Epigenetic Connection of the Calcitonin Gene-Related Peptide and Its Potential in Migraine
Abstract
:1. Introduction
2. Calcitonin Gene-Related Peptide—The Gene and the Protein
3. Calcitonin Gene-Related Peptide in Migraine
4. Epigenetic Connections of CGRP
4.1. CGRP and DNA Methylation
4.2. CGRP and Histone Modifications
4.3. CGRP and Non-Coding RNAs
4.3.1. Micro RNAs
4.3.2. Circular RNAs
4.3.3. Long Non-Coding RNAs
5. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tajti, J.; Szok, D.; Nyári, A.; Vécsei, L. CGRP and CGRP-Receptor as Targets of Migraine Therapy: Brain Prize-2021. CNS Neurol. Dis. Drug Targets 2022, 21, 460–478. [Google Scholar] [CrossRef] [PubMed]
- Vécsei, L.; Szok, D.; Csáti, A.; Tajti, J. CGRP antagonists and antibodies for the treatment of migraine. Expert Opin. Investig. Drugs 2015, 24, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Barbanti, P.; Aurilia, C.; Cevoli, S.; Egeo, G.; Fofi, L.; Messina, R.; Salerno, A.; Torelli, P.; Albanese, M.; Carnevale, A.; et al. Long-term (48 weeks) effectiveness, safety, and tolerability of erenumab in the prevention of high-frequency episodic and chronic migraine in a real world: Results of the EARLY 2 study. Headache 2021, 61, 1351–1363. [Google Scholar] [CrossRef] [PubMed]
- Barbanti, P.; Egeo, G.; Aurilia, C.; d’Onofrio, F.; Albanese, M.; Cetta, I.; Di Fiore, P.; Zucco, M.; Filippi, M.; Bono, F.; et al. Fremanezumab in the prevention of high-frequency episodic and chronic migraine: A 12-week, multicenter, real-life, cohort study (the FRIEND study). J. Headache Pain 2022, 23, 46. [Google Scholar] [CrossRef]
- Vernieri, F.; Altamura, C.; Brunelli, N.; Costa, C.M.; Aurilia, C.; Egeo, G.; Fofi, L.; Favoni, V.; Pierangeli, G.; Lovati, C.; et al. Galcanezumab for the prevention of high frequency episodic and chronic migraine in real life in Italy: A multicenter prospective cohort study (the GARLIT study). J. Headache Pain 2021, 22, 35. [Google Scholar] [CrossRef]
- Mahon, R.; Huels, J.; Hacking, V.; Cooney, P.; Danyliv, A.; Vudumula, U.; Vadapalle, S.; Vo, P.; Maniyar, F.H.; Palmer, S.; et al. Economic evaluations in migraine: Systematic literature review and a novel approach. J. Med. Econ. 2020, 23, 864–876. [Google Scholar] [CrossRef]
- Sutherland, H.G.; Albury, C.L.; Griffiths, L.R. Advances in genetics of migraine. J. Headache Pain 2019, 20, 72. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Eising, E.; Datson, N.; van den Maagdenberg, A.M.; Ferrari, M.D. Epigenetic mechanisms in migraine: A promising avenue? BMC Med. 2013, 11, 26. [Google Scholar] [CrossRef]
- Nattagh-Eshtivani, E.; Sani, M.A.; Dahri, M.; Ghalichi, F.; Ghavami, A.; Arjang, P.; Tarighat-Esfanjani, A. The role of nutrients in the pathogenesis and treatment of migraine headaches: Review. Biomed. Pharmacother. 2018, 102, 317–325. [Google Scholar] [CrossRef]
- Fila, M.; Chojnacki, C.; Chojnacki, J.; Blasiak, J. Is an “Epigenetic Diet” for Migraines Justified? The Case of Folate and DNA Methylation. Nutrients 2019, 11, 2763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danaher, R.J.; Zhang, L.; Donley, C.J.; Laungani, N.A.; Hui, S.E.; Miller, C.S.; Westlund, K.N. Histone deacetylase inhibitors prevent persistent hypersensitivity in an orofacial neuropathic pain model. Mol. Pain 2018, 14, 1744806918796763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Penas, C.; Navarro, X. Epigenetic Modifications Associated to Neuroinflammation and Neuropathic Pain After Neural Trauma. Front. Cell. Neurosci. 2018, 12, 158. [Google Scholar] [CrossRef] [Green Version]
- Park, K.Y.; Fletcher, J.R.; Raddant, A.C.; Russo, A.F. Epigenetic regulation of the calcitonin gene-related peptide gene in trigeminal glia. Cephalalgia Int. J. Headache 2011, 31, 614–624. [Google Scholar] [CrossRef] [Green Version]
- Hansen, R.R.; Vacca, V.; Pitcher, T.; Clark, A.K.; Malcangio, M. Role of extracellular calcitonin gene-related peptide in spinal cord mechanisms of cancer-induced bone pain. Pain 2016, 157, 666–676. [Google Scholar] [CrossRef] [Green Version]
- Priller, J.; Haas, C.A.; Reddington, M.; Kreutzberg, G.W. Calcitonin gene-related peptide and ATP induce immediate early gene expression in cultured rat microglial cells. Glia 1995, 15, 447–457. [Google Scholar] [CrossRef] [PubMed]
- Reddington, M.; Priller, J.; Treichel, J.; Haas, C.; Kreutzberg, G.W. Astrocytes and microglia as potential targets for calcitonin gene related peptide in the central nervous system. Can. J. Physiol. Pharmacol. 1995, 73, 1047–1049. [Google Scholar] [CrossRef]
