Curvicollide D Isolated from the Fungus Amesia sp. Kills African Trypanosomes by Inhibiting Transcription
Abstract
:1. Introduction
2. Results
2.1. Isolation and Structural Characterization of Curvicollide D
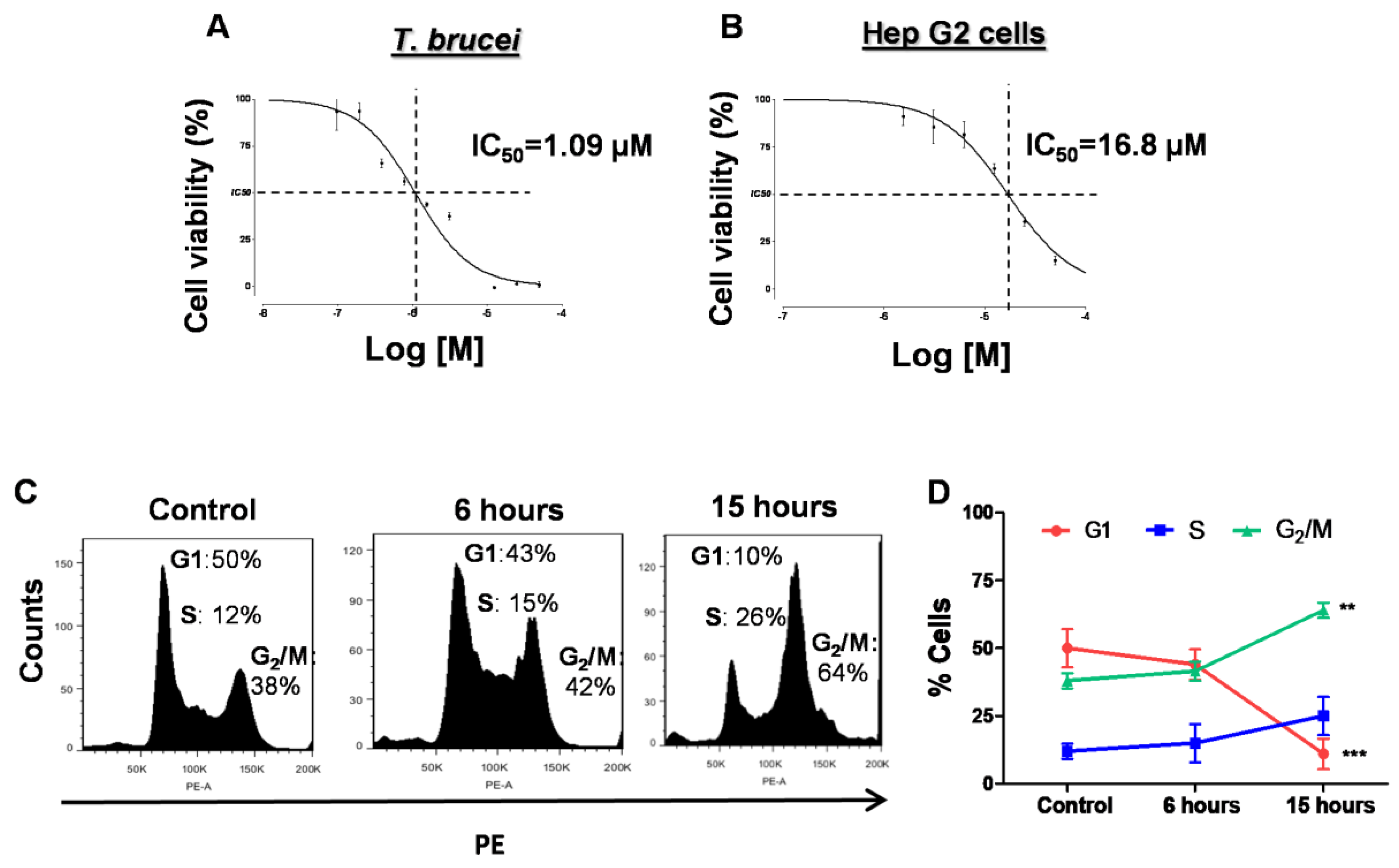
2.2. Curvicollide D Inhibits the Growth of T. brucei Bloodstream Forms and Causes Cell Cycle Arrest at G2/M Phase
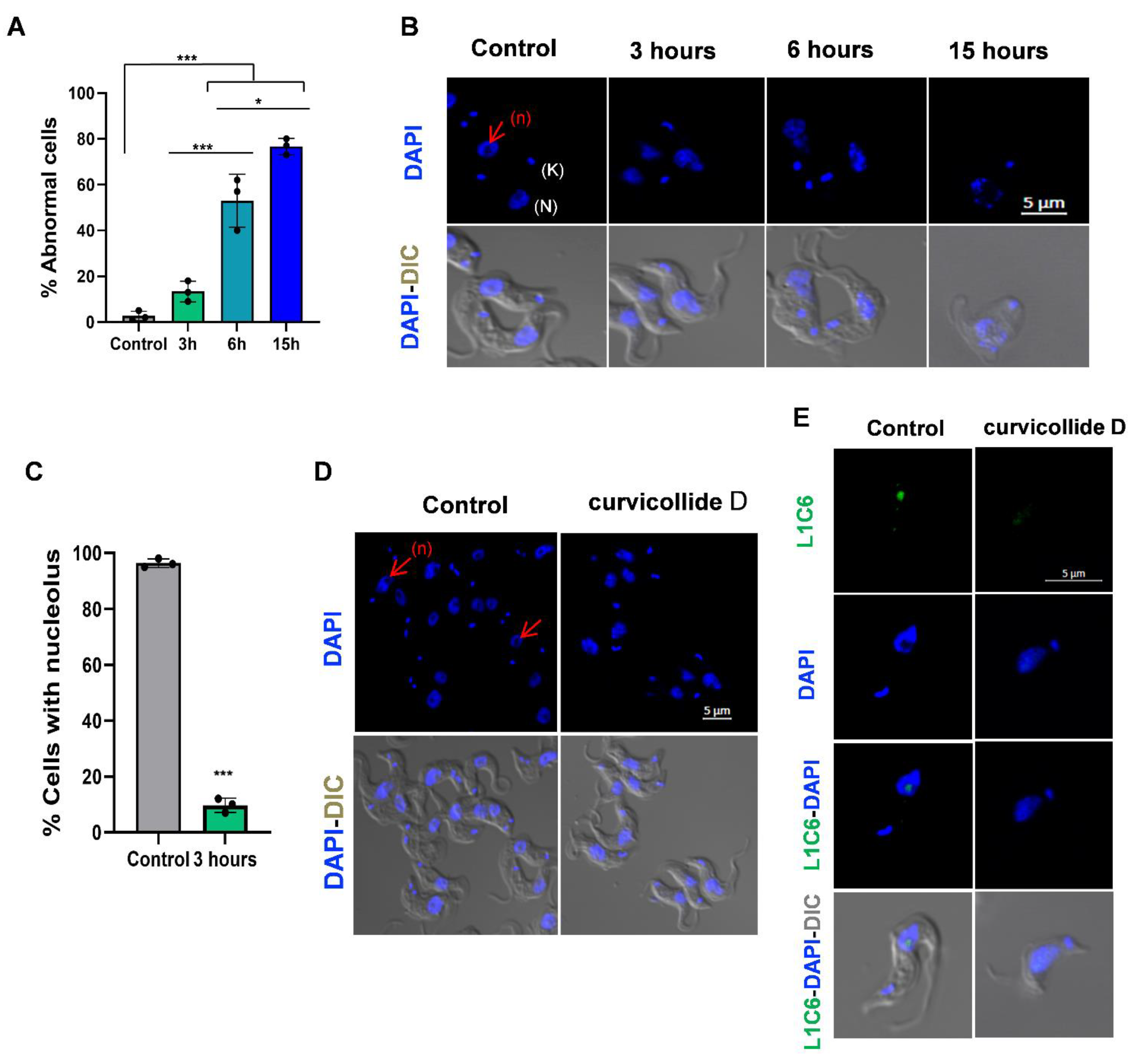
2.3. Curvicollide D Induces Morphological Changes and Nucleolus Disassembly in T. brucei
2.4. Curvicollide D Inhibits T. brucei RNA Polymerases I and II
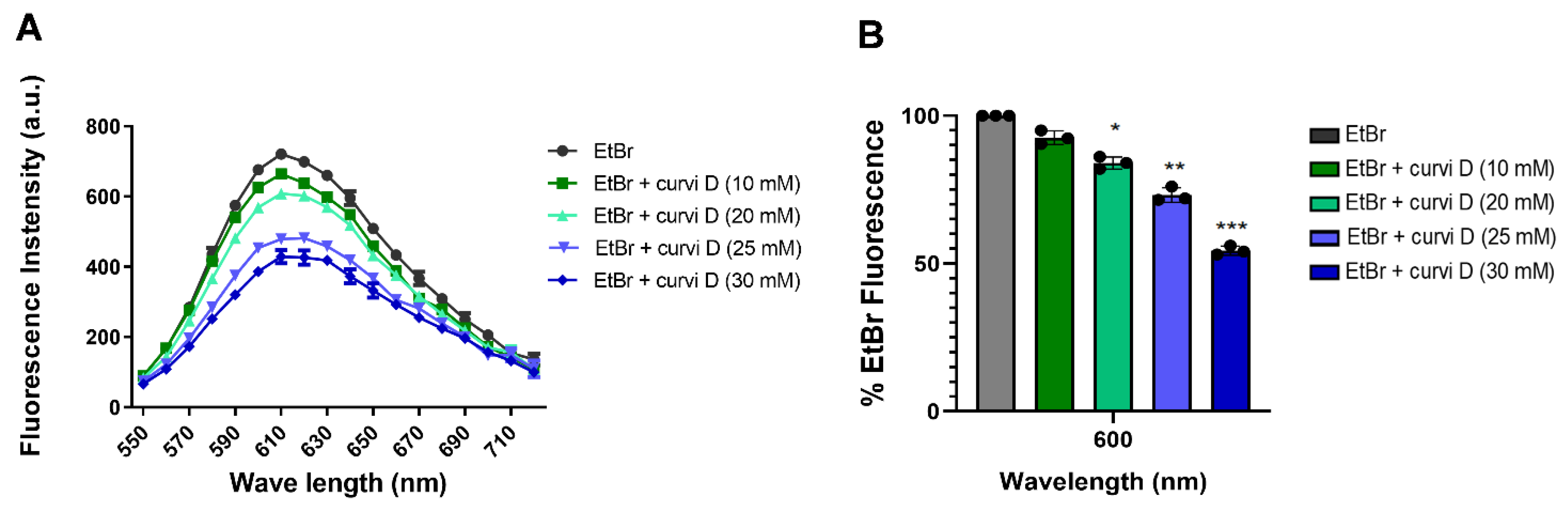
2.5. Curvicollide D Binds to Duplex DNA and Displaces Ethidium Bromide
3. Discussion
4. Materials and Methods
4.1. General Experimental Procedures
4.2. Isolation and Characterization of the Producing Strain
4.3. Fermentation Process
4.4. Isolation of Curvicollide D
4.5. Cell Culture
4.6. Resazurin-Based Assay
4.7. Analysis of Cell Cycle
4.8. Confocal Microscopy
4.9. RNA Isolation and Reverse Transcription
4.10. q-PCR Experiments
Primers Used for q-PCR
4.11. Fluorescence Intercalator Displacement Assay (FID)
