The Mediator Complex: A Central Coordinator of Plant Adaptive Responses to Environmental Stresses
Abstract
:1. Introduction
2. Conserved and Unique Features of Transcription and Mediator Complex in Plants
3. Mediator Complex in Signaling of JA, a Stress-Responsive Hormone
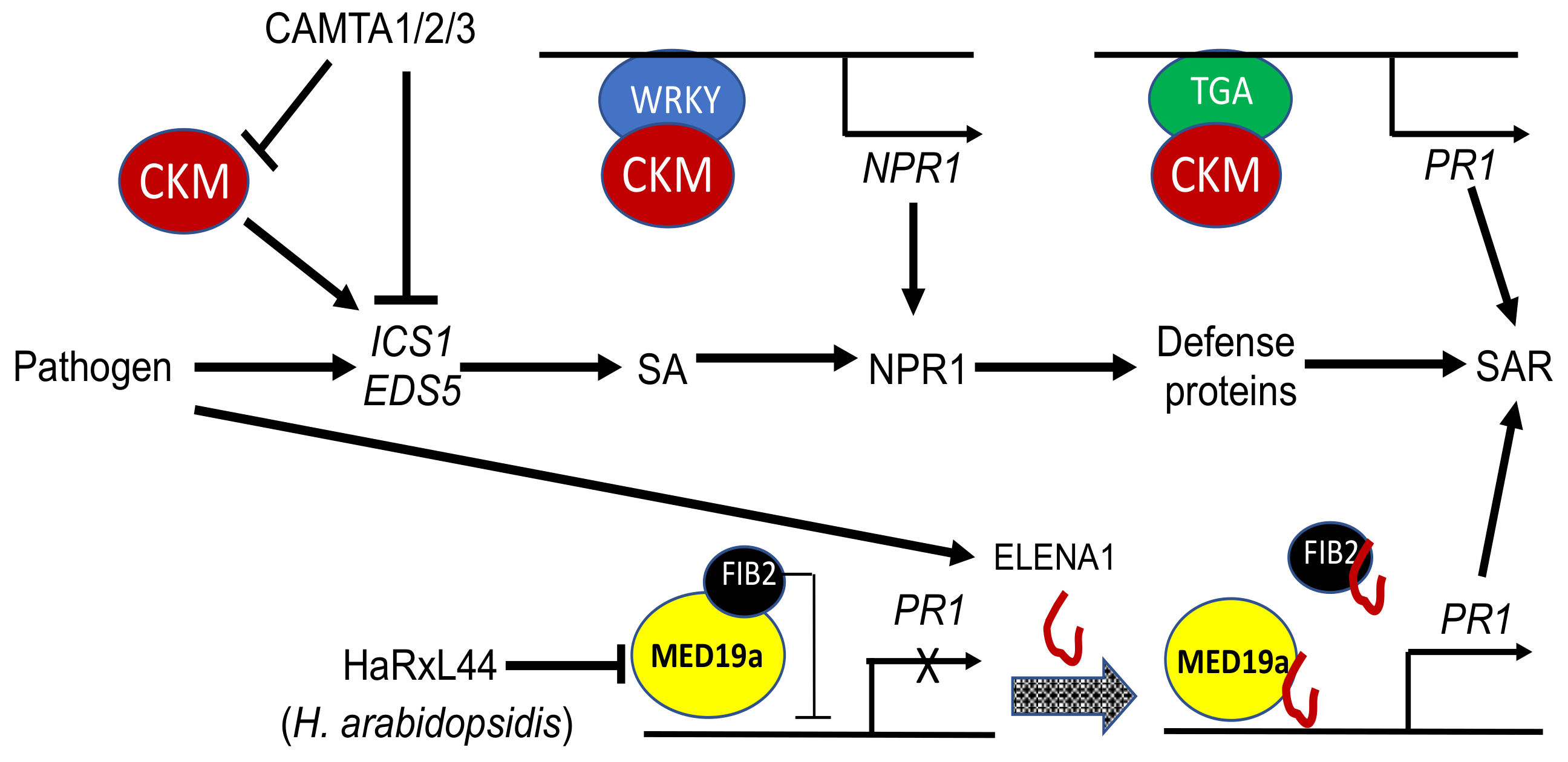
4. Function of the Mediator Complex in Broad Plant Biotic Interactions
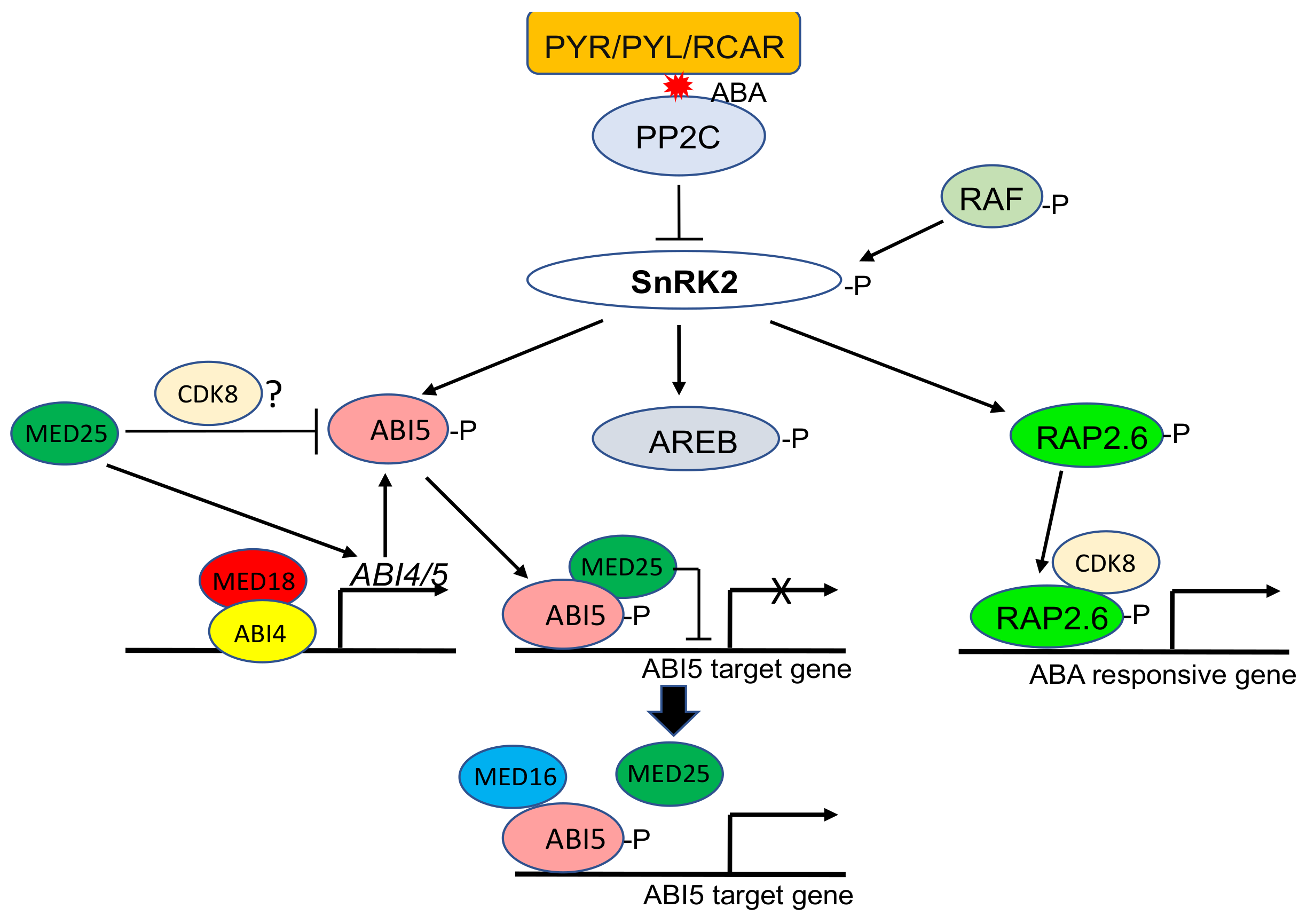
5. Mediator Complex as a Central Regulator of ABA-Mediated Stress Responses
6. Roles of the Mediator Complex in Plant Responses to Abiotic Stresses
7. Regulation of Phenylpropanoid Biosynthesis by Plant Mediator Complex
8. Summary and Prospects
Author Contributions
Funding
Conflicts of Interest
References
- Cox, M.M.; Doudna, J.; O’Donnell, M. Molecular Biology: Principle and Practice; Freenman and Company: New York, NY, USA, 2012. [Google Scholar]
- Vannini, A.; Cramer, P. Conservation between the RNA Polymerase I, II, and III Transcription Initiation Machineries. Mol. Cell 2012, 45, 439–446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Mendoza, A.; Sebé-Pedrós, A. Origin and evolution of eukaryotic transcription factors. Curr. Opin. Genet. Dev. 2019, 58–59, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.I.; Young, R.A. Transcriptional Regulation and Its Misregulation in Disease. Cell 2013, 152, 1237–1251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, L.; Nogales, E.; Ciferri, C. Structure and function of SWI/SNF chromatin remodeling complexes and mechanistic implications for transcription. Prog. Biophys. Mol. Biol. 2010, 102, 122–128. [Google Scholar] [CrossRef] [Green Version]
- Fukasawa, R.; Iida, S.; Tsutsui, T.; Hirose, Y.; Ohkuma, Y. Mediator complex cooperatively regulates transcription of retinoic acid target genes with Polycomb Repressive Complex 2 during neuronal differentiation. J. Biochem. 2015, 158, 373–384. [Google Scholar] [CrossRef]
- Soutourina, J. Transcription regulation by the Mediator complex. Nat. Rev. Mol. Cell Biol. 2017, 19, 262–274. [Google Scholar] [CrossRef]
- Conaway, R.C.; Conaway, J.W. Origins and activity of the Mediator complex. Semin. Cell Dev. Biol. 2011, 22, 729–734. [Google Scholar] [CrossRef] [Green Version]
- Robinson, P.J.; Trnka, M.J.; Pellarin, R.; Greenberg, C.H.; Bushnell, D.A.; Davis, R.; Burlingame, A.L.; Sali, A.; Kornberg, R.D. Molecular architecture of the yeast Mediator complex. eLife 2015, 4, e08719. [Google Scholar] [CrossRef]
- Poss, Z.C.; Ebmeier, C.C.; Taatjes, D.J. The Mediator complex and transcription regulation. Crit. Rev. Biochem. Mol. Biol. 2013, 48, 575–608. [Google Scholar] [CrossRef] [Green Version]
- Osman, S.; Mohammad, E.; Lidschreiber, M.; Stuetzer, A.; Bazsó, F.L.; Maier, K.C.; Urlaub, H.; Cramer, P. The Cdk8 kinase module regulates interaction of the mediator complex with RNA polymerase II. J. Biol. Chem. 2021, 296, 100734. [Google Scholar] [CrossRef]
- Bäckström, S.; Elfving, N.; Nilsson, R.; Wingsle, G.; Björklund, S. Purification of a Plant Mediator from Arabidopsis thaliana Identifies PFT1 as the Med25 Subunit. Mol. Cell 2007, 26, 717–729. [Google Scholar] [CrossRef]
- Dolan, W.L.; Chapple, C. Conservation and Divergence of Mediator Structure and Function: Insights from Plants. Plant Cell Physiol. 2016, 58, 4–21. [Google Scholar] [CrossRef]
- An, C.; Mou, Z. The function of the Mediator complex in plant immunity. Plant Signal. Behav. 2013, 8, e23182. [Google Scholar] [CrossRef] [Green Version]
- Buendia-Monreal, M.; Gillmor, C.S. Mediator: A key regulator of plant development. Dev. Biol. 2016, 419, 7–18. [Google Scholar] [CrossRef]