- Guo, X.; Chen, D.; An, S.; Wang, Z. ChIP-seq Profiling Identifies Histone Deacetylase 2 Targeting Genes Involved in Immune and Inflammatory Regulation Induced by Calcitonin Gene-Related Peptide in Microglial Cells. J. Immunol. Res. 2020, 2020, 4384696. [Google Scholar] [CrossRef]
- Rosenfeld, M.G.; Amara, S.G.; Evans, R.M. Alternative RNA processing: Determining neuronal phenotype. Science 1984, 225, 1315–1320. [Google Scholar] [CrossRef]
- Amara, S.G.; Arriza, J.L.; Leff, S.E.; Swanson, L.W.; Evans, R.M.; Rosenfeld, M.G. Expression in brain of a messenger RNA encoding a novel neuropeptide homologous to calcitonin gene-related peptide. Science 1985, 229, 1094–1097. [Google Scholar] [CrossRef]
- Mulderry, P.K.; Ghatei, M.A.; Spokes, R.A.; Jones, P.M.; Pierson, A.M.; Hamid, Q.A.; Kanse, S.; Amara, S.G.; Burrin, J.M.; Legon, S.; et al. Differential expression of alpha-CGRP and beta-CGRP by primary sensory neurons and enteric autonomic neurons of the rat. Neuroscience 1988, 25, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Broad, P.M.; Symes, A.J.; Thakker, R.V.; Craig, R.K. Structure and methylation of the human calcitonin/alpha-CGRP gene. Nucleic Acids Res. 1989, 17, 6999–7011. [Google Scholar] [CrossRef]
- Park, K.Y.; Russo, A.F. Control of the calcitonin gene-related peptide enhancer by upstream stimulatory factor in trigeminal ganglion neurons. J. Biol. Chem. 2008, 283, 5441–5451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poyner, D.R.; Sexton, P.M.; Marshall, I.; Smith, D.M.; Quirion, R.; Born, W.; Muff, R.; Fischer, J.A.; Foord, S.M. International Union of Pharmacology. XXXII. The mammalian calcitonin gene-related peptides, adrenomedullin, amylin, and calcitonin receptors. Pharm. Rev. 2002, 54, 233–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walker, C.S.; Conner, A.C.; Poyner, D.R.; Hay, D.L. Regulation of signal transduction by calcitonin gene-related peptide receptors. Trends Pharmacol. Sci. 2010, 31, 476–483. [Google Scholar] [CrossRef] [PubMed]
- Sabharwal, R.; Mason, B.N.; Kuburas, A.; Abboud, F.M.; Russo, A.F.; Chapleau, M.W. Increased receptor activity-modifying protein 1 in the nervous system is sufficient to protect against autonomic dysregulation and hypertension. J. Cereb. Blood Flow Metab. 2019, 39, 690–703. [Google Scholar] [CrossRef]
- Kee, Z.; Kodji, X.; Brain, S.D. The Role of Calcitonin Gene Related Peptide (CGRP) in Neurogenic Vasodilation and Its Cardioprotective Effects. Front. Physiol. 2018, 9, 1249. [Google Scholar] [CrossRef] [Green Version]
- Ray, J.C.; Kapoor, M.; Stark, R.J.; Wang, S.J.; Bendtsen, L.; Matharu, M.; Hutton, E.J. Calcitonin gene related peptide in migraine: Current therapeutics, future implications and potential off-target effects. J. Neurol. Neurosurg. Psychiatry 2021, 92, 1325–1334. [Google Scholar] [CrossRef]
- Amara, S.G.; Evans, R.M.; Rosenfeld, M.G. Calcitonin/calcitonin gene-related peptide transcription unit: Tissue-specific expression involves selective use of alternative polyadenylation sites. Mol. Cell. Biol. 1984, 4, 2151–2160. [Google Scholar] [CrossRef]
- Coleman, T.P.; Roesser, J.R. RNA secondary structure: An important cis-element in rat calcitonin/CGRP pre-messenger RNA splicing. Biochemistry 1998, 37, 15941–15950. [Google Scholar] [CrossRef]
- Amara, S.G.; Jonas, V.; Rosenfeld, M.G.; Ong, E.S.; Evans, R.M. Alternative RNA processing in calcitonin gene expression generates mRNAs encoding different polypeptide products. Nature 1982, 298, 240–244. [Google Scholar] [CrossRef] [PubMed]
- Hillyard, C.J.; Myers, C.; Abeyasekera, G.; Stevvensvenson, J.C.; Craig, R.K.; MacIntyre, I. Katacalcin: A new plasma calcium-lowering hormone. Lancet 1983, 1, 846–848. [Google Scholar] [CrossRef]
- Giscard-Dartevelle, S.; Ghillani, P.; Taboulet, J.; Troalen, F.; Segond, N.; Lasmoles, F. Specific immunostaining for CCP II, a novel calcitonin carboxyl terminal peptide encoded by the calcitonin/CGRP gene. J. Histochem. Cytochem. 1993, 41, 1605–1610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breeze, A.L.; Harvey, T.S.; Bazzo, R.; Campbell, I.D. Solution structure of human calcitonin gene-related peptide by 1H NMR and distance geometry with restrained molecular dynamics. Biochemistry 1991, 30, 575–582. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, K.A.; Schmidt, R.; von Mentzer, B.; Haglund, U.; Roberts, E.; Walpole, C. Turn structures in CGRP C-terminal analogues promote stable arrangements of key residue side chains. Biochemistry 2001, 40, 8317–8325. [Google Scholar] [CrossRef]