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kennedy, P.G.E.; Rodgers, J. Clinical and Neuropathogenetic Aspects of Human African Trypanosomiasis. Front. Immunol. 2019, 10, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Available online: https://www.who.int/news-room/fact-sheets/detail/trypanosomiasis-human-african-(sleeping-sickness) (accessed on 25 April 2022).
- Yaro, M.; Munyard, K.A.; Stear, M.J.; Groth, D.M. Combatting African Animal Trypanosomiasis (AAT) in livestock: The potential role of trypanotolerance. Vet. Parasitol. 2016, 225, 43–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Babokhov, P.; Sanyaolu, A.O.; Oyibo, W.A.; Fagbenro-Beyioku, A.F.; Iriemenam, N.C. A current analysis of chemotherapy strategies for the treatment of human African trypanosomiasis. Pathog. Glob. Health 2013, 107, 242–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Salcedo, J.A.; Unciti-Broceta, J.D.; Valverde-Pozo, J.; Soriano, M. New Approaches to Overcome Transport Related Drug Resistance in Trypanosomatid Parasites. Front. Pharmacol. 2016, 7, 351. [Google Scholar] [CrossRef] [PubMed]
- Unciti-Broceta, J.D.; Arias, J.L.; Maceira, J.; Soriano, M.; Ortiz-González, M.; Hernández-Quero, J.; Muñóz-Torres, M.; de Koning, H.P.; Magez, S.; Garcia-Salcedo, J.A. Specific Cell Targeting Therapy Bypasses Drug Resistance Mechanisms in African Trypanosomiasis. PLoS Pathog. 2015, 11, e1004942. [Google Scholar] [CrossRef] [PubMed]
- Deeks, E.D. Fexinidazole: First Global Approval. Drugs 2019, 79, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Mesu, V.; Kalonji, W.M.; Bardonneau, C.; Mordt, O.V.; Blesson, S.; Simon, F.; Delhomme, S.; Bernhard, S.; Kuziena, W.; Lubaki, J.F.; et al. Oral fexinidazole for late-stage African Trypanosoma brucei gambiense trypanosomiasis: A pivotal multicentre, randomised, non-inferiority trial. Lancet 2018, 391, 144–154. [Google Scholar] [CrossRef]
- Lutje, V.; Probyn, K.; Seixas, J.; Bergman, H.; Villanueva, G. Chemotherapy for second-stage human African trypanosomiasis: Drugs in use. Cochrane Database Syst. Rev. 2021, 12, Cd015374. [Google Scholar] [CrossRef]
- EMA. Available online: https://www.ema.europa.eu/en/fexinidazole-winthrop-h-w-2320 (accessed on 26 April 2022).