- Yang, Y.; Li, L.; Qu, L.-J. Plant Mediator complex and its critical functions in transcription regulation. J. Integr. Plant Biol. 2015, 58, 106–118. [Google Scholar] [CrossRef]
- Zhai, Q.; Li, C. The plant Mediator complex and its role in jasmonate signaling. J. Exp. Bot. 2019, 70, 3415–3424. [Google Scholar] [CrossRef]
- Huang, Y.; Kendall, T.; Forsythe, E.S.; Dorantes-Acosta, A.; Li, S.; Caballero-Pérez, J.; Chen, X.; Arteaga-Vázquez, M.; Beilstein, M.A.; Mosher, R.A. Ancient Origin and Recent Innovations of RNA Polymerase IV and V. Mol. Biol. Evol. 2015, 32, 1788–1799. [Google Scholar] [CrossRef] [Green Version]
- Knutson, B.A. Emergence and expansion of TFIIB-like factors in the plant kingdom. Gene 2013, 526, 30–38. [Google Scholar] [CrossRef] [Green Version]
- Ning, H.; Yang, S.; Fan, B.; Zhu, C.; Chen, Z. Expansion and Functional Diversification of TFIIB-like Factors in Plants. Int. J. Mol. Sci. 2021, 22, 1078. [Google Scholar] [CrossRef]
- Eychenne, T.; Novikova, E.; Barrault, M.-B.; Alibert, O.; Boschiero, C.; Peixeiro, N.; Cornu, D.; Redeker, V.; Kuras, L.; Nicolas, P.; et al. Functional interplay between Mediator and TFIIB in preinitiation complex assembly in relation to promoter architecture. Genes Dev. 2016, 30, 2119–2132. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, D.; Bowen, A.J.; Carroll, T.S.; Conlan, R.S. The Transcription Corepressor LEUNIG Interacts with the Histone Deacetylase HDA19 and Mediator Components MED14 (SWP) and CDK8 (HEN3) To Repress Transcription. Mol. Cell. Biol. 2007, 27, 5306–5315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Chen, X. HUA ENHANCER3 reveals a role for a cyclin-dependent protein kinase in the specification of floral organ identity in Arabidopsis. Development 2004, 131, 3147–3156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourbon, H.-M. Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res. 2008, 36, 3993–4008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, J.; Wei, L.; Chen, S.; Cai, X.; Su, Y.; Li, L.; Chen, S.; He, X. The CBP/p300 histone acetyltransferases function as plant-specific MEDIATOR subunits in Arabidopsis. J. Integr. Plant Biol. 2020, 63, 755–771. [Google Scholar] [CrossRef]
- An, C.; Li, L.; Zhai, Q.; You, Y.; Deng, L.; Wu, F.; Chen, R.; Jiang, H.; Wang, H.; Chen, Q.; et al. Mediator subunit MED25 links the jasmonate receptor to transcriptionally active chromatin. Proc. Natl. Acad. Sci. USA 2017, 114, E8930–E8939. [Google Scholar] [CrossRef] [Green Version]
- Acevedo, M.L.; Kraus, W.L. Mediator and p300/CBP-Steroid Receptor Coactivator Complexes Have Distinct Roles, but Function Synergistically, during Estrogen Receptor α-Dependent Transcription with Chromatin Templates. Mol. Cell. Biol. 2003, 23, 335–348. [Google Scholar] [CrossRef] [Green Version]
- Wallberg, A.E.; Yamamura, S.; Malik, S.; Spiegelman, B.M.; Roeder, R.G. Coordination of p300-Mediated Chromatin Remodeling and TRAP/Mediator Function through Coactivator PGC-1α. Mol. Cell 2003, 12, 1137–1149. [Google Scholar] [CrossRef]
- Carlsten, J.O.; Zhu, X.; López, M.D.; Samuelsson, T.; Gustafsson, C.M. Loss of the Mediator subunit Med20 affects transcription of tRNA and other non-coding RNA genes in fission yeast. Biochim. Biophys. Acta 2016, 1859, 339–347. [Google Scholar] [CrossRef]
- Uthe, H.; Vanselow, J.T.; Schlosser, A. Proteomic Analysis of the Mediator Complex Interactome in Saccharomyces cerevisiae. Sci. Rep. 2017, 7, 43584. [Google Scholar] [CrossRef]
- Lai, Z.; Schluttenhofer, C.M.; Bhide, K.; Shreve, J.; Thimmapuram, J.; Lee, S.Y.; Yun, D.-J.; Mengiste, T. MED18 interaction with distinct transcription factors regulates multiple plant functions. Nat. Commun. 2014, 5, 3064. [Google Scholar] [CrossRef] [Green Version]