- Wattiez, A.S.; Sowers, L.P.; Russo, A.F. Calcitonin gene-related peptide (CGRP): Role in migraine pathophysiology and therapeutic targeting. Expert Opin. Ther. Targets 2020, 24, 91–100. [Google Scholar] [CrossRef]
- Alevizaki, M.; Shiraishi, A.; Rassool, F.V.; Ferrier, G.J.; MacIntyre, I.; Legon, S. The calcitonin-like sequence of the beta CGRP gene. FEBS Lett. 1986, 206, 47–52. [Google Scholar] [CrossRef] [Green Version]
- Mulderry, P.K.; Ghatei, M.A.; Bishop, A.E.; Allen, Y.S.; Polak, J.M.; Bloom, S.R. Distribution and chromatographic characterisation of CGRP-like immunoreactivity in the brain and gut of the rat. Regul. Pept. 1985, 12, 133–143. [Google Scholar] [CrossRef]
- Hautakangas, H.; Winsvold, B.S.; Ruotsalainen, S.E.; Bjornsdottir, G.; Harder, A.V.E.; Kogelman, L.J.A.; Thomas, L.F.; Noordam, R.; Benner, C.; Gormley, P.; et al. Genome-wide analysis of 102,084 migraine cases identifies 123 risk loci and subtype-specific risk alleles. Nat. Genet. 2022, 54, 152–160. [Google Scholar] [CrossRef]
- Dong, Y.L.; Vegiraju, S.; Gangula, P.R.; Kondapaka, S.B.; Wimalawansa, S.J.; Yallampalli, C. Expression and regulation of calcitonin gene-related Peptide receptor in rat placentas. Biol. Reprod. 2002, 67, 1321–1326. [Google Scholar] [CrossRef] [Green Version]
- Kraenzlin, M.E.; Ch’ng, J.L.; Mulderry, P.K.; Ghatei, M.A.; Bloom, S.R. Infusion of a novel peptide, calcitonin gene-related peptide (CGRP) in man. Pharmacokinet. Eff. Gastric Acid Secret. Gastrointest. Horm. Regul. Pept. 1985, 10, 189–197. [Google Scholar] [CrossRef]
- Doi, Y.; Kudo, H.; Nishino, T.; Kayashima, K.; Kiyonaga, H.; Nagata, T.; Nara, S.; Morita, M.; Fujimoto, S. Synthesis of calcitonin gene-related peptide (CGRP) by rat arterial endothelial cells. Histol. Histopathol. 2001, 16, 1073–1079. [Google Scholar] [CrossRef] [PubMed]
- Linscheid, P.; Seboek, D.; Schaer, D.J.; Zulewski, H.; Keller, U.; Müller, B. Expression and secretion of procalcitonin and calcitonin gene-related peptide by adherent monocytes and by macrophage-activated adipocytes. Crit. Care Med. 2004, 32, 1715–1721. [Google Scholar] [CrossRef] [PubMed]
- Messlinger, K.; Russo, A.F. Current understanding of trigeminal ganglion structure and function in headache. Cephalalgia Int. J. Headache 2019, 39, 1661–1674. [Google Scholar] [CrossRef] [PubMed]
- Goadsby, P.J.; Edvinsson, L.; Ekman, R. Vasoactive peptide release in the extracerebral circulation of humans during migraine headache. Ann. Neurol. 1990, 28, 183–187. [Google Scholar] [CrossRef]
- Moriarty, M.; Mallick-Searle, T.; Barch, C.A.; Oas, K. Monoclonal Antibodies to CGRP or Its Receptor for Migraine Prevention. J. Nurse Pract. 2019, 15, 717–724.e711. [Google Scholar] [CrossRef]
- Goadsby, P.J.; Holland, P.R.; Martins-Oliveira, M.; Hoffmann, J.; Schankin, C.; Akerman, S. Pathophysiology of Migraine: A Disorder of Sensory Processing. Physiol. Rev. 2017, 97, 553–622. [Google Scholar] [CrossRef]
- Hargreaves, R. New migraine and pain research. Headache 2007, 47 (Suppl. S1), S26–S43. [Google Scholar] [CrossRef]
- Spekker, E.; Tanaka, M.; Szabó, Á.; Vécsei, L. Neurogenic Inflammation: The Participant in Migraine and Recent Advancements in Translational Research. Biomedicines 2021, 10, 76. [Google Scholar] [CrossRef]
- Tvedskov, J.F.; Lipka, K.; Ashina, M.; Iversen, H.K.; Schifter, S.; Olesen, J. No increase of calcitonin gene-related peptide in jugular blood during migraine. Ann. Neurol. 2005, 58, 561–568. [Google Scholar] [CrossRef]
- Lassen, L.H.; Haderslev, P.A.; Jacobsen, V.B.; Iversen, H.K.; Sperling, B.; Olesen, J. CGRP may play a causative role in migraine. Cephalalgia Int. J. Headache 2002, 22, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Vandervorst, F.; Van Deun, L.; Van Dycke, A.; Paemeleire, K.; Reuter, U.; Schoenen, J.; Versijpt, J. CGRP monoclonal antibodies in migraine: An efficacy and tolerability comparison with standard prophylactic drugs. J. Headache Pain 2021, 22, 128. [Google Scholar] [CrossRef] [PubMed]
- Durham, P.L. Diverse Physiological Roles of Calcitonin Gene-Related Peptide in Migraine Pathology: Modulation of Neuronal-Glial-Immune Cells to Promote Peripheral and Central Sensitization. Curr. Pain Headache Rep. 2016, 20, 48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raddant, A.C.; Russo, A.F. Reactive oxygen species induce procalcitonin expression in trigeminal ganglia glia. Headache 2014, 54, 472–484. [Google Scholar] [CrossRef] [Green Version]