- Annang, F.; Perez-Moreno, G.; Garcia-Hernandez, R.; Cordon-Obras, C.; Martin, J.; Tormo, J.R.; Rodriguez, L.; de Pedro, N.; Gomez-Perez, V.; Valente, M.; et al. High-throughput screening platform for natural product-based drug discovery against 3 neglected tropical diseases: Human African trypanosomiasis, leishmaniasis, and Chagas disease. J. Biomol. Screen. 2015, 20, 82–91. [Google Scholar] [CrossRef] [Green Version]
- Pena, I.; Pilar Manzano, M.; Cantizani, J.; Kessler, A.; Alonso-Padilla, J.; Bardera, A.I.; Alvarez, E.; Colmenarejo, G.; Cotillo, I.; Roquero, I.; et al. New compound sets identified from high throughput phenotypic screening against three kinetoplastid parasites: An open resource. Sci. Rep. 2015, 5, 8771. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, T.; Reker, D.; Schneider, P.; Schneider, G. Counting on natural products for drug design. Nat. Chem. 2016, 8, 531–541. [Google Scholar] [CrossRef] [PubMed]
- Stiles, M.; Winkler, R.R.; Chang, Y.-L.; Traynor, L. Stereochemical Assignments for β-Ketols Formed by Aldol Addition of Three Simple Ketones to p-Nitrobenzaldehyde. J. Am. Chem. Soc. 1964, 86, 3337–3342. [Google Scholar] [CrossRef]
- House, H.O.; Crumrine, D.S.; Teranishi, A.Y.; Olmstead, H.D. Chemistry of carbanions. XXIII. Use of metal complexes to control the aldol condensation. J. Am. Chem. Soc. 1973, 95, 3310–3324. [Google Scholar] [CrossRef]
- Heathcock, C.H.; Pirrung, M.C.; Sohn, J.E. Acyclic stereoselection. 4. Assignment of stereostructure to beta.-hydroxycarbonyl compounds by carbon-13 nuclear magnetic resonance. J. Org. Chem. 1979, 44, 4294–4299. [Google Scholar] [CrossRef]
- Che, Y.; Gloer, J.B.; Wicklow, D.T. Curvicollides A−C: New Polyketide-Derived Lactones from a Sclerotium-Colonizing Isolate of Podospora curvicolla (NRRL 25778). Org. Lett. 2004, 6, 1249–1252. [Google Scholar] [CrossRef]
- von Kiedrowski, V.; Quentin, F.; Hiersemann, M. Total Synthesis and Structural Assignment of Curvicollide C. Org. Lett. 2017, 19, 4391–4394. [Google Scholar] [CrossRef]
- Unciti-Broceta, J.D.; Maceira, J.; Morales, S.; García-Pérez, A.; Muñóz-Torres, M.E.; Garcia-Salcedo, J.A. Nicotinamide inhibits the lysosomal cathepsin b-like protease and kills African trypanosomes. J. Biol. Chem. 2013, 288, 10548–10557. [Google Scholar] [CrossRef] [Green Version]
- Devaux, S.; Kelly, S.; Lecordier, L.; Wickstead, B.; Perez-Morga, D.; Pays, E.; Vanhamme, L.; Gull, K. Diversification of function by different isoforms of conventionally shared RNA polymerase subunits. Mol. Biol. Cell 2007, 18, 1293–1301. [Google Scholar] [CrossRef] [Green Version]
- Boulon, S.; Westman, B.J.; Hutten, S.; Boisvert, F.M.; Lamond, A.I. The nucleolus under stress. Mol. Cell 2010, 40, 216–227. [Google Scholar] [CrossRef]
- Cestari, I.; Stuart, K. Transcriptional Regulation of Telomeric Expression Sites and Antigenic Variation in Trypanosomes. Curr. Genom. 2018, 19, 119–132. [Google Scholar] [CrossRef]