- Liao, C.-J.; Lai, Z.; Lee, S.; Yun, D.J.; Mengiste, T. Arabidopsis HOOKLESS1 regulates responses to pathogens and abscisic acid through interaction with MED18 and acetylation of WRKY33 and ABI5 chromatin. Plant Cell 2016, 28, 1662–1681. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Shi, C.; Fu, W.; Gu, X.; Qi, Z.; Xu, W.; Xia, G. Arabidopsis MED18 Interaction with RNA Pol IV and V Subunit NRPD2a in Transcriptional Regulation of Plant Immune Responses. Front. Plant Sci. 2021, 12, 692036. [Google Scholar] [CrossRef]
- Howe, G.A.; Major, I.T.; Koo, A.J. Modularity in Jasmonate Signaling for Multistress Resilience. Annu. Rev. Plant Biol. 2018, 69, 387–415. [Google Scholar] [CrossRef] [Green Version]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef]
- Breeze, E. Master MYCs: MYC2, the Jasmonate Signaling “Master Switch”. Plant Cell 2019, 31, 9–10. [Google Scholar] [CrossRef] [Green Version]
- Du, M.; Zhao, J.; Tzeng, D.T.; Liu, Y.; Deng, L.; Yang, T.; Zhai, Q.; Wu, F.; Huang, Z.; Zhou, M.; et al. MYC2 Orchestrates a Hierarchical Transcriptional Cascade That Regulates Jasmonate-Mediated Plant Immunity in Tomato. Plant Cell 2017, 29, 1883–1906. [Google Scholar] [CrossRef] [Green Version]
- Chico, J.M.; Chini, A.; Fonseca, S.; Solano, R. JAZ repressors set the rhythm in jasmonate signaling. Curr. Opin. Plant Biol. 2008, 11, 486–494. [Google Scholar] [CrossRef]
- Chini, A.; Fonseca, S.; Fernández, G.; Adie, B.; Chico, J.M.; Lorenzo, O.; García-Casado, G.; López-Vidriero, I.; Lozano, F.M.; Ponce, M.R.; et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007, 448, 666–671. [Google Scholar] [CrossRef]
- Pauwels, L.; Barbero, G.F.; Geerinck, J.; Tilleman, S.; Grunewald, W.; Pérez, A.C.; Chico, J.M.; Bossche, R.V.; Sewell, J.; Gil, E.; et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 2010, 464, 788–791. [Google Scholar] [CrossRef] [Green Version]
- Katsir, L.; Schilmiller, A.L.; Staswick, P.E.; He, S.Y.; Howe, G.A. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. USA 2008, 105, 7100–7105. [Google Scholar] [CrossRef] [Green Version]
- Sheard, L.B.; Tan, X.; Mao, H.; Withers, J.; Ben-Nissan, G.; Hinds, T.R.; Kobayashi, Y.; Hsu, F.-F.; Sharon, M.; Browse, J.; et al. Jasmonate perception by inositol-phosphate-potentiated COI1–JAZ co-receptor. Nature 2010, 468, 400–405. [Google Scholar] [CrossRef] [PubMed]
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.; Nomura, K.; He, S.Y.; Howe, G.A.; Browse, J. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 2007, 448, 661–665. [Google Scholar] [CrossRef] [PubMed]
- Leydon, A.R.; Wang, W.; Gala, H.P.; Gilmour, S.; Juarez-Solis, S.; Zahler, M.L.; Zemke, J.E.; Zheng, N.; Nemhauser, J.L. Repression by the Arabidopsis TOPLESS corepressor requires association with the core mediator complex. eLife 2021, 10, e66739. [Google Scholar] [CrossRef] [PubMed]
- Causier, B.; Ashworth, M.; Guo, W.; Davies, B. The TOPLESS Interactome: A Framework for Gene Repression in Arabidopsis. Plant Physiol. 2011, 158, 423–438. [Google Scholar] [CrossRef] [Green Version]
- Causier, B.; Lloyd, J.; Stevens, L.; Davies, B. TOPLESS co-repressor interactions and their evolutionary conservation in plants. Plant Signal. Behav. 2012, 7, 325–328. [Google Scholar] [CrossRef] [Green Version]
- Martin-Arevalillo, R.; Nanao, M.H.; Larrieu, A.; Vinos-Poyo, T.; Mast, D.; Galvan-Ampudia, C.; Brunoud, G.; Vernoux, T.; Dumas, R.; Parcy, F. Structure of the Arabidopsis TOPLESS corepressor provides insight into the evolution of transcriptional repression. Proc. Natl. Acad. Sci. USA 2017, 114, 8107–8112. [Google Scholar] [CrossRef] [Green Version]