- Sowers, L.P.; Wang, M.; Rea, B.J.; Taugher, R.J.; Kuburas, A.; Kim, Y.; Wemmie, J.A.; Walker, C.S.; Hay, D.L.; Russo, A.F. Stimulation of Posterior Thalamic Nuclei Induces Photophobic Behavior in Mice. Headache 2020, 60, 1961–1981. [Google Scholar] [CrossRef]
- Raddant, A.C.; Russo, A.F. Calcitonin gene-related peptide in migraine: Intersection of peripheral inflammation and central modulation. Expert Rev. Mol. Med. 2011, 13, e36. [Google Scholar] [CrossRef] [Green Version]
- Russell, F.A.; King, R.; Smillie, S.J.; Kodji, X.; Brain, S.D. Calcitonin gene-related peptide: Physiology and pathophysiology. Physiol. Rev. 2014, 94, 1099–1142. [Google Scholar] [CrossRef] [Green Version]
- Lyko, F. The DNA methyltransferase family: A versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 2018, 19, 81–92. [Google Scholar] [CrossRef]
- Lister, R.; Pelizzola, M.; Dowen, R.H.; Hawkins, R.D.; Hon, G.; Tonti-Filippini, J.; Nery, J.R.; Lee, L.; Ye, Z.; Ngo, Q.M.; et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 2009, 462, 315–322. [Google Scholar] [CrossRef] [Green Version]
- Illingworth, R.S.; Bird, A.P. CpG islands—‘A rough guide’. FEBS Lett. 2009, 583, 1713–1720. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Zhang, Y. TET-mediated active DNA demethylation: Mechanism, function and beyond. Nat. Rev. Genet. 2017, 18, 517–534. [Google Scholar] [CrossRef]
- Winsvold, B.S.; Palta, P.; Eising, E.; Page, C.M.; van den Maagdenberg, A.M.; Palotie, A.; Zwart, J.A. Epigenetic DNA methylation changes associated with headache chronification: A retrospective case-control study. Cephalalgia Int. J. Headache 2018, 38, 312–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basedau, H.; Sturm, L.M.; Mehnert, J.; Peng, K.P.; Schellong, M.; May, A. Migraine monoclonal antibodies against CGRP change brain activity depending on ligand or receptor target—An fMRI study. eLife 2022, 11, e77146. [Google Scholar] [CrossRef] [PubMed]
- Gerring, Z.F.; McRae, A.F.; Montgomery, G.W.; Nyholt, D.R. Genome-wide DNA methylation profiling in whole blood reveals epigenetic signatures associated with migraine. BMC Genom. 2018, 19, 69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rubino, E.; Boschi, S.; Giorgio, E.; Pozzi, E.; Marcinnò, A.; Gallo, E.; Roveta, F.; Grassini, A.; Brusco, A.; Rainero, I. Analysis of the DNA methylation pattern of the promoter region of calcitonin gene-related peptide 1 gene in patients with episodic migraine: An exploratory case-control study. Neurobiol. Pain 2022, 11, 100089. [Google Scholar] [CrossRef]
- Zamanian Azodi, M.; Rezaei Tavirani, M.; Robati, R.M. Introducing Genes With Significant Role in Migraine: An Interactomic Approach. Basic Clin. Neurosci. 2019, 10, 363–372. [Google Scholar] [CrossRef] [PubMed]
- Labruijere, S.; Stolk, L.; Verbiest, M.; de Vries, R.; Garrelds, I.M.; Eilers, P.H.; Danser, A.H.; Uitterlinden, A.G.; MaassenVanDenBrink, A. Methylation of migraine-related genes in different tissues of the rat. PLoS ONE 2014, 9, e87616. [Google Scholar] [CrossRef] [Green Version]
- Wan, D.; Hou, L.; Zhang, X.; Han, X.; Chen, M.; Tang, W.; Liu, R.; Dong, Z.; Yu, S. DNA methylation of RAMP1 gene in migraine: An exploratory analysis. J. Headache Pain 2015, 16, 90. [Google Scholar] [CrossRef] [Green Version]
- Stucky, N.L.; Gregory, E.; Winter, M.K.; He, Y.Y.; Hamilton, E.S.; McCarson, K.E.; Berman, N.E. Sex differences in behavior and expression of CGRP-related genes in a rodent model of chronic migraine. Headache 2011, 51, 674–692. [Google Scholar] [CrossRef]
- Zill, P.; Baghai, T.C.; Schüle, C.; Born, C.; Früstück, C.; Büttner, A.; Eisenmenger, W.; Varallo-Bedarida, G.; Rupprecht, R.; Möller, H.J.; et al. DNA methylation analysis of the angiotensin converting enzyme (ACE) gene in major depression. PLoS ONE 2012, 7, e40479. [Google Scholar] [CrossRef] [Green Version]
- Dell’Osso, B.; D’Addario, C.; Carlotta Palazzo, M.; Benatti, B.; Camuri, G.; Galimberti, D.; Fenoglio, C.; Scarpini, E.; Di Francesco, A.; Maccarrone, M.; et al. Epigenetic modulation of BDNF gene: Differences in DNA methylation between unipolar and bipolar patients. J. Affect Disord. 2014, 166, 330–333. [Google Scholar] [CrossRef] [PubMed]