- Silva Pereira, S.; Jackson, A.P.; Figueiredo, L.M. Evolution of the variant surface glycoprotein family in African trypanosomes. Trends Parasitol. 2022, 38, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Michaeli, S. Trans-splicing in trypanosomes: Machinery and its impact on the parasite transcriptome. Future Microbiol. 2011, 6, 459–474. [Google Scholar] [CrossRef] [PubMed]
- Gilinger, G.; Bellofatto, V. Trypanosome spliced leader RNA genes contain the first identified RNA polymerase II gene promoter in these organisms. Nucleic Acids Res. 2001, 29, 1556–1564. [Google Scholar] [CrossRef] [Green Version]
- Günzl, A.; Ullu, E.; Dörner, M.; Fragoso, S.P.; Hoffmann, K.F.; Milner, J.D.; Morita, Y.; Nguu, E.K.; Vanacova, S.; Wünsch, S.; et al. Transcription of the Trypanosoma brucei spliced leader RNA gene is dependent only on the presence of upstream regulatory elements. Mol. Biochem. Parasitol. 1997, 85, 67–76. [Google Scholar] [CrossRef]
- Tse, W.C.; Boger, D.L. Sequence-selective DNA recognition: Natural products and nature’s lessons. Chem. Biol. 2004, 11, 1607–1617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewis, M.A.; Long, E.C. Fluorescent intercalator displacement analyses of DNA binding by the peptide-derived natural products netropsin, actinomycin, and bleomycin. Bioorganic Med. Chem. 2006, 14, 3481–3490. [Google Scholar] [CrossRef] [PubMed]
- Del Villar-Guerra, R.; Gray, R.D.; Trent, J.O.; Chaires, J.B. A rapid fluorescent indicator displacement assay and principal component/cluster data analysis for determination of ligand-nucleic acid structural selectivity. Nucleic Acids Res. 2018, 46, e41. [Google Scholar] [CrossRef] [Green Version]
- Sanchez-Martin, V.; Schneider, D.A.; Ortiz-Gonzalez, M.; Soriano-Lerma, A.; Linde-Rodriguez, A.; Perez-Carrasco, V.; Gutierrez-Fernandez, J.; Cuadros, M.; González, C.; Soriano, M.; et al. Targeting ribosomal G-quadruplexes with naphthalene-diimides as RNA polymerase I inhibitors for colorectal cancer treatment. Cell Chem. Biol. 2021, 28, 1590–1601.e4. [Google Scholar] [CrossRef]
- Firn, R.D.; Jones, C.G. Natural products—A simple model to explain chemical diversity. Nat. Prod. Rep. 2003, 20, 382–391. [Google Scholar] [CrossRef]
- Fleming, A. On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzae. Bull. World Health Organ. 2001, 79, 780–790. [Google Scholar] [CrossRef]
- Behie, S.W.; Bonet, B.; Zacharia, V.M.; McClung, D.J.; Traxler, M.F. Molecules to Ecosystems: Actinomycete Natural Products In Situ. Front. Microbiol. 2017, 7, 2149. [Google Scholar] [CrossRef] [Green Version]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro da Cunha, B.; Fonseca, L.P.; Calado, C.R.C. Antibiotic Discovery: Where Have We Come from, Where Do We Go? Antibiotics 2019, 8, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cragg, G.M.; Newman, D.J. Natural products: A continuing source of novel drug leads. Biochim. Biophys. Acta 2013, 6, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuma, A.A.; Santos, J.O.; Mendes, I.; de Souza, W.; Machado, C.R.; Motta, M.C.M. Chaetocin-A histone methyltransferase inhibitor-Impairs proliferation, arrests cell cycle and induces nucleolar disassembly in Trypanosoma cruzi. Acta Trop. 2017, 170, 149–160. [Google Scholar] [CrossRef]
