMed]
- Somogyi-Ganss, E.; Nakayama, Y.; Iwasaki, K.; Nakano, Y.; Stolf, D.; McKee, M.D.; Ganss, B. Comparative temporospatial expression profiling of murine amelotin protein during amelogenesis. Cells Tissues Organs 2012, 195, 535–549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, J.C.; Ryu, O.H.; Chen, J.J.; Uchida, T.; Wakida, K.; Murakami, C.; Jiang, H.; Qian, Q.; Zhang, C.; Ottmers, V.; et al. Localization of EMSP1 expression during tooth formation and cloning of mouse cDNA. J. Dent. Res. 2000, 79, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.C.; Zhang, C.; Sun, X.; Yang, Y.; Cao, X.; Ryo, O.; Simmer, J.P. Characterization of the mouse and human PRSS17 genes, their relationships to other serine proteases, and the expression of PRSS17 in the developing mouse incisors. Gene 2000, 251, 1–8. [Google Scholar] [CrossRef]
- Ryu, O.; Hu, J.C.; Yamakoshi, Y.; Villemain, J.L.; Cao, X.; Zhang, C.; Bartlett, J.D.; Simmer, J.P. Porcine kallikrein-4 activation, glycosylation, activity, and expression in prokaryotic and eukaryotic hosts. Eur. J. Oral Sci. 2002, 110, 358–365. [Google Scholar] [CrossRef]
- Wang, S.K.; Zhang, H.; Chavez, M.B.; Hu, Y.; Seymen, F.; Koruyucu, M.; Kasimoglu, Y.; Colvin, C.D.; Kolli, T.N.; Tan, M.H.; et al. Dental malformations associated with biallelic MMP20 mutations. Mol. Genet. Genomic Med. 2020, 8, e1307. [Google Scholar] [CrossRef]
- Wright, J.T.; Carrion, I.A.; Morris, C. The molecular basis of hereditary enamel defects in humans. J. Dent. Res. 2015, 94, 52–61. [Google Scholar] [CrossRef] [Green Version]
- Gibson, C.W.; Yuan, Z.A.; Hall, B.; Longenecker, G.; Chen, E.; Thyagarajan, T.; Sreenath, T.; Wright, J.T.; Decker, S.; Piddington, R.; et al. Amelogenin-deficient mice display an amelogenesis imperfecta phenotype. J. Biol. Chem. 2001, 276, 31871–31875. [Google Scholar] [CrossRef] [Green Version]
- Caterina, J.J.; Skobe, Z.; Shi, J.; Ding, Y.; Simmer, J.P.; Birkedal-Hansen, H.; Bartlett, J.D. Enamelysin (matrix metalloproteinase 20)-deficient mice display an amelogenesis imperfecta phenotype. J. Biol. Chem. 2002, 277, 49598–49604. [Google Scholar] [CrossRef] [Green Version]
- Bartlett, J.D.; Smith, C.E. Modulation of cell-cell junctional complexes by matrix metalloproteinases. J. Dent. Res. 2013, 92, 10–17. [Google Scholar] [CrossRef] [Green Version]
- Bartlett, J.C.; Skobe, Z.; Lee, D.H.; Wright, J.T.; Li, Y.; Kulkarni, A.B.; Gibson, C.W. A developmental comparison of matrix metalloproteinase-20 and amelogenin null mouse enamel. Eur. J. Oral Sci. 2006, 114 (Suppl. 1), 18–23. [Google Scholar] [CrossRef] [PubMed]
- Fukumoto, S.; Kiba, T.; Hall, B.; Iehara, N.; Nakamura, T.; Longenecker, G.; Krebsbach, P.H.; Nanci, A.; Kulkarni, A.B.; Yamada, Y. Ameloblastin is a cell adhesion molecule required for maintaining the differentiation state of ameloblasts. J. Cell Biol. 2004, 167, 973–983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masuya, H.; Shimizu, K.; Sezutsu, H.; Sakuraba, Y.; Nagano, J.; Shimizu, A.; Fujimoto, N.; Kawai, A.; Miura, I.; Kaneda, H.; et al. Enamelin (Enam) is essential for amelogenesis: ENU-induced mouse mutants as models for different clinical subtypes of human amelogenesis imperfecta (AI). Hum. Mol. Genet. 2005, 14, 575–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simmer, J.P.; Hu, Y.; Lertlam, R.; Yamakoshi, Y.; Hu, J.C. Hypomaturation enamel defects in Klk4 knockout/lacZ knock-in mice. J. Biol. Chem. 2009, 284, 1910–19121. [Google Scholar] [CrossRef] [Green Version]