- Ito, J.; Fukaki, H.; Onoda, M.; Li, L.; Li, C.; Tasaka, M.; Furutani, M. Auxin-dependent compositional change in Mediator in ARF7- and ARF19-mediated transcription. Proc. Natl. Acad. Sci. USA 2016, 113, 6562–6567. [Google Scholar] [CrossRef] [Green Version]
- Zhai, Q.; Deng, L.; Li, C. Mediator subunit MED25: At the nexus of jasmonate signaling. Curr. Opin. Plant Biol. 2020, 57, 78–86. [Google Scholar] [CrossRef]
- Chen, R.; Jiang, H.; Li, L.; Zhai, Q.; Qi, L.; Zhou, W.; Liu, X.; Li, H.; Zheng, W.; Sun, J.; et al. The Arabidopsis Mediator Subunit MED25 Differentially Regulates Jasmonate and Abscisic Acid Signaling through Interacting with the MYC2 and ABI5 Transcription Factors. Plant Cell 2012, 24, 2898–2916. [Google Scholar] [CrossRef] [Green Version]
- Çevik, V.; Kidd, B.N.; Zhang, P.; Hill, C.; Kiddle, S.; Denby, K.; Holub, E.B.; Cahill, D.; Manners, J.M.; Schenk, P.; et al. MEDIATOR25 Acts as an Integrative Hub for the Regulation of Jasmonate-Responsive Gene Expression in Arabidopsis. Plant Physiol. 2012, 160, 541–555. [Google Scholar] [CrossRef] [Green Version]
- Ou, B.; Yin, K.-Q.; Liu, S.-N.; Yang, Y.; Gu, T.; Hui, J.M.W.; Zhang, L.; Miao, J.; Kondou, Y.; Matsui, M.; et al. A High-Throughput Screening System for Arabidopsis Transcription Factors and Its Application to Med25-Dependent Transcriptional Regulation. Mol. Plant 2011, 4, 546–555. [Google Scholar] [CrossRef]
- You, Y.; Zhai, Q.; An, C.; Li, C. LEUNIG_HOMOLOG Mediates MYC2-Dependent Transcriptional Activation in Cooperation with the Coactivators HAC1 and MED25. Plant Cell 2019, 31, 2187–2205. [Google Scholar] [CrossRef] [Green Version]
- Chung, H.S.; Cooke, T.F.; DePew, C.L.; Patel, L.C.; Ogawa, N.; Kobayashi, Y.; Howe, G.A. Alternative splicing expands the repertoire of dominant JAZ repressors of jasmonate signaling. Plant J. 2010, 63, 613–622. [Google Scholar] [CrossRef] [Green Version]
- Mach, J. Alternative Splicing Produces a JAZ Protein That Is Not Broken Down in Response to Jasmonic Acid. Plant Cell 2009, 21, 14. [Google Scholar] [CrossRef] [Green Version]
- Wu, F.; Deng, L.; Zhai, Q.; Zhao, J.; Chen, Q.; Li, C. Mediator Subunit MED25 Couples Alternative Splicing of JAZ Genes with Fine-Tuning of Jasmonate Signaling. Plant Cell 2019, 32, 429–448. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Du, M.; Deng, L.; Shen, J.; Fang, M.; Chen, Q.; Lu, Y.; Wang, Q.; Li, C.; Zhai, Q. MYC2 Regulates the Termination of Jasmonate Signaling via an Autoregulatory Negative Feedback Loop. Plant Cell 2019, 31, 106–127. [Google Scholar] [CrossRef] [Green Version]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [Green Version]
- Durrant, W.E.; Dong, X. Systemic acquired resistance. Annu. Rev. Phytopathol. 2004, 42, 185–209. [Google Scholar] [CrossRef]
- Zipfel, C.; Kunze, G.; Chinchilla, D.; Caniard, A.; Jones, J.D.G.; Boller, T.; Felix, G. Perception of the Bacterial PAMP EF-Tu by the Receptor EFR Restricts Agrobacterium-Mediated Transformation. Cell 2006, 125, 749–760. [Google Scholar] [CrossRef]
- Seo, J.S.; Sun, H.-X.; Park, B.S.; Huang, C.-H.; Yeh, S.-D.; Jung, C.; Chua, N.-H. ELF18-INDUCED LONG-NONCODING RNA Associates with Mediator to Enhance Expression of Innate Immune Response Genes in Arabidopsis. Plant Cell 2017, 29, 1024–1038. [Google Scholar] [CrossRef] [Green Version]
- Seo, J.S.; Diloknawarit, P.; Park, B.S.; Chua, N.-H. ELF18-INDUCED LONG NONCODING RNA 1 evicts fibrillarin from mediator subunit to enhance PATHOGENESIS-RELATED GENE 1 (PR1) expression. New Phytol. 2019, 221, 2067–2079. [Google Scholar] [CrossRef]