- Vodnala, S.K.; Ferella, M.; Lundén-Miguel, H.; Betha, E.; van Reet, N.; Amin, D.N.; Oberg, B.; Andersson, B.; Kristensson, K.; Wigzell, H.; et al. Preclinical assessment of the treatment of second-stage African trypanosomiasis with cordycepin and deoxycoformycin. PLoS Negl. Trop. Dis. 2009, 3, e495. [Google Scholar] [CrossRef]
- DNDI. Available online: https://dndi.org/scientific-articles/2009/drug-screening-for-kinetoplastid-diseases-a-training-manual-for-screening-in-neglected-diseases/ (accessed on 25 April 2022).
- Mukherjee, A.; Sasikala, W.D. Drug-DNA intercalation: From discovery to the molecular mechanism. Adv. Protein Chem. Struct. Biol. 2013, 92, 1–62. [Google Scholar] [CrossRef]
- Figgitt, D.; Denny, W.; Chavalitshewinkoon, P.; Wilairat, P.; Ralph, R. In vitro study of anticancer acridines as potential antitrypanosomal and antimalarial agents. Antimicrob. Agents Chemother. 1992, 36, 1644–1647. [Google Scholar] [CrossRef] [Green Version]
- Roy Chowdhury, A.; Bakshi, R.; Wang, J.; Yildirir, G.; Liu, B.; Pappas-Brown, V.; Tolun, G.; Griffith, J.D.; Shapiro, T.A.; Jensen, R.E.; et al. The killing of African trypanosomes by ethidium bromide. PLoS Pathog. 2010, 6, e1001226. [Google Scholar] [CrossRef] [Green Version]
- da Silva Oliveira, G.L.; de Freitas, R.M. Diminazene aceturate--An antiparasitic drug of antiquity: Advances in pharmacology & therapeutics. Pharmacol. Res. 2015, 102, 138–157. [Google Scholar] [CrossRef]
- Wu, N.; Wu, X.W.; Agama, K.; Pommier, Y.; Du, J.; Li, D.; Gu, L.Q.; Huang, Z.S.; An, L.K. A novel DNA topoisomerase I inhibitor with different mechanism from camptothecin induces G2/M phase cell cycle arrest to K562 cells. Biochemistry 2010, 49, 10131–10136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schonn, I.; Hennesen, J.; Dartsch, D.C. Cellular responses to etoposide: Cell death despite cell cycle arrest and repair of DNA damage. Apoptosis 2010, 15, 162–172. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Lee, Y.S.; Kim, D.K. Doxorubicin exerts cytotoxic effects through cell cycle arrest and Fas-mediated cell death. Pharmacology 2009, 84, 300–309. [Google Scholar] [CrossRef] [PubMed]
- Staker, B.L.; Hjerrild, K.; Feese, M.D.; Behnke, C.A.; Burgin, A.B., Jr.; Stewart, L. The mechanism of topoisomerase I poisoning by a camptothecin analog. Proc. Natl. Acad. Sci. USA 2002, 99, 15387–15392. [Google Scholar] [CrossRef] [Green Version]
- Sheader, K.; Vaughan, S.; Minchin, J.; Hughes, K.; Gull, K.; Rudenko, G. Variant surface glycoprotein RNA interference triggers a precytokinesis cell cycle arrest in African trypanosomes. Proc. Natl. Acad. Sci. USA 2005, 102, 8716–8721. [Google Scholar] [CrossRef] [Green Version]