- Wazen, R.M.; Moffat, P.; Francis Zalzal, S.; Yamada, Y.; Nanci, A. A mouse model expressing a truncated form of ameloblastin exhibits dental and junctional epithelium defects. Matrix Biol. 2009, 28, 292–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, C.E.; Richardson, A.C.; Hu, Y.; Bartlett, J.D.; Hu, J.C.; Simmer, J.P. Effect of kallikrein 4 loss on enamel mineralization: Comparison with mice lacking matrix metalloproteinase 20. J. Biol. Chem. 2011, 286, 18149–18160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brookes, S.J.; Barron, M.J.; Boot-Handford, R.; Kirkham, J.; Dixon, M.J. Endoplasmic reticulum stress in amelogenesis imperfecta and phenotypic rescue using 4-phenylbutyrate. Hum. Mol. Genet. 2014, 23, 2468–2480. [Google Scholar] [CrossRef] [Green Version]
- Brookes, S.J.; Barron, M.J.; Smith, C.E.L.; Poulter, J.A.; Mighell, A.J.; Inglehearn, C.F.; Brown, C.J.; Rodd, H.; Kirkham, J.; Dixon, M. Amelogenesis imperfecta caused by N-terminal enamelin point mutation in mice and men is driven by endoplasmic reticulum stress. Hum. Mol. Genet. 2017, 26, 1863–1876. [Google Scholar] [CrossRef] [Green Version]
- Nakayama, Y.; Holcroft, J.; Ganss, B. Enamel hypomineralization and structural defects in Amelotin-deficient mice. J. Dent. Res. 2015, 94, 697–705. [Google Scholar] [CrossRef]
- Hu, Y.; Smith, C.E.; Richardson, A.S.; Bartlett, J.D.; Hu, J.C.-C.; Simmer, J.P. MMP20, KLK4, and MMP20/KLK4 double null mice define roles for matrix proteases during dental enamel formation. Mol. Genet. Genomic Med. 2016, 4, 178–196. [Google Scholar] [CrossRef] [Green Version]
- Shin, M.; Chavez, M.B.; Ikeda, A.; Foster, B.L.; Bartlett, J.D. MMP20 overexpression disrupts molar ameloblast polarity and migration. J. Dent. Res. 2018, 97, 820–827. [Google Scholar] [CrossRef] [PubMed]
- Lacruz, R.S.; Brookes, S.J.; Wen, X.; Jimenez, J.M.; Vikman, S.; Hu, P.; White, S.N.; Lyngstadaas, S.P.; Okamoto, C.T.; Smith, C.E.; et al. Adaptor protein 2-mediated clathrin-dependent endocytosis, and related gene activities, are a prominent feature during maturation stage amelogenesis. J. Bone Min. Res. 2013, 28, 672–687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pham, C.-D.; Smith, C.E.; Hu, Y.; Hi, J.C.-C.; Simmer, J.P.; Chun, Y.-H.P. Endocytosis and enamel formation. Front. Physiol. 2017, 8, 529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dassule, H.; Lewis, P.; Bei, M.; McMahon, A.P. Sonic hedgehog regulates growth and morphogenesis of the tooth. Development 2000, 127, 4775–4785. [Google Scholar] [CrossRef]
- Gritli-Linde, A.; Bei, M.; Maas, R.; Zhang, X.M.; Linde, A.; McMahon, A.P. Shh signaling within the dental epithelium is necessary for cell proliferation, growth and polarization. Development 2002, 129, 5323–5337. [Google Scholar] [CrossRef] [Green Version]
- Bei, M.; Stowell, S.; Maas, R. Msx2 controls ameloblast terminal differentiation. Dev. Dyn. 2004, 231, 758–765. [Google Scholar] [CrossRef]
- Wang, X.-P.; Suomalainen, M.; Jorgez, C.J.; Matzuk, M.M.; Werner, S.; Thesleff, I. Follistatin regulates enamel patterning in mouse incisors by asymmetrically inhibiting BMP signaling and ameloblast differentiation. Dev. Cell 2004, 7, 719–730. [Google Scholar] [CrossRef] [Green Version]
- Xie, X.; Liu, C.; Zhang, H.; Jani, P.H.; Lu, Y.; Wang, X.; Zhang, B.; Qin, C. Abrogation of epithelial BMP2 and BMP4 causes amelogenesis imperfecta by reducing MMP20 and KLK4 expression. Sci. Rep. 2016, 6, 25364. [Google Scholar] [CrossRef] [Green Version]
- Chu, Q.; Gao, Y.; Gao, X.; Dong, Z.; Song, W.; Xu, Z.; Xiang, L.; Wang, Y.; Zhang, L.; Li, M.; et al. Ablation of Runx2 in ameloblasts suppresses enamel maturation in tooth development. Sci. Rep. 2018, 8, 9594. [Google Scholar] [CrossRef] [Green Version]