- Caillaud, M.-C.; Asai, S.; Rallapalli, G.; Piquerez, S.; Fabro, G.; Jones, J.D.G. A Downy Mildew Effector Attenuates Salicylic Acid-Triggered Immunity in Arabidopsis by Interacting with the Host Mediator Complex. PLoS Biol. 2013, 11, e1001732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, J.; Sun, Y.; Orduna, A.R.; Jetter, R.; Li, X. The Mediator kinase module serves as a positive regulator of salicylic acid accumulation and systemic acquired resistance. Plant J. 2019, 98, 842–852. [Google Scholar] [CrossRef] [PubMed]
- Dong, X. NPR1, all things considered. Curr. Opin. Plant Biol. 2004, 7, 547–552. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Sun, T.; Ao, K.; Peng, Y.; Zhang, Y.; Li, X.; Zhang, Y. Opposite Roles of Salicylic Acid Receptors NPR1 and NPR3/NPR4 in Transcriptional Regulation of Plant Immunity. Cell 2018, 173, 1454–1467.e15. [Google Scholar] [CrossRef]
- Manohar, M.; Tian, M.; Moreau, M.; Park, S.-W.; Choi, H.W.; Fei, Z.; Friso, G.; Asif, M.; Manosalva, P.; von Dahl, C.C.; et al. Identification of multiple salicylic acid-binding proteins using two high throughput screens. Front. Plant Sci. 2015, 5, 777. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Zhang, D.; Chu, J.Y.; Boyle, P.; Wang, Y.; Brindle, I.D.; De Luca, V.; Després, C. The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 2012, 1, 639–647. [Google Scholar] [CrossRef] [Green Version]
- Després, C.; Chubak, C.; Rochon, A.; Clark, R.; Bethune, T.; Desveaux, D.; Fobert, P.R. The Arabidopsis NPR1 Disease Resistance Protein Is a Novel Cofactor That Confers Redox Regulation of DNA Binding Activity to the Basic Domain/Leucine Zipper Transcription Factor TGA1. Plant Cell 2003, 15, 2181–2191. [Google Scholar] [CrossRef] [Green Version]
- Despres, C.; DeLong, C.; Glaze, S.; Liu, E.; Fobert, P.R. The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 2000, 12, 279–290. [Google Scholar] [CrossRef] [Green Version]
- Yu, D.; Chen, C.; Chen, Z. Evidence for an important role of WRKY DNA binding proteins in the regulation of NPR1 gene expression. Plant Cell 2001, 13, 1527–1540. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Mohan, R.; Zhang, Y.; Li, M.; Chen, H.; Palmer, I.A.; Chang, M.; Qi, G.; Spoel, S.H.; Mengiste, T.; et al. NPR1 Promotes Its Own and Target Gene Expression in Plant Defense by Recruiting CDK8. Plant Physiol. 2019, 181, 289–304. [Google Scholar] [CrossRef]
- Van Kan, J.A.L. Licensed to kill: The lifestyle of a necrotrophic plant pathogen. Trends Plant Sci. 2006, 11, 247–253. [Google Scholar] [CrossRef]
- Govrin, E.M.; Levine, A. The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Curr. Biol. 2000, 10, 751–757. [Google Scholar] [CrossRef] [Green Version]
- Ferrari, S.; Plotnikova, J.M.; De Lorenzo, G.; Ausubel, F.M. Arabidopsis local resistance to Botrytis cinerea involves salicylic acid and camalexin and requires EDS4 and PAD2, but not SID2, EDS5 or PAD4. Plant J. Cell Mol. Biol. 2003, 35, 193–205. [Google Scholar] [CrossRef] [Green Version]
- Penninckx, I.A.; Thomma, B.P.; Buchala, A.; Metraux, J.P.; Broekaert, W.F. Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 1998, 10, 2103–2113. [Google Scholar] [CrossRef] [Green Version]
- Thomma, B.P.H.J.; Eggermont, K.; Penninckx, I.A.M.A.; Mauch-Mani, B.; Vogelsang, R.; Cammue, B.P.A.; Broekaert, W.F. Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. USA 1998, 95, 15107–15111. [Google Scholar] [CrossRef] [Green Version]
- Thomma, B.P.; Eggermont, K.; Tierens, K.F.M.-J.; Broekaert, W.F. Requirement of Functional Ethylene-Insensitive 2Gene for Efficient Resistance of Arabidopsis to Infection by Botrytis cinerea. Plant Physiol. 1999, 121, 1093–1101. [Google Scholar] [CrossRef] [Green Version]