- Musso, L.; Mazzini, S.; Rossini, A.; Castagnoli, L.; Scaglioni, L.; Artali, R.; Di Nicola, M.; Zunino, F.; Dallavalle, S. c-MYC G-quadruplex binding by the RNA polymerase I inhibitor BMH-21 and analogues revealed by a combined NMR and biochemical Approach. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 615–629. [Google Scholar] [CrossRef]
- Drygin, D.; Siddiqui-Jain, A.; O’Brien, S.; Schwaebe, M.; Lin, A.; Bliesath, J.; Ho, C.B.; Proffitt, C.; Trent, K.; Whitten, J.P.; et al. Anticancer activity of CX-3543: A direct inhibitor of rRNA biogenesis. Cancer Res. 2009, 69, 7653–7661. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Di Antonio, M.; McKinney, S.; Mathew, V.; Ho, B.; O’Neil, N.J.; Santos, N.D.; Silvester, J.; Wei, V.; Garcia, J.; et al. CX-5461 is a DNA G-quadruplex stabilizer with selective lethality in BRCA1/2 deficient tumours. Nat. Commun. 2017, 8, 14432. [Google Scholar] [CrossRef]
- Kerry, L.E.; Pegg, E.E.; Cameron, D.P.; Budzak, J.; Poortinga, G.; Hannan, K.M.; Hannan, R.D.; Rudenko, G. Selective inhibition of RNA polymerase I transcription as a potential approach to treat African trypanosomiasis. PLoS Negl. Trop. Dis. 2017, 11, e0005432. [Google Scholar] [CrossRef]
- Bills, G.F.; Christensen, M.; Powell, M.; Thorn, G. Saprobic Soil Fungi; Elsevier Academic Press: Amsterdam, The Netherlands, 2004; pp. 271–302. [Google Scholar]
- Gonzalez-Menendez, V.; Martin, J.; Siles, J.A.; Gonzalez-Tejero, M.R.; Reyes, F.; Platas, G.; Tormo, J.R.; Genilloud, O. Biodiversity and chemotaxonomy of Preussia isolates from the Iberian Peninsula. Mycol. Prog. 2017, 16, 713–728. [Google Scholar] [CrossRef]
- NITE. Available online: http://www.nbrc.nite.go.jp/ (accessed on 25 April 2022).
- Blaxter, M.; Mann, J.; Chapman, T.; Thomas, F.; Whitton, C.; Floyd, R.; Abebe, E. Defining operational taxonomic units using DNA barcode data. Philos. Trans. R Soc. B Biol. Sci. 2005, 360, 1935–1943. [Google Scholar] [CrossRef] [PubMed]
- Peláez, F.; Cabello, A.; Platas, G.; Díez, M.T.; González del Val, A.; Basilio, A.; Martán, I.; Vicente, F.; Bills, G.E.; Giacobbe, R.A.; et al. The discovery of enfumafungin, a novel antifungal compound produced by an endophytic Hormonema species biological activity and taxonomy of the producing organisms. Syst. Appl. Microbiol. 2000, 23, 333–343. [Google Scholar] [CrossRef]
- del Castillo, T.; Marales-Sanfrutos, J.; Santoyo-González, F.; Magez, S.; Lopez-Jaramillo, F.J.; Garcia-Salcedo, J.A. Monovinyl sulfone β-cyclodextrin. A flexible drug carrier system. ChemMedChem 2014, 9, 383–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Salcedo, J.A.; Nolan, D.P.; Gijón, P.; Gómez-Rodriguez, J.; Pays, E. A protein kinase specifically associated with proliferative forms of Trypanosoma brucei is functionally related to a yeast kinase involved in the co-ordination of cell shape and division. Mol. Microbiol. 2002, 45, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Stanne, T.M.; Rudenko, G. Active VSG expression sites in Trypanosoma brucei are depleted of nucleosomes. Eukaryot. Cell 2010, 9, 136–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