- Katagiri, T.; Watanabe, T. Bone morphogenetic proteins. Cold Spring Harb. Perspect. Biol. 2016, 8, a021899. [Google Scholar] [CrossRef] [Green Version]
- Gomez-Puerto, M.C.; Lyengar, P.V.; García de Vinuesa, A.; ten Dijke, P.; Sanchez-Duffhues, G. Bone morphogenetic protein receptor signal transduction in human disease. J. Pathol. 2019, 247, 9–20. [Google Scholar] [CrossRef]
- Zhao, G.-Q. Consequences of knocking out BMP signaling in the mouse. Genesis 2003, 35, 43–56. [Google Scholar] [CrossRef] [PubMed]
- Åberg, T.; Wozney, J.; Thesleff, I. Expression patterns of Bone morphogenetic proteins (Bmps) in the developing mouse tooth suggest roles in morphogenesis and differentiation. Dev. Dyn. 1997, 210, 383–396. [Google Scholar] [CrossRef]
- Yamashiro, T.; Tummers, M.; Thesleff, I. Expression of Bone morphogenetic protein and Msx genes during root formation. J. Dent. Res. 2003, 82, 172–176. [Google Scholar] [CrossRef]
- Bitgood, M.J.; McMahon, A.P. Hedgehog and Bmp genes are coexpressed at many sites of cell-cell interaction in the mouse embryo. Dev. Biol. 1995, 172, 126–138. [Google Scholar] [CrossRef] [Green Version]
- Koyama, E.; Yamaai, T.; Iseki, S.; Ohuchi, H.; Nohno, T.; Yoshioka, H.; Hayashi, Y.; Leatherman, J.L.; Golden, E.B.; Noji, S.; et al. Polarizing activity, Sonic hedgehog, and tooth development in embryonic and postnatal mouse. Dev. Dyn. 1996, 206, 59–72. [Google Scholar] [CrossRef]
- Iseki, S.; Araga, A.; Ohuchi, H.; Nohno, T.; Yoshioka, H.; Hayashi, F.; Noji, S. Sonic hedgehog is expressed in epithelial cells during development of whisker, hair, and tooth. Biochem. Biophys. Res. Commun. 1996, 218, 688–693. [Google Scholar] [CrossRef]
- Jernvall, J.; Thesleff, I. Reiterative signaling and patterning during mammalian tooth morphogenesis. Mech. Dev. 2000, 92, 19–29. [Google Scholar] [CrossRef]
- Gritli-Linde, A.; Lewis, P.; McMahon, A.P.; Linde, A. The whereabouts of a morphogen: Direct evidence for short- and graded long-range activity of Hedgehog signaling peptides. Dev. Biol. 2001, 236, 364–386. [Google Scholar] [CrossRef]
- Lin, H.M.; Nakamura, H.; Noda, T.; Ozawa, H. Localization of H+-ATPAse and carbonic anhydrase II in ameloblasts at maturation. Calcif. Tissue Int. 1994, 55, 38–45. [Google Scholar] [CrossRef]
- Toyosawa, S.; Ogawa, Y.; Inagaki, T.; Ijuhin, N. Immunohistochemical localization of carbonic anhydrase isoenzyme II in rat incisor epithelial cells at various stages of amelogenesis. Cell Tissue Res. 1996, 285, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.E.; Nanci, A.; Moffat, P. Evidence by signal peptide trap technology for the expression of carbonic anhydrase 6 in rat incisor enamel organs. Eur. J. Oral Sci. 2006, 114 (Suppl. 1), 147–153. [Google Scholar] [CrossRef]
- Josephsen, K.; Takano, Y.; Frische, S.; Praetorius, J.; Nielsen, L.; Aoba, T.; Fejerkov, O. Ion transporters in secretory and cyclically modulating ameloblasts: A new hypothesis for cellular control of preeruptive enamel maturation. Am. J. Physiol. Cell Physiol. 2010, 299, Cl299–Cl307. [Google Scholar] [CrossRef] [Green Version]
- Reibring, C.-G.; El Shahawy, M.; Hallberg, K.; Kannius-Janson, M.; Nilsson, J.; Parkkila, S.; Sly, W.S.; Waheed, A.; Linde, A.; Gritli-Linde, A. Expression patterns and subcellular localization of carbonic anhydrases are developmentally regulated during tooth formation. PLoS ONE 2014, 9, e96007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cook, N.R.; Row, P.E.; Davidson, H.W. Lysosome associated membrane protein 1 (Lamp1) traffics directly from the TGN to early endosomes. Traffic 2004, 5, 685–699. [Google Scholar] [CrossRef] [PubMed]