- Bollati, V.; Galimberti, D.; Pergoli, L.; Dalla Valle, E.; Barretta, F.; Cortini, F.; Scarpini, E.; Bertazzi, P.A.; Baccarelli, A. DNA methylation in repetitive elements and Alzheimer disease. Brain Behav. Immun. 2011, 25, 1078–1083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shahid, Z.; Simpson, B.; Miao, K.H.; Singh, G. Genetics, Histone Code; StatPearls; StatPearls Publishing LLC.: Treasure Island, FL, USA, 2022. [Google Scholar]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- DesJarlais, R.; Tummino, P.J. Role of Histone-Modifying Enzymes and Their Complexes in Regulation of Chromatin Biology. Biochemistry 2016, 55, 1584–1599. [Google Scholar] [CrossRef]
- Yoshida, M.; Kudo, N.; Kosono, S.; Ito, A. Chemical and structural biology of protein lysine deacetylases. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2017, 93, 297–321. [Google Scholar] [CrossRef] [Green Version]
- D’Ydewalle, C.; Bogaert, E.; Van Den Bosch, L. HDAC6 at the Intersection of Neuroprotection and Neurodegeneration. Traffic 2012, 13, 771–779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janke, C.; Montagnac, G. Causes and Consequences of Microtubule Acetylation. Curr. Biol. CB 2017, 27, R1287–R1292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bertels, Z.; Singh, H.; Dripps, I.; Siegersma, K.; Tipton, A.F.; Witkowski, W.D.; Sheets, Z.; Shah, P.; Conway, C.; Mangutov, E.; et al. Neuronal complexity is attenuated in preclinical models of migraine and restored by HDAC6 inhibition. eLife 2021, 10, e63076. [Google Scholar] [CrossRef]
- Yamanaka, G.; Suzuki, S.; Morishita, N.; Takeshita, M.; Kanou, K.; Takamatsu, T.; Suzuki, S.; Morichi, S.; Watanabe, Y.; Ishida, Y.; et al. Role of Neuroinflammation and Blood-Brain Barrier Permutability on Migraine. Int. J. Mol. Sci. 2021, 22, 8929. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, Y.; Yuan, J.; Li, N.; Pei, S.; Xu, J.; Luo, X.; Mao, C.; Liu, J.; Yu, T.; et al. Macrophage/microglial Ezh2 facilitates autoimmune inflammation through inhibition of Socs3. J. Exp. Med. 2018, 215, 1365–1382. [Google Scholar] [CrossRef]
- Cai, Y.; Zhang, Y.; Loh, Y.P.; Tng, J.Q.; Lim, M.C.; Cao, Z.; Raju, A.; Lieberman Aiden, E.; Li, S.; Manikandan, L.; et al. H3K27me3-rich genomic regions can function as silencers to repress gene expression via chromatin interactions. Nat. Commun. 2021, 12, 719. [Google Scholar] [CrossRef]
- Yadav, R.; Weng, H.R. EZH2 regulates spinal neuroinflammation in rats with neuropathic pain. Neuroscience 2017, 349, 106–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, Q.; Sun, C.; Li, R.; Chen, S.; Gu, X.; An, S.; Wang, Z. Calcitonin gene-related peptide regulates spinal microglial activation through the histone H3 lysine 27 trimethylation via enhancer of zeste homolog-2 in rats with neuropathic pain. J. Neuroinflam. 2021, 18, 117. [Google Scholar] [CrossRef] [PubMed]
- Romanos, J.; Benke, D.; Pietrobon, D.; Zeilhofer, H.U.; Santello, M. Astrocyte dysfunction increases cortical dendritic excitability and promotes cranial pain in familial migraine. Sci. Adv. 2020, 6, eaaz1584. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Blaeser, A.S.; Levy, D. Astrocytes mediate migraine-related intracranial meningeal mechanical hypersensitivity. Pain 2021, 162, 2386–2396. [Google Scholar] [CrossRef]
- Sun, C.; An, Q.; Li, R.; Chen, S.; Gu, X.; An, S.; Wang, Z. Calcitonin gene-related peptide induces the histone H3 lysine 9 acetylation in astrocytes associated with neuroinflammation in rats with neuropathic pain. CNS Neurosci. Ther. 2021, 27, 1409–1424. [Google Scholar] [CrossRef]
- Hashikawa-Hobara, N.; Mishima, S.; Okujima, C.; Shitanishi, Y.; Hashikawa, N. Npas4 impairs fear memory via phosphorylated HDAC5 induced by CGRP administration in mice. Sci. Rep. 2021, 11, 7006. [Google Scholar] [CrossRef]
- Hashikawa-Hobara, N.; Yoneyama, Y.; Fujiwara, K.; Hashikawa, N. Intranasal calcitonin gene-related peptide administration impairs fear memory retention in mice through the PKD/p-HDAC5/Npas4 pathway. Sci. Rep. 2022, 12, 1450. [Google Scholar] [CrossRef]
- Lee, H.; Zhang, Z.; Krause, H.M. Long Noncoding RNAs and Repetitive Elements: Junk or Intimate Evolutionary Partners? Trends in genetics. TIG 2019, 35, 892–902. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Wu, W.; Chen, Q.; Chen, M. Non-Coding RNAs and their Integrated Networks. J. Integr. Bioinform. 2019, 16, 1515. [Google Scholar] [CrossRef]