- Thomma, B.P.H.J.; Nelissen, I.; Eggermont, K.; Broekaert, W.F. Deficiency in phytoalexin production causes enhanced susceptibility of Arabidopsis thaliana to the fungus Alternaria brassicicola. Plant J. 1999, 19, 163–171. [Google Scholar] [CrossRef]
- Kidd, B.N.; Edgar, C.I.; Kumar, K.K.; Aitken, E.A.; Schenk, P.M.; Manners, J.M.; Kazan, K. The Mediator Complex Subunit PFT1 Is a Key Regulator of Jasmonate-Dependent Defense in Arabidopsis. Plant Cell 2009, 21, 2237–2252. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Yao, J.; Du, X.; Zhang, Y.; Sun, Y.; Rollins, J.A.; Mou, Z. The Arabidopsis Mediator Complex Subunit16 Is a Key Component of Basal Resistance against the Necrotrophic Fungal Pathogen Sclerotinia sclerotiorum. Plant Physiol. 2015, 169, 856–872. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Yang, R.; Chen, H. The Arabidopsis thaliana Mediator subunit MED8 regulates plant immunity to Botrytis Cinerea through interacting with the basic helix-loop-helix (bHLH) transcription factor FAMA. PLoS ONE 2018, 13, e0193458. [Google Scholar] [CrossRef] [Green Version]
- Feng, Q.; Li, L.; Liu, Y.; Shao, X.; Li, X. Jasmonate regulates the FAMA/mediator complex subunit 8-THIOGLUCOSIDE GLUCOHYDROLASE 1 cascade and myrosinase activity. Plant Physiol. 2021, 187, 963–980. [Google Scholar] [CrossRef]
- Mao, G.; Meng, X.; Liu, Y.; Zheng, Z.; Chen, Z.; Zhang, S. Phosphorylation of a WRKY Transcription Factor by Two Pathogen-Responsive MAPKs Drives Phytoalexin Biosynthesis in Arabidopsis. Plant Cell 2011, 23, 1639–1653. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Z.; Abu Qamar, S.; Chen, Z.; Mengiste, T. Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 2006, 48, 592–605. [Google Scholar] [CrossRef] [PubMed]
- Dhawan, R.; Luo, H.; Foerster, A.M.; AbuQamar, S.; Du, H.-N.; Briggs, S.D.; Scheid, O.M.; Mengiste, T. HISTONE MONOUBIQUITINATION1 Interacts with a Subunit of the Mediator Complex and Regulates Defense against Necrotrophic Fungal Pathogens in Arabidopsis. Plant Cell 2009, 21, 1000–1019. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Schluttenhoffer, C.M.; Wang, P.; Fu, F.; Thimmapuram, J.; Zhu, J.-K.; Lee, S.Y.; Yun, D.-J.; Mengiste, T. CYCLIN-DEPENDENT KINASE8 Differentially Regulates Plant Immunity to Fungal Pathogens through Kinase-Dependent and -Independent Functions in Arabidopsis. Plant Cell 2014, 26, 4149–4170. [Google Scholar] [CrossRef] [Green Version]
- Chen, K.; Li, G.; Bressan, R.A.; Song, C.; Zhu, J.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2019, 62, 25–54. [Google Scholar] [CrossRef] [Green Version]
- Fàbregas, N.; Yoshida, T.; Fernie, A.R. Role of Raf-like kinases in SnRK2 activation and osmotic stress response in plants. Nat. Commun. 2020, 11, 6184. [Google Scholar] [CrossRef]
- Katsuta, S.; Masuda, G.; Bak, H.; Shinozawa, A.; Kamiyama, Y.; Umezawa, T.; Takezawa, D.; Yotsui, I.; Taji, T.; Sakata, Y. Arabidopsis Raf-like kinases act as positive regulators of subclass III SnRK2 in osmostress signaling. Plant J. 2020, 103, 634–644. [Google Scholar] [CrossRef]
- Soma, F.; Takahashi, F.; Suzuki, T.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Plant Raf-like kinases regulate the mRNA population upstream of ABA-unresponsive SnRK2 kinases under drought stress. Nat. Commun. 2020, 11, 1373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, P.; Chong, L.; Wu, F.; Hsu, C.; Li, C.; Zhu, J.; Zhu, Y. Mediator tail module subunits MED16 and MED25 differentially regulate abscisic acid signaling in Arabidopsis. J. Integr. Plant Biol. 2020, 63, 802–815. [Google Scholar] [CrossRef] [PubMed]