- Wartosch, L.; Bright, N.A.; Luzio, J.P. Lysosomes. Curr. Biol. 2015, 25, R315–R316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shearer, L.S.; Petersen, N.O. Distribution and co-localization of endosome markers in cells. Helyon 2019, 5, e02375. [Google Scholar] [CrossRef] [Green Version]
- Michie, K.A.; Bermeister, A.; Robertson, N.O.; Goodchild, S.C.; Curmi, P.M.G. Two sides of the coin: Ezrin/Radixin/Moesin and Merlin control membrane structure and contact inhibition. Int. J. Mol. Sci. 2019, 20, 1996. [Google Scholar] [CrossRef] [Green Version]
- Niesch, A.L.; Fehon, R.G. Ezrin, Eadixin, Moesin: Key regulators of membrane-cortex interactions and signaling. Curr. Opin. Cell Biol. 2011, 23, 377–382. [Google Scholar] [CrossRef] [Green Version]
- Jian, L.; Phang, J.M.; Yu, J.; Harrop, S.J.; Sokolova, A.V.; Duff, A.P.; Wilk, K.E.; Alkhamici, H.; Breit, S.N.; Valenzuela, S.M.; et al. CLIC proteins, ezrin, radixin, moesin and the coupling of membranes to the actin cytoskeleton: A smoking gun? Biochim. Biophys. Acta 2014, 1838, 643–657. [Google Scholar] [CrossRef] [Green Version]
- Kawaguchi, K.; Yoshida, S.; Hatano, R.; Asano, S. Pathophysiological roles of Ezrin/Radixin/Moesin. Biol. Pharm. Bull. 2017, 40, 381–390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamura, H.; Ozawa, H. Immunolocalization of CD44 and the Ezrin-Radixin-Moesin (ERM) family in the stratum intermedium and papillary layer of the mouse enamel organ. J. Histochem. Cytochem. 1997, 45, 1481–1492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsukita, S.; Furuse, M.; Itoh, M. Structural and signaling molecules come together at tight junctions. Curr. Opin. Cell Biol. 1999, 11, 628–633. [Google Scholar] [CrossRef]
- Vasioukhin, V.; Fuchs, E. Actin dynamics and cell-cell adhesion in epithelia. Curr. Opin. Cell Biol. 2001, 13, 76–84. [Google Scholar] [CrossRef]
- Inai, T.; Sengoku, A.; Hirose, E.; Iida, H.; Shibata, Y. Differentiatil expression of the tight junction proteins, Claudin-1, Claudin-4, Occludin, ZO-1, and PAR3, in the ameloblasts of rat upper incisor. Anat. Rec. 2008, 291, 577–585. [Google Scholar] [CrossRef]
- Takano, Y.; Ozawa, H. Ultrastructural and cytochemical observations on the alternating morphologic changes of the ameloblasts at the stage of enamel maturation. Arch. Histol. Jpn. 1980, 43, 385–399. [Google Scholar] [CrossRef] [Green Version]
- Narisawa, S.; Fröhlander, N.; Millán, J.L. Inactivation of two mouse alkaline phosphatase genes and establishment of a model of infantile hypophosphatasia. Dev. Dyn. 1997, 208, 432–446. [Google Scholar] [CrossRef]
- Fedde, K.N.; Blair, L.; Silverstein, J.; Coburn, S.P.; Ryan, L.M.; Weinstein, R.S.; Waymire, K.; Narisawa, S.; Millán, J.L.; MacGregor, G.R.; et al. Alkaline phosphatase knockout mice recapitulate the metabolic and skeletal defects of infantile hypophosphatasia. J. Bone Miner. Res. 1999, 14, 2015–2026. [Google Scholar] [CrossRef] [Green Version]
- Hessle, L.; Johnson, K.A.; Anderson, H.C.; Narisawa, S.; Sali, A.; Goding, J.W.; Terkeltaub, R.; Millan, J.L. Tissue-nonspecific alkaline phosphatase and plasma cell membrane glycoprotein-1 are central antagonistic regulators of bone mineralization. Proc. Natl. Acad. Sci. USA 2002, 99, 9445–9449. [Google Scholar] [CrossRef] [Green Version]