- Wahid, F.; Shehzad, A.; Khan, T.; Kim, Y.Y. MicroRNAs: Synthesis, mechanism, function, and recent clinical trials. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2010, 1803, 1231–1243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greco, R.; De Icco, R.; Demartini, C.; Zanaboni, A.M.; Tumelero, E.; Sances, G.; Allena, M.; Tassorelli, C. Plasma levels of CGRP and expression of specific microRNAs in blood cells of episodic and chronic migraine subjects: Towards the identification of a panel of peripheral biomarkers of migraine? J. Headache Pain 2020, 21, 122. [Google Scholar] [CrossRef] [PubMed]
- De Icco, R.; Fiamingo, G.; Greco, R.; Bottiroli, S.; Demartini, C.; Zanaboni, A.M.; Allena, M.; Guaschino, E.; Martinelli, D.; Putortì, A.; et al. Neurophysiological and biomolecular effects of erenumab in chronic migraine: An open label study. Cephalalgia Int. J. Headache 2020, 40, 1336–1345. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Y.; Zhu, Y.Y. MiR-30a relieves migraine by degrading CALCA. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 2022–2028. [Google Scholar] [CrossRef] [PubMed]
- Andersen, H.H.; Duroux, M.; Gazerani, P. Serum MicroRNA Signatures in Migraineurs During Attacks and in Pain-Free Periods. Mol. Neurobiol. 2016, 53, 1494–1500. [Google Scholar] [CrossRef]
- Chen, S.; Gu, Y.; Dai, Q.; He, Y.; Wang, J. Spinal miR-34a regulates inflammatory pain by targeting SIRT1 in complete Freund’s adjuvant mice. Biochem. Biophys. Res. Commun. 2019, 516, 1196–1203. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, X.M.; Zong, D.D.; Ji, X.Y.; Jiang, H.; Zhang, F.Z.; He, S.D. miR-34a-5p up-regulates the IL-1β/COX2/PGE2 inflammation pathway and induces the release of CGRP via inhibition of SIRT1 in rat trigeminal ganglion neurons. FEBS Open Biol. 2021, 11, 300–311. [Google Scholar] [CrossRef]
- Dong, Y.; Li, P.; Ni, Y.; Zhao, J.; Liu, Z. Decreased microRNA-125a-3p contributes to upregulation of p38 MAPK in rat trigeminal ganglions with orofacial inflammatory pain. PLoS ONE 2014, 9, e111594. [Google Scholar] [CrossRef] [Green Version]
- Wen, Q.; Wang, Y.; Pan, Q.; Tian, R.; Zhang, D.; Qin, G.; Zhou, J.; Chen, L. MicroRNA-155-5p promotes neuroinflammation and central sensitization via inhibiting SIRT1 in a nitroglycerin-induced chronic migraine mouse model. J. Neuroinflam. 2021, 18, 287. [Google Scholar] [CrossRef]
- Li, J.; Peng, J.; Wang, C.; Deng, H.; Li, Y. Calcitonin gene-related peptide suppresses isoprenaline-induced cardiomyocyte apoptosis through regulation of microRNA-1 and microRNA-133a expression. Zhong Nan Da Xue Xue Bao Yi Xue Ban 2011, 36, 964–971. [Google Scholar] [CrossRef]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nature reviews. Genetics 2019, 20, 675–691. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, Z. Efficient backsplicing produces translatable circular mRNAs. mRNA 2015, 21, 172–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrett, S.P.; Wang, P.L.; Salzman, J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. eLife 2015, 4, e07540. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.H.; Wu, W.P.; Xiong, X.D. Circular RNAs: Promising Biomarkers for Age-related Diseases. Aging Dis. 2020, 11, 1585–1593. [Google Scholar] [CrossRef] [PubMed]
- Deng, T.; Yang, L.; Zheng, Z.; Li, Y.; Ren, W.; Wu, C.; Guo, L. Calcitonin gene-related peptide induces IL-6 expression in RAW264.7 macrophages mediated by mmu_circRNA_007893. Mol. Med. Rep. 2017, 16, 9367–9374. [Google Scholar] [CrossRef] [Green Version]
- Leira, Y.; Domínguez, C.; Ameijeira, P.; López-Arias, E.; Ávila-Gómez, P.; Pérez-Mato, M.; Sobrino, T.; Campos, F.; Blanco, J.; Leira, R. Mild systemic inflammation enhances response to OnabotulinumtoxinA in chronic migraineurs. Sci. Rep. 2021, 11, 1092. [Google Scholar] [CrossRef]
- Ren, W.; Yang, L.; Deng, T.; Wu, C.; Li, Y.; Wu, J.; Huang, Z.; Du, F.; Guo, L. Calcitonin gene-related peptide regulates FOSL2 expression and cell proliferation of BMSCs via mmu_circRNA_003795. Mol. Med. Rep. 2019, 19, 3732–3742. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Sun, D.; Wang, Y.; Ren, F.; Pang, S.; Wang, D.; Xu, S. FOSL2 positively regulates TGF-β1 signalling in non-small cell lung cancer. PLoS ONE 2014, 9, e112150. [Google Scholar] [CrossRef]
- Kamato, D.; Burch, M.L.; Piva, T.J.; Rezaei, H.B.; Rostam, M.A.; Xu, S.; Zheng, W.; Little, P.J.; Osman, N. Transforming growth factor-β signalling: Role and consequences of Smad linker region phosphorylation. Cell. Signal. 2013, 25, 2017–2024. [Google Scholar] [CrossRef]