- Bossi, F.; Cordoba, E.; Dupré, P.; Mendoza, M.S.; Román, C.S.; León, P. The Arabidopsis ABA-INSENSITIVE (ABI) 4 factor acts as a central transcription activator of the expression of its own gene, and for the induction of ABI5 and SBE2.2 genes during sugar signaling. Plant J. 2009, 59, 359–374. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Huang, P.; Guo, P.; Chong, L.; Yu, G.; Sun, X.; Hu, T.; Li, Y.; Hsu, C.; Tang, K.; et al. CDK8 is associated with RAP2.6 and SnRK2.6 and positively modulates abscisic acid signaling and drought response in Arabidopsis. New Phytol. 2020, 228, 1573–1590. [Google Scholar] [CrossRef]
- Zhu, Q.; Zhang, J.; Gao, X.; Tong, J.; Xiao, L.; Li, W.; Zhang, H. The Arabidopsis AP2/ERF transcription factor RAP2.6 participates in ABA, salt and osmotic stress responses. Gene 2010, 457, 1–12. [Google Scholar] [CrossRef]
- Warren, G.; McKown, R.; Marin, A.; Teutonico, R. Isolation of Mutations Affecting the Development of Freezing Tolerance in Arabidopsis thaliana (L.) Heynh. Plant Physiol. 1996, 111, 1011–1019. [Google Scholar] [CrossRef] [Green Version]
- Knight, H.; Mugford, S.G.; Ülker, B.; Gao, D.; Thorlby, G.; Knight, M.R. Identification of SFR6, a key component in cold acclimation acting post-translationally on CBF function. Plant J. 2009, 58, 97–108. [Google Scholar] [CrossRef]
- Knight, H.; Veale, E.L.; Warren, G.J.; Knight, M.R. The sfr6 Mutation in Arabidopsis Suppresses Low-Temperature Induction of Genes Dependent on the CRT/DRE Sequence Motif. Plant Cell 1999, 11, 875–886. [Google Scholar] [CrossRef] [Green Version]
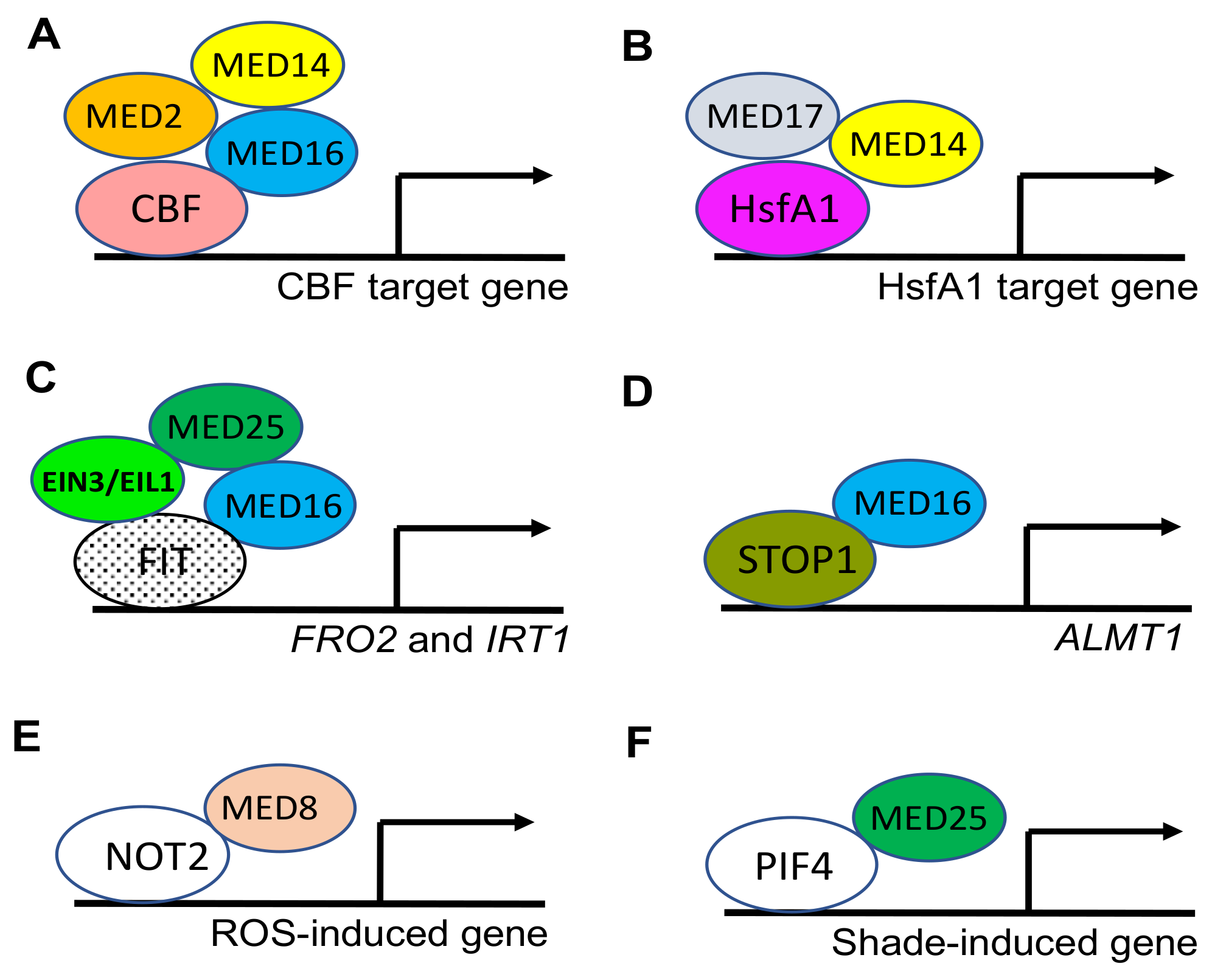
- Hemsley, P.A.; Hurst, C.H.; Kaliyadasa, E.; Lamb, R.; Knight, M.R.; De Cothi, E.A.; Steele, J.F.; Knight, H. The Arabidopsis Mediator Complex Subunits MED16, MED14, and MED2 Regulate Mediator and RNA Polymerase II Recruitment to CBF-Responsive Cold-Regulated Genes. Plant Cell 2014, 26, 465–484. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.; Dominguez-Ferreras, A.; Kaliyadasa, E.; Huang, W.-J.; Antony, E.; Stevenson, T.; Lehmann, S.; Schäfer, P.; Knight, M.R.; Ntoukakis, V.; et al. Mediator Subunits MED16, MED14, and MED2 Are Required for Activation of ABRE-Dependent Transcription in Arabidopsis. Front. Plant Sci. 2021, 12, 649720. [Google Scholar] [CrossRef]
- Ohama, N.; Moo, T.L.; Chua, N. Differential requirement of MED14/17 recruitment for activation of heat inducible genes. New Phytol. 2020, 229, 3360–3376. [Google Scholar] [CrossRef]
- Guo, M.; Liu, J.-H.; Ma, X.; Luo, D.-X.; Gong, Z.-H.; Lu, M.-H. The Plant Heat Stress Transcription Factors (HSFs): Structure, Regulation, and Function in Response to Abiotic Stresses. Front. Plant Sci. 2016, 7, 114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Von Koskull-Döring, P.; Scharf, K.-D.; Nover, L. The diversity of plant heat stress transcription factors. Trends Plant Sci. 2007, 12, 452–457. [Google Scholar] [CrossRef]
- Connolly, E.L.; Fett, J.; Guerinot, M.L. Expression of the IRT1 Metal Transporter Is Controlled by Metals at the Levels of Transcript and Protein Accumulation. Plant Cell 2002, 14, 1347–1357. [Google Scholar] [CrossRef] [Green Version]
- Eide, D.; Broderius, M.; Fett, J.; Guerinot, M.L. A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc. Natl. Acad. Sci. USA 1996, 93, 5624–5628. [Google Scholar] [CrossRef] [Green Version]
- Robinson, N.J.; Procter, C.M.; Connolly, E.L.; Guerinot, M.L. A ferric-chelate reductase for iron uptake from soils. Nature 1999, 397, 694–697. [Google Scholar] [CrossRef]
- Wang, N.; Cui, Y.; Liu, Y.; Fan, H.; Du, J.; Huang, Z.; Yuan, Y.; Wu, H.; Ling, H.-Q. Requirement and Functional Redundancy of Ib Subgroup bHLH Proteins for Iron Deficiency Responses and Uptake in Arabidopsis thaliana. Mol. Plant 2013, 6, 503–513. [Google Scholar] [CrossRef] [Green Version]