- Waymire, K.G.; Mahuren, J.D.; Jaje, J.M.; Guilarte, T.R.; Coburn, S.P.; MacGregor, G.R. Mice lacking tissue non-specific alkaline phosphatase die from seizures due to defective metabolism of vitamin B-6. Nat. Genet. 1995, 11, 45–51. [Google Scholar] [CrossRef]
- Beertsen, W.T.; Vandenbos, T.; Everts, V. Root development in mice lacking functional tissue non-specific alkaline phosphatase gene: Inhibition of acellular cementum formation. J. Dent. Res. 1999, 78, 1221–1229. [Google Scholar] [CrossRef] [PubMed]
- Yadav, M.C.; Cardoso de Oliveira, R.; Foster, B.L.; Fong, H.; Cory, E.; Narisawa, S.; Sah, R.L.; Somerman, M.; Whyte, M.P.; Millán, J.L. Enzyme replacement prevents enamel defects in hypophosphatasia mice. J. Bone Min. Res. 2012, 27, 1722–1734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gasque, K.C.S.; Foster, B.L.; Kuss, P.; Yadav, M.C.; Liu, J.; Kiffer-Moreira, T.; van Elsas, A.; Hatch, N.; Somerman, M.J.; Millán, J.L. Improvement of the skeletal and dental hypophosphatasia phenotype in Apl-/- mice by administration of soluble (non-targeted) chimeric alkaline phosphatase. Bone 2015, 72, 137–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takuwa, Y.; Ohse, C.; Wang, E.A.; Wosney, J.M.; Yamashita, K. Bone morphogenetic protein-2 stimulates alkaline phosphatase activity and collagen synthesis is cultured osteoblastic cells, MC3T3-E1. Biochem. Biophys. Res. Commun. 1991, 174, 96–101. [Google Scholar] [CrossRef]
- Symons, N.B.B. Alkaline phosphatase activity in the developing teeth of the rat. J. Anat. 1955, 89, 238–245. [Google Scholar]
- Tenorio, D.; Germain, J.P.; Hughes, F.J. Histochemical studies of acid and alkaline phosphatases in rat tooth germs with undecalcified resin-embedded specimens. J. Histochem. Cytochem. 1992, 40, 1229–1233. [Google Scholar] [CrossRef] [Green Version]
- Gomez, S.; Boyde, A. Correlated alkaline phosphatase histochemistry and quantitative backscattered electron imaging in the study of rat incisor ameloblasts and enamel mineralization. Microsc. Res. Tech. 1994, 29, 29–36. [Google Scholar] [CrossRef]
- Hotton, D.; Mauro, N.; Lezot, F.; Forest, N.; Berdal, A. Differential expression and activity of tissue non-specific alkaline phosphatase (TNAP) in rat odontogenic cells in vivo. J. Histochem. Cytochem. 1999, 47, 1541–1552. [Google Scholar] [CrossRef] [Green Version]
- McKee, M.D.; Yadav, M.C.; Foster, B.L.; Somerman, M.J.; Farquharson, C.; Millán, J.L. Compounded PHOSPHO1/ALPL deficiencies reduce dentin mineralization. J. Dent. Res. 2013, 92, 721–727. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Yan, X.; Pandya, M.; Luan, X.; Diekwisch, T.G.H. Daughters of the enamel organ: Development, fate, and function of the stratum intermedium, stellate reticulum, and outer enamel epithelium. Stem Cells Dev. 2016, 25, 1580–1590. [Google Scholar] [CrossRef] [Green Version]
- Izumi, M.; Fujio, Y.; Kunisada, K.; Negoro, S.; Tone, E.; Funamoto, M.; Osugi, T.; Oshima, Y.; Nakaoka, Y.; Kishimoto, T.; et al. Bone morphogenetic protein-2 inhibits serum deprivation-induced apoptosis of neonatal cardiac myocytes through activation of the Smad1 pathway. J. Biol. Chem. 2001, 276, 31133–31141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashique, A.M.; Fu, K.; Richman, J.M. Endogenous bone morphogenetic proteins regulate outgrowth and epithelial survival during avian lip fusion. Development 2002, 129, 4647–4660. [Google Scholar] [CrossRef] [PubMed]
- Yoon, B.S.; Ovchinnikov, D.A.; Yoshiii, I.; Mishina, Y.; Behringer, R.R.; Lyons, K.M. Bmpr1 and Bmpr2 have overlapping functions and are essential for chondrogenesis in vivo. Proc. Natl. Acad. Sci. USA 2005, 102, 5062–5067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaltcheva, M.M.; Anderson, M.J.; Harfe, B.D.; Lewandoski, M. BMPs are direct triggers of intedigital programmed cell death. Dev. Biol. 2016, 411, 266–276. [Google Scholar] [CrossRef] [PubMed]