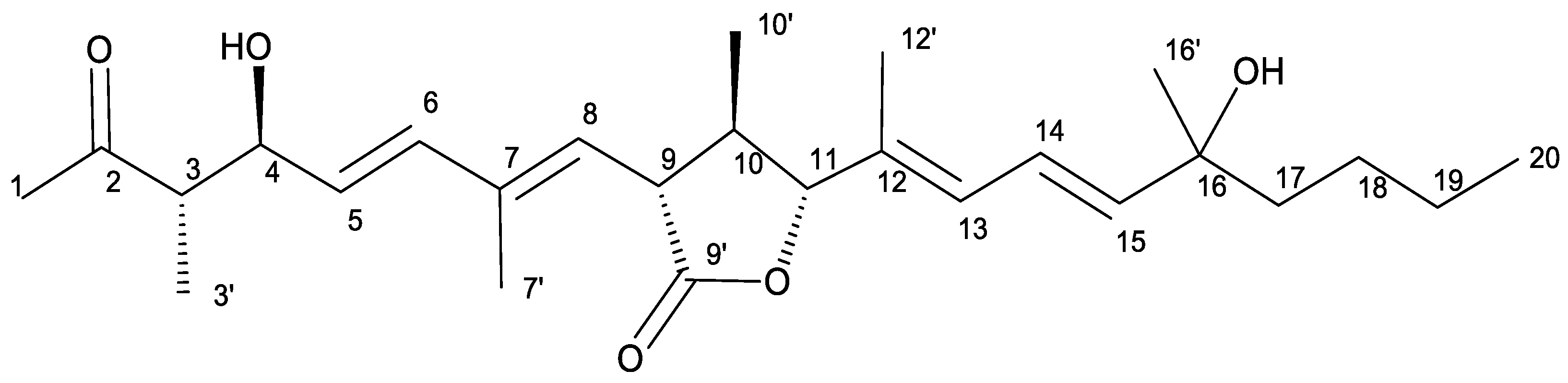
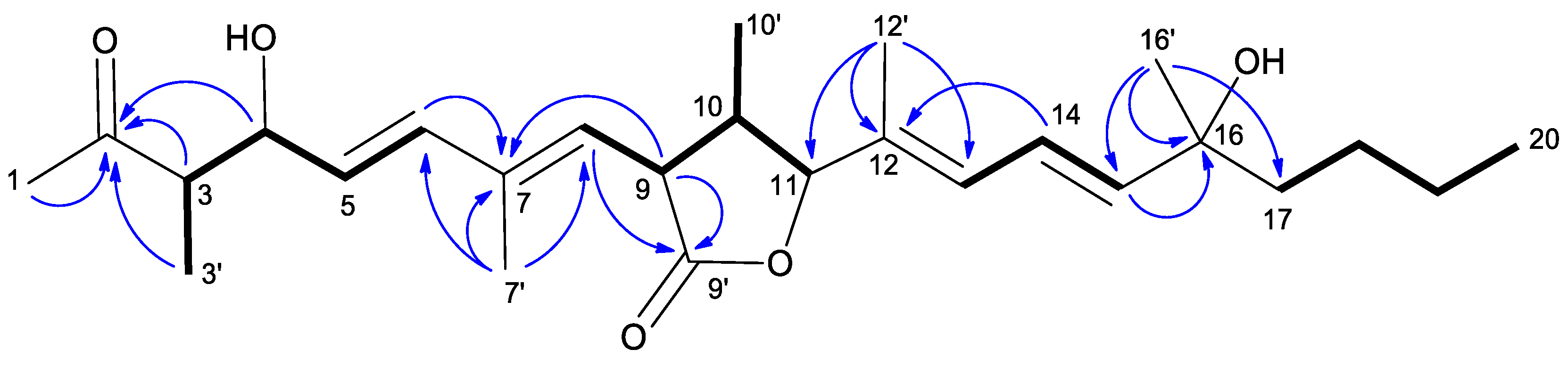
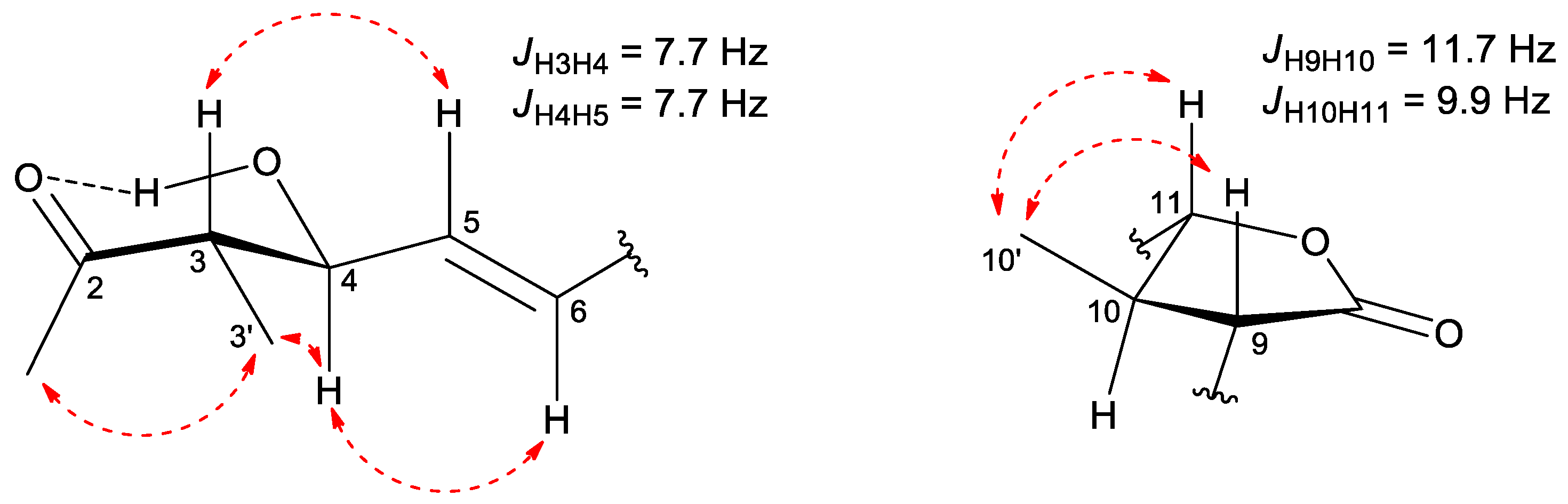

| Curvicollide D | ||
|---|---|---|
| Position | δC, Type | δH, mult. (J in Hz) |
| 1 | 29.8, CH3 | 2.22, s |
| 2 | 213.2, C | |
| 3 | 52.3, CH | 2.69, dq (7.7, 7.2) |
| 3′ | 14.0, CH3 | 1.10, d (7.2) |
| 4 | 75.1, CH | 4.25, dd (7.7, 7.7) |
| 5 | 129.0, CH | 5.65, dd (15.6, 7.7) |
| 6 | 136.4, CH | 6.32, d (15.6) |
| 7 | 138.5, C | |
| 7′ | 13.2, CH3 | 1.83, d (0.9) |
| 8 | 126.0, CH | 5.38, d (9.2) |
| 9 | 48.4, CH | 3.26, dd (11.7, 9.2) |
| 9′ | 176.2, C | |
| 10 | 42.7, CH | 2.18, m |
| 10′ | 14.8, CH3 | 1.06, d (6.6) |
| 11 | 90.4, CH | 4.37, d (9.9) |
| 12 | 131.1, C | |
| 12′ | 11.6, CH3 | 1.78, d (0.6) |
| 13 | 129.4, CH | 6.11, d (10.8) |
| 14 | 122.2, CH | 6.46, dd (15.3, 10.8) |
| 15 | 143.0, CH | 5.84, d (15.3) |
| 16 | 73.1, C | |
| 16′ | 28.0, CH3 | 1.32, s |
| 17 | 42.6, CH2 | 1.56, m |
| 18 | 26.2, CH2 | 1.30, m |
| 19 | 23.1, CH2 | 1.31, m |
| 20 | 14.0, CH3 | 0.90, t (6.7) |
| Forward Primer Sequence (5′ → 3′) | Reverse Primer Sequence (5′ → 3′) |
|---|---|
| (1) rDNAProm_938s: 5′-ATAAAAGGGAGTTATAGCGT-3′ | rDNAProm_1048as: 5′-GTACAACACAATCCGTTAAG-3′ [60] |
| (2) rDNASp 27s _27s: 5′-TATGTGTATGTGTGTTGTGTTA-3′ | rDNASp108as_108as: 5′-ATGCAAAATAGGAGACTACA-3′ [60] |
| (3) ESProm_330s: 5′-GAGATTGTGAGGGTTAGGA-3′ | ESProm_662as: 5′-CATCATCCTGCGTCGTTC-3′ [60] |
| (4) ES_6-F: 5′-GTACAAGCTACGAAAACGTG-3′ | ES_6-R: 5′-CCACTCCCACTGGAAACTTA-3′ |
| (5) SL_sp_1245s: 5′-CTTTGTTTCCCATAAGTCTAC-3′ | SL_sp_1306As: 5′-AGACACTTGCCATATTTTACT-3′ [60] |
| (6) SL_spacer_644s: 5′-GCAGCAATAACAGCGAGCATAC-3′ | SL_spacer_763As: 5′-GGACGGTTGAGCTGAGTGTAAT-3′ [60] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ortiz-Gonzalez, M.; Pérez-Victoria, I.; Ramirez-Macias, I.; de Pedro, N.; Linde-Rodriguez, A.; González-Menéndez, V.; Sanchez-Martin, V.; Martín, J.; Soriano-Lerma, A.; Genilloud, O.; et al. Curvicollide D Isolated from the Fungus Amesia sp. Kills African Trypanosomes by Inhibiting Transcription. Int. J. Mol. Sci. 2022, 23, 6107. https://doi.org/10.3390/ijms23116107
Ortiz-Gonzalez M, Pérez-Victoria I, Ramirez-Macias I, de Pedro N, Linde-Rodriguez A, González-Menéndez V, Sanchez-Martin V, Martín J, Soriano-Lerma A, Genilloud O, et al. Curvicollide D Isolated from the Fungus Amesia sp. Kills African Trypanosomes by Inhibiting Transcription. International Journal of Molecular Sciences. 2022; 23(11):6107. https://doi.org/10.3390/ijms23116107
Chicago/Turabian StyleOrtiz-Gonzalez, Matilde, Ignacio Pérez-Victoria, Inmaculada Ramirez-Macias, Nuria de Pedro, Angel Linde-Rodriguez, Víctor González-Menéndez, Victoria Sanchez-Martin, Jesús Martín, Ana Soriano-Lerma, Olga Genilloud, and et al. 2022. "Curvicollide D Isolated from the Fungus Amesia sp. Kills African Trypanosomes by Inhibiting Transcription" International Journal of Molecular Sciences 23, no. 11: 6107. https://doi.org/10.3390/ijms23116107