- Ishizaki, K.; Takeshima, T.; Fukuhara, Y.; Araki, H.; Nakaso, K.; Kusumi, M.; Nakashima, K. Increased plasma transforming growth factor-beta1 in migraine. Headache 2005, 45, 1224–1228. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Zheng, Z.; Ren, W.; Deng, T.; Li, Y.; Yang, L.; Wu, J.; Huang, Z.; Li, Z.; Guo, L. Mm9_circ_009056 enhances osteogenesis by targeting BMP7 via CGRP-mediated miR-22-3p. Biochem. Biophys. Biophys. Res. Commun. 2018, 501, 199–205. [Google Scholar] [CrossRef] [PubMed]
- Röther, S.; Meister, G. Small RNAs derived from longer non-coding RNAs. Biochimie 2011, 93, 1905–1915. [Google Scholar] [CrossRef] [PubMed]
- Blasiak, J.; Hyttinen, J.M.T.; Szczepanska, J.; Pawlowska, E.; Kaarniranta, K. Potential of Long Non-Coding RNAs in Age-Related Macular Degeneration. Int. J. Mol. Sci. 2021, 22, 9178. [Google Scholar] [CrossRef]
- Xu, M.; Yan, Y.; Zhu, M.; Wang, Z.; Zhang, X.; Zhang, D. Effects of long non-coding RNA Gm14461 on pain transmission in trigeminal neuralgia. J. Inflamm. 2020, 17, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biondi, D.M. Is migraine a neuropathic pain syndrome? Curr. Pain Headache Rep. 2006, 10, 167–178. [Google Scholar] [CrossRef]
- Xiong, W.; Huang, L.; Shen, Y.; Guan, S.; He, L.; Tong, Z.; Tan, M.; Liu, L.; Gao, Y. Effects of lncRNA uc.48+ siRNA on the release of CGRP in the spinal cords of rats with diabetic neuropathic pain. Int. J. Clin. Exp. Pathol. 2017, 10, 9960–9969. [Google Scholar]
- Sun, R.M.; Wei, J.; Wang, S.S.; Xu, G.Y.; Jiang, G.Q. Upregulation of lncRNA-NONRATT021203.2 in the dorsal root ganglion contributes to cancer-induced pain via CXCL9 in rats. Biochem. Biophys. Res. Commun. 2020, 524, 983–989. [Google Scholar] [CrossRef]
- Zhou, J. LncRNA MIAT promotes hypoxia-induced H9C2 cell pyroptosis via binding to SF1 to inhibit CGRP transcription. Exp. Physiol. 2022, 107, 58–67. [Google Scholar] [CrossRef]
- Zhou, H.L.; Lou, H. Repression of prespliceosome complex formation at two distinct steps by Fox-1/Fox-2 proteins. Mol. Cell. Biol. 2008, 28, 5507–5516. [Google Scholar] [CrossRef] [Green Version]
- Hu, F.; Yang, J.; Chen, X.; Shen, Y.; Chen, K.; Fu, X.; Guo, S.; Jiang, Z. LncRNA 1700020I14Rik/miR-297a/CGRP axis suppresses myocardial cell apoptosis in myocardial ischemia-reperfusion injury. Mol. Immunol. 2020, 122, 54–61. [Google Scholar] [CrossRef]
- Borkum, J.M. Migraine Triggers, Oxidative Stress, and the Thyroid. Headache 2016, 56, 784–785. [Google Scholar] [CrossRef] [PubMed]
- Borkum, J.M. Migraine Triggers and Oxidative Stress: A Narrative Review and Synthesis. Headache 2016, 56, 12–35. [Google Scholar] [CrossRef] [PubMed]
- Borkum, J.M. The Migraine Attack as a Homeostatic, Neuroprotective Response to Brain Oxidative Stress: Preliminary Evidence for a Theory. Headache 2018, 58, 118–135. [Google Scholar] [CrossRef] [PubMed]
- Borkum, J.M. Brain Energy Deficit as a Source of Oxidative Stress in Migraine: A Molecular Basis for Migraine Susceptibility. Neurochem. Res. 2021, 46, 1913–1932. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-Y.; Lim, J.-S.; Oh, D.J.; Kong, I.G.; Choi, H.G. Increased risk of neurodegenerative dementia in women with migraines: A nested case-control study using a national sample cohort. Medicine 2019, 98, e14467. [Google Scholar] [CrossRef]
- Pozo-Rosich, P.; Coppola, G.; Pascual, J.; Schwedt, T.J. How does the brain change in chronic migraine? Developing disease biomarkers. Cephalalgia Int. J. Headache 2021, 41, 613–630. [Google Scholar] [CrossRef]
- Borkum, J.M. Harnessing migraines for neural regeneration. Neural Regen. Res. 2018, 13, 609–615. [Google Scholar] [CrossRef]
- Aizawa, H.; Sun, W.; Sugiyama, K.; Itou, Y.; Aida, T.; Cui, W.; Toyoda, S.; Terai, H.; Yanagisawa, M.; Tanaka, K. Glial glutamate transporter GLT-1 determines susceptibility to spreading depression in the mouse cerebral cortex. Glia 2020, 68, 2631–2642. [Google Scholar] [CrossRef]
- Bartley, J. Could glial activation be a factor in migraine? Med. Hypotheses 2009, 72, 255–257. [Google Scholar] [CrossRef]
- Vila-Pueyo, M.; Page, K.; Murdock, P.R.; Loraine, H.J.; Woodrooffe, A.J.; Johnson, K.W.; Goadsby, P.J.; Holland, P.R. The selective 5-HT(1F) receptor agonist lasmiditan inhibits trigeminal nociceptive processing: Implications for migraine and cluster headache. Br. J. Pharmacol. 2022, 179, 358–370. [Google Scholar] [CrossRef] [PubMed]