- Andl, T.; Ahn, K.; Kairo, A.; Chu, E.Y.; Wine-Lee, L.; Reddy, S.T.; Croft, N.T.; Cebra-Thoma, J.A.; Metzger, D.; Chambon, P.; et al. Epithelial Bmpr1a regulates differentiation and proliferation in postnatal hair follicles and is essential for tooth development. Development 2004, 131, 2257–2268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Lin, M.; Wang, Y.; Cserjesi, P.; Chen, Z.; Chen, Y.P. Bmpr1a is required in mesenchymal tissue and has limited redundant function with Bmpr1b in tooth and palate development. Dev. Biol. 2011, 349, 451–461. [Google Scholar] [CrossRef] [Green Version]
- Jia, S.; Zhou, J.; Gao, Y.; Baek, J.A.; Martin, J.F.; Lan, Y.; Jiang, R. Roles of Bmp4 during tooth morphogenesis and sequential tooth formation. Development 2013, 140, 423–432. [Google Scholar] [CrossRef] [Green Version]
- Omi, M.; Kulkarni, A.K.; Raichur, A.; Fox, M.; Uptergrove, A.; Zhang, H.; Mishina, Y. BMP-Smad signaling regulates postnatal crown dentinogenesis in mouse molar. J. BMR. Plus 2019, 4, e10249. [Google Scholar] [CrossRef] [Green Version]
- Gluhak-Heinrich, J.; Guo, D.; Yang, W.; Harris, M.A.; Lichter, A.; Kream, B.; Zhang, J.; Feng, J.Q.; Harris, S.E. New roles and mechanism of action of BMP4 in post-natal tooth differentiation. Bone 2010, 46, 1533–1545. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.; Harris, M.A.; Cui, Y.; Mishina, Y.; Gluhak-Heinrisch, J. Bmp2 is required for odontoblast differentiation and pulp vasculogenesis. J. Dent. Res. 2012, 91, 58–64. [Google Scholar] [CrossRef]
- Jani, P.; Liu, C.; Zhang, H.; Younes, K.; Benson, D.M. The role of Bone morphogenetic proteins 2 and 4 in mouse dentinogenesis. Arch. Oral. Biol. 2018, 90, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Plikus, M.V.; Zeichner-David, M.; Mayer, J.A.; Reyna, J.; Bringas, P.; Thewissen, J.G.M.; Snead, M.L.; Chai, Y.; Chuong, C.-M. Morphoregulation of teeth: Modulating the number, size, shape and differentiation by tuning Bmp activity. Evo. Devo. 2005, 7, 440–457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Li, L.; Zheng, Y.; Yuan, G.; Yang, G.; He, F.; Chen, Y. BMP activity is required for tooth development from the lamina to the bud stage. J. Dent. Res. 2012, 9, 690–695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coin, R.; Haikel, Y.; Ruch, J.V. Effects of apatite, transforming growth factor beta-1, bone morphogeneric protein-2 and interleukin-7 on ameloblast differentiation in vitro. Eur. J. Oral Sci. 1999, 107, 487–495. [Google Scholar] [CrossRef] [PubMed]
- Simmer, J.P.; Papagerakis, P.; Smith, C.E.; Fisher, D.C.; Rountrey, A.N.; Zheng, L.; Hu, J.C.-C. Regulation of dental enamel shape and hardness. J. Dent. Res. 2010, 89, 1024–1038. [Google Scholar] [CrossRef] [PubMed]
- Azpiazu, D.; Gonzalo, S.; Villa-Bellosta, R. Tissue non-specific alkaline phosphatase and vascular calcification: A potential therapeutic target. Curr. Cardiol. Rev. 2019, 15, 91–95. [Google Scholar] [CrossRef]
- Villa-Suárez, J.M.; García-Fontana, C.; Andujar-Vera, F.; González-Salvatierra, S.; de Haro-Muñoz, T.; Contreras-Bolívar, V.; García-Fontana, B.; Muñoz-Torres, M. Hypophosphatasia: A unique disorder of bone mineralization. Int. J. Mol. Sci. 2021, 22, 4303. [Google Scholar] [CrossRef]
- Yoshioka, T.; Inomata, K. Cytochemical localization of alkaline phosphatase in the spinal cord and ependyma of newborn and adult rats. Acta Histochem. Cytochem. 1983, 16, 77–83. [Google Scholar] [CrossRef] [Green Version]
- Nagata, A.; Komoda, T.; Sakagishi, Y. Relationship between the uptake of calcium or phosphorus and alkaline phosphatase activity induced by certain modulators in rat organs. Calcif. Tissue Int. 1989, 45, 173–181. [Google Scholar] [CrossRef]
- Angelov, D.N. Distribution of activity of alkaline phosphatase and Mg-dependent adenosine triphosphatase in the cranial dura mater-arachnoid interface zone of the rat. Cell Tissue Res. 1990, 260, 595–600. [Google Scholar] [CrossRef]
- Hui, M.; Hu, M.; Tenenbaum, H.C. Changes in cell adhesion and cell proliferation associated with expression of tissue non-specific alkaline phosphatase. Cell Tissue Res. 1993, 274, 429–437. [Google Scholar] [CrossRef] [PubMed]
- Fraser, D. Hypophosphatasia. Am. J. Med. 1957, 22, 730–746. [Google Scholar] [CrossRef]
- Whyte, M.P. Hypophosphatasia and the role of alkaline phosphatase in skeletal mineralization. Endocr. Rev. 1994, 15, 439–461. [Google Scholar] [PubMed]
- Hu, J.C.-C.; Plaetke, R.; Mornet, E.; Zhang, C.; Sun, X.; Thomas, H.F.; Simmer, J.P. Characterization of a family with dominant hypophosphatasia. Eur. J. Oral Sci. 2000, 108, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Gibson, C.W.; Yuan, Z.A.; Li, Y.; Daly, B.; Suggs, C.; Aragon, M.A.; Alawi, F.; Kulkarni, A.B.; Wright, J.T. Transgenic mice that express normal and mutated Amelogenins. J. Dent. Res. 2007, 86, 331–335. [Google Scholar] [CrossRef] [PubMed]
- Gibson, M.; Perrimon, N. Extrusion and death of DPP/BMP-compromised epithelial cells in the developing Drosophila wing. Science 2005, 307, 1785–1789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, J.; Dahmann, C. Extrusion of cells with inappropriate Dpp signaling from Drosophila wing disc epithelia. Science 2005, 307, 1789–1790. [Google Scholar] [CrossRef] [Green Version]
- Moe, H. Physiological cell death of secretory ameloblasts in the rat incisor. Cell Tissue Res. 1979, 197, 443–451. [Google Scholar] [CrossRef]
- Wakita, M.; Hinrichsen, K. Ultrastructure of the ameloblast-stratum intermedium border during ameloblast differentiation. Acta Anat. 1980, 108, 10–29. [Google Scholar] [CrossRef]
- Barron, M.J.; Brookes, S.J.; Draper, C.E.; Garrod, D.; Kirkham, J.; Shore, R.C.; Dixon, M.J. The cell adhesion molecule nectin-1 is critical for normal enamel formation in mice. Hum. Mol. Genet. 2008, 17, 3509–3520. [Google Scholar] [CrossRef] [Green Version]
- Satokata, I.; Ma, L.; Ohshima, H.; Bei, M.; Woo, I.; Nishizawa, K.; Maeda, T.; Takano, Y.; Uchiyama, M.; Heany, S.; et al. Msx2 deficiency in mice causes pleiotropic defects in bone growth and ectodermal organ formation. Nat. Genet. 2000, 24, 391–395. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Miyoshi, J.; Takai, Y.; Thesleff, I. Cooperation of Nectin-1 and Nectin-3 is required for normal ameloblast function and crown shape development in mouse teeth. Dev. Dyn. 2010, 239, 2558–2569. [Google Scholar] [CrossRef] [PubMed]
- Harfe, B.D.; Scherz, P.J.; Nissim, S.; Tian, H.; McMahon, A.P.; Tabin, C. Evidence for expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell 2004, 118, 517–528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bandyopadhyay, A.; Tsuji, K.; Cox, K.; Harfe, B.D.; Rosen, V.; Tabin, C.J. Genetic analysis of the roles of BMP2, BMP4, and BMP7 in limb patterning and skeletogenesis. PLoS Genet. 2006, 12, e216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsuji, K.; Bandyopadhyay, A.; Harfe, B.D.; Cox, K.; Kakar, S.; Gerstenfeld, L.; Einhorn, T.; Tabin, C.J.; Rosen, V. BMP2 activity, although dispensable for bone formation, is required for the initiation of fracture healing. Nat. Genet. 2006, 38, 1424–1429. [Google Scholar] [CrossRef]