- Zobdeh, F.; Ben Kraiem, A.; Attwood, M.M.; Chubarev, V.N.; Tarasov, V.V.; Schiöth, H.B.; Mwinyi, J. Pharmacological treatment of migraine: Drug classes, mechanisms of action, clinical trials and new treatments. Br. J. Pharmacol. 2021, 178, 4588–4607. [Google Scholar] [CrossRef] [PubMed]
- Russo, A.F. Calcitonin gene-related peptide (CGRP): A new target for migraine. Annu. Rev. Pharm. Toxicol. 2015, 55, 533–552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zecca, C.; Cargnin, S.; Schankin, C.; Giannantoni, N.M.; Viana, M.; Maraffi, I.; Riccitelli, G.C.; Sihabdeen, S.; Terrazzino, S.; Gobbi, C. Clinic and genetic predictors in response to erenumab. Eur. J. Neurol. 2022, 29, 1209–1217. [Google Scholar] [CrossRef]
- Austin, L.A.; Heath, H., III. Calcitonin: Physiology and pathophysiology. N. Engl. J. Med. 1981, 304, 269–278. [Google Scholar] [CrossRef]
- Twery, M.J.; Obie, J.F.; Cooper, C.W. Ability of calcitonins to alter food and water consumption in the rat. Peptides 1982, 3, 749–755. [Google Scholar] [CrossRef]



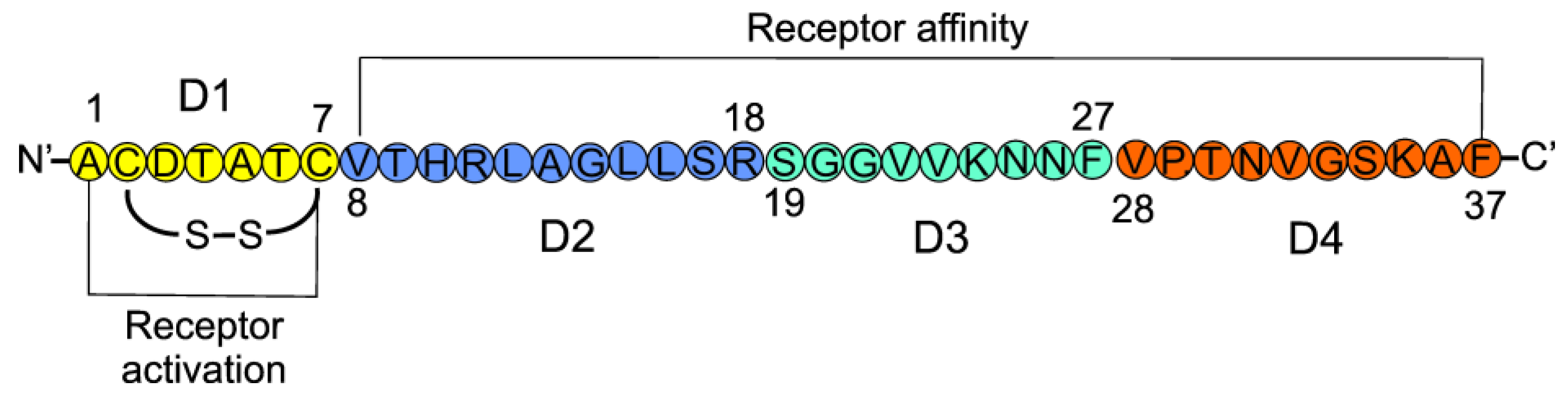
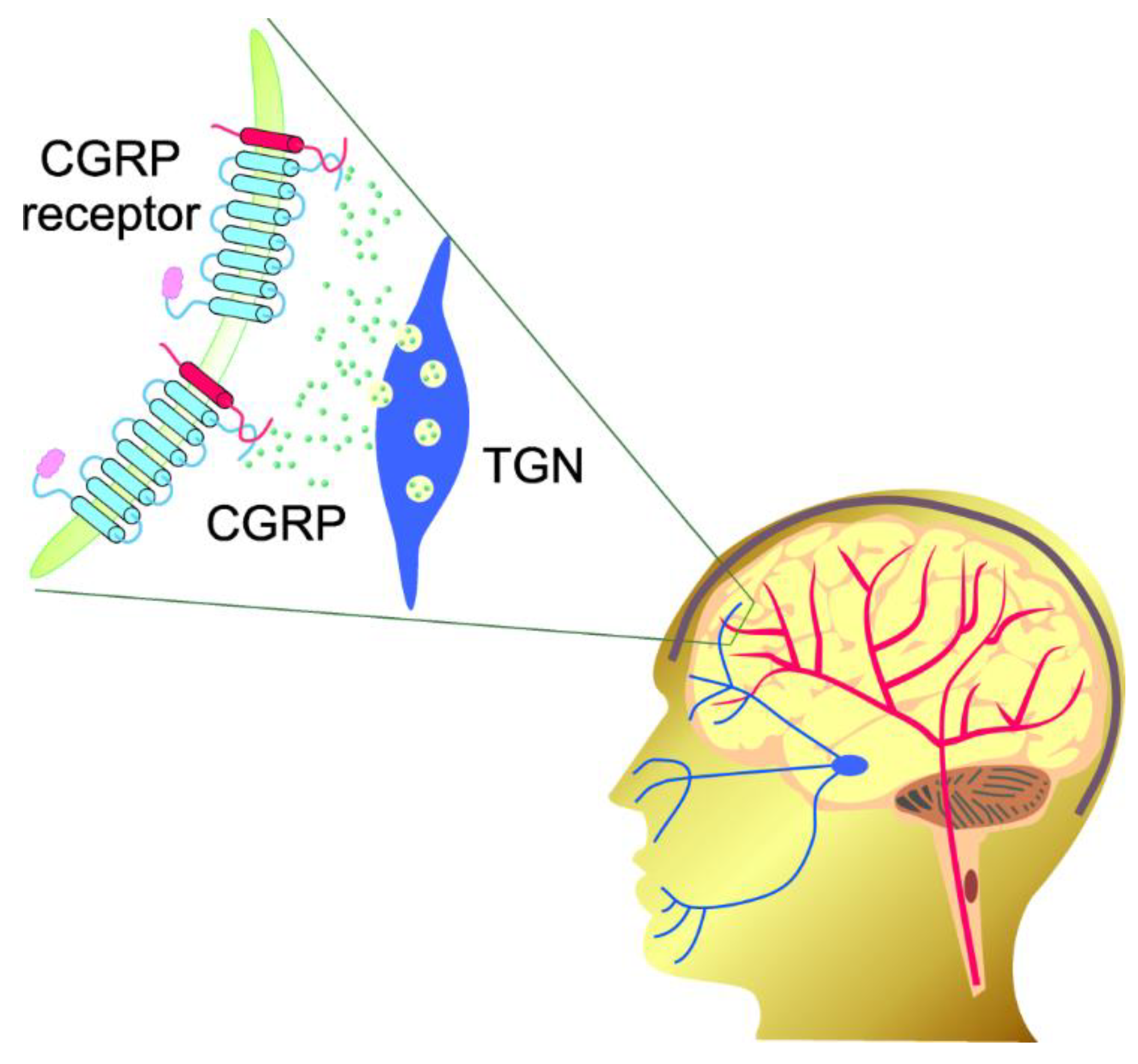

| Epigenetic Modification | Effect | Reference |
|---|---|---|
| DNA methylation | Hypomethylation of two CpG dinucleotides in the promoter of CALCA in sporadic migraine patients | [65] |
| Low DNA methylation trend in the promoter of RAMP1 in migraineurs with higher methylation levels in patients with family history, lower methylation in female migraineurs | [68] | |
| Histone modifications | Blockade of the CGRP receptor by HDAC6 inhibition, resulting in reversing of NTG-induced chronic allodynia in mice | [79] |
| Co-expression of CGRP-induced abnormal histone modifications, including H3K27me3, with proinflammatory mediators in the spinal dorsal horn and cultured microglial cells | [80,81,84] | |
| Altered HDAC2 enrichment in over 1200 genes in microglial cells by CGRP, with most of the genes belonging to immune- and inflammation-related pathways | [18] | |
| H3K9ac signal in rat cells expressing CALCA, but lack of such signals in cells non-expressing CALCA; that effect of histone acetylation required DNA methylation to be manifested | [14] | |
| CGRP-induced acetylation of H3K9 in astrocytes linked with neuroinflammation in rats with neuropathic pain and H3K9ac enrichment on gene promoters in astroglial cells | [87] | |
| Micro RNAs | CGRP plasma levels positively correlated with the expression of miR-382-5p, and miR-34a-5p in plasma of migraineurs | [93] |
| Overexpression of miR-30a caused downregulation of CALCA, while knockdown of miR-30 resulted in CALCA upregulation; miR-30a might bind CALCA in its 3′-UTR to degrade it | [95] | |
| miR-125a-3p regulated CGRP in inflammatory pain; | [99] | |
| increased levels of CGRP induced by NTG were abolished by miR-155-5p antagomir; miR-155-5p agomir aggravated neuroinflammation and central sensitization and increased CGRP expression | [100] | |
| Circular RNAs | CGRP promoted mmu_circRNA_007893 expression in RAW264.7 macrophages | [107] |
| CGRP regulated migraine-related FOSL2 expression mediated by miR-504-3p sponging by mmu_circRNA_003795 | [109,112] | |
| Aberrant expression of mm9_circ_009056 circRNA in CGRP-induced MC3T3 cells | [113] | |
| Long non-coding RNAs | Gm14461 lncRNA knockdown increased, whereas Gm14461 overexpression decreased CGRP levels in primary mouse trigeminal ganglion neurons | [116] |
| Overexpression of lncRNA-NONRATT021203.2 in rats with induced CIP; lncRNA-NONRATT021203.2 targeted CXCL9, increased in CIP rats; CXCL9 was expressed in the CGRP-positive dorsal root ganglion neurons that colocalized with lncRNA-NONRATT021203.2 | [119] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fila, M.; Sobczuk, A.; Pawlowska, E.; Blasiak, J. Epigenetic Connection of the Calcitonin Gene-Related Peptide and Its Potential in Migraine. Int. J. Mol. Sci. 2022, 23, 6151. https://doi.org/10.3390/ijms23116151
Fila M, Sobczuk A, Pawlowska E, Blasiak J. Epigenetic Connection of the Calcitonin Gene-Related Peptide and Its Potential in Migraine. International Journal of Molecular Sciences. 2022; 23(11):6151. https://doi.org/10.3390/ijms23116151
Chicago/Turabian StyleFila, Michal, Anna Sobczuk, Elzbieta Pawlowska, and Janusz Blasiak. 2022. "Epigenetic Connection of the Calcitonin Gene-Related Peptide and Its Potential in Migraine" International Journal of Molecular Sciences 23, no. 11: 6151. https://doi.org/10.3390/ijms23116151