- Selever, J.; Liu, W.; Lu, M.F.; Behringer, R.R.; Martin, J.F. Bmp4 in limb bud mesoderm regulates digit pattern by controlling AER development. Dev. Biol. 2004, 276, 268–279. [Google Scholar] [CrossRef]
- Liu, W.; Selever, J.; Wang, D.; Lu, M.F.; Moses, K.A.; Schwartz, R.J.; Martin, J.F. Bmp4 signaling is required for outflow-tract septation and branchial-arch artery remodelling. Proc. Natl. Acad. Sci. USA 2004, 101, 4489–4494. [Google Scholar] [CrossRef] [Green Version]
- Soriano, P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999, 21, 70–71. [Google Scholar] [CrossRef]
- El Shahawy, M.; Reibring, C.-G.; Neben, C.L.; Hallberg, K.; Marangoni, P.; Harfe, B.D.; Klein, O.D.; Linde, A.; Gritli-Linde, A. Cell fate specification in the lingual epithelium is controlled by antagonistic activities of Sonic hedgehog and retinoic acid. PLoS Genet. 2017, 13, e1006914. [Google Scholar] [CrossRef] [Green Version]
- Reibring, C.G.; Hallberg, K.; Linde, A.; Gritli-Linde, A. Distinct and overlapping expression patterns of the Homer family of scaffolding proteins and their encoding genes in developing murine cephalic tissues. Int. J. Mol. Sci. 2020, 21, 1264. [Google Scholar] [CrossRef] [Green Version]
- Angerer, L.M.; Angerer, R.C. In situ hybridization to cellular RNA with radiolabelled RNA probes. In In Situ Hybridization. A Practical Approach; Wilkinson, D.G., Ed.; Oxford University Press: Oxford, UK, 1992; pp. 15–32. [Google Scholar]
- Cerny, R.; Slaby, I.; Hammarström, L.; Wurtz, T. A novel gene expressed in rat ameloblasts codes for proteins with cell binding domains. J. Bone Miner. Res. 1996, 11, 883–891. [Google Scholar] [CrossRef] [PubMed]
- Fong, C.D.; Cerny, R.; Hammarström, L.; Slaby, I. Sequential expression of an amelin gene in mesenchymal and epithelial cells during odontogenesis. Eur. J. Oral Sci. 1998, 106, 324–330. [Google Scholar] [CrossRef] [PubMed]
- Yamashiro, T.; Åberg, T.; Levanon, D.; Groner, Y.; Thesleff, I. Expression of Runx1, -2 and -3 during tooth, palate and craniofacial bone development. Mech. Dev. 2002, 119S, S107–S110. [Google Scholar] [CrossRef]
- Vaziri Sani, F.; Hallberg, K.; Harfe, B.D.; McMahon, A.P.; Linde, A.; Gritli-Linde, A. Fate-mapping of the epithelial seam during palatal fusion rules out epithelial-mesenchymal transformation. Dev. Biol. 2005, 285, 490–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reibring, C.-G.; El Shahawy, M.; Hallberg, K.; Harfe, B.D.; Linde, A.; Gritli-Linde, A. Loss of BMP2 and BMP4 Signaling in the Dental Epithelium Causes Defective Enamel Maturation and Aberrant Development of Ameloblasts. Int. J. Mol. Sci. 2022, 23, 6095. https://doi.org/10.3390/ijms23116095
Reibring C-G, El Shahawy M, Hallberg K, Harfe BD, Linde A, Gritli-Linde A. Loss of BMP2 and BMP4 Signaling in the Dental Epithelium Causes Defective Enamel Maturation and Aberrant Development of Ameloblasts. International Journal of Molecular Sciences. 2022; 23(11):6095. https://doi.org/10.3390/ijms23116095
Chicago/Turabian StyleReibring, Claes-Göran, Maha El Shahawy, Kristina Hallberg, Brian D. Harfe, Anders Linde, and Amel Gritli-Linde. 2022. "Loss of BMP2 and BMP4 Signaling in the Dental Epithelium Causes Defective Enamel Maturation and Aberrant Development of Ameloblasts" International Journal of Molecular Sciences 23, no. 11: 6095. https://doi.org/10.3390/ijms23116095