- Yuan, Y.; Wu, H.; Wang, N.; Li, J.; Zhao, W.; Du, J.; Wang, D.; Ling, H.-Q. FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res. 2008, 18, 385–397. [Google Scholar] [CrossRef]
- Yuan, Y.X.; Zhang, J.; Wang, D.W.; Ling, H.Q. AtbHLH29 of Arabidopsis thaliana is a functional ortholog of tomato FER involved in controlling iron acquisition in strategy I plants. Cell Res. 2005, 15, 613–621. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Wu, H.; Wang, N.; Fan, H.; Chen, C.; Cui, Y.; Liu, H.; Ling, H. Mediator subunit 16 functions in the regulation of iron uptake gene expression in Arabidopsis. New Phytol. 2014, 203, 770–783. [Google Scholar] [CrossRef]
- Yang, Y.; Ou, B.; Zhang, J.; Si, W.; Gu, H.; Qin, G.; Qu, L.-J. The Arabidopsis Mediator subunit MED16 regulates iron homeostasis by associating with EIN3/EIL1 through subunit MED25. Plant J. 2014, 77, 838–851. [Google Scholar] [CrossRef]
- Lingam, S.; Mohrbacher, J.; Brumbarova, T.; Potuschak, T.; Fink-Straube, C.; Blondet, E.; Genschik, P.; Bauer, P. Interaction between the bHLH Transcription Factor FIT and ETHYLENE INSENSITIVE3/ETHYLENE INSENSITIVE3-LIKE1 Reveals Molecular Linkage between the Regulation of Iron Acquisition and Ethylene Signaling in Arabidopsis. Plant Cell 2011, 23, 1815–1829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raya-González, J.; Ojeda-Rivera, J.O.; Mora-Macias, J.; Oropeza-Aburto, A.; Ruiz-Herrera, L.F.; López-Bucio, J.; Herrera-Estrella, L. MEDIATOR16 orchestrates local and systemic responses to phosphate scarcity in Arabidopsis roots. New Phytol. 2020, 229, 1278–1288. [Google Scholar] [CrossRef]
- Ward, J.T.; Lahner, B.; Yakubova, E.; Salt, D.E.; Raghothama, K.G. The Effect of Iron on the Primary Root Elongation of Arabidopsis during Phosphate Deficiency. Plant Physiol. 2008, 147, 1181–1191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balzergue, C.; Dartevelle, T.; Godon, C.; Laugier, E.; Meisrimler, C.; Teulon, J.-M.; Creff, A.; Bissler, M.; Brouchoud, C.; Hagège, A. Low phosphate activates STOP1-ALMT1 to rapidly inhibit root cell elongation. Nat. Commun. 2017, 8, 15300. [Google Scholar] [CrossRef] [Green Version]
- He, H.; Denecker, J.; Van Der Kelen, K.; Willems, P.; Pottie, R.; Phua, S.Y.; Hannah, M.A.; Vertommen, D.; Van Breusegem, F.; Mhamdi, A. The Arabidopsis mediator complex subunit 8 regulates oxidative stress responses. Plant Cell 2021, 33, 2032–2057. [Google Scholar] [CrossRef]
- Hagkarim, N.C.; Grand, R. The Regulatory Properties of the Ccr4–Not Complex. Cells 2020, 9, 2379. [Google Scholar] [CrossRef]
- Sun, W.; Han, H.; Deng, L.; Sun, C.; Xu, Y.; Lin, L.; Ren, P.; Zhao, J.; Zhai, Q.; Li, C. Mediator Subunit MED25 Physically Interacts with PHYTOCHROME INTERACTING FACTOR4 to Regulate Shade-Induced Hypocotyl Elongation in Tomato. Plant Physiol. 2020, 184, 1549–1562. [Google Scholar] [CrossRef]
- Pham, V.N.; Kathare, P.K.; Huq, E. Phytochromes and Phytochrome Interacting Factors. Plant Physiol. 2017, 176, 1025–1038. [Google Scholar] [CrossRef] [Green Version]
- Hornitschek, P.; Kohnen, M.V.; Lorrain, S.; Rougemont, J.; Ljung, K.; López-Vidriero, I.; Franco-Zorrilla, J.M.; Solano, R.; Trevisan, M.; Pradervand, S.; et al. Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 2012, 71, 699–711. [Google Scholar] [CrossRef] [Green Version]
- Umezawa, T. The cinnamate/monolignol pathway. Phytochem. Rev. 2009, 9, 1–17. [Google Scholar] [CrossRef]
- Liu, Q.; Luo, L.; Zheng, L. Lignins: Biosynthesis and Biological Functions in Plants. Int. J. Mol. Sci. 2018, 19, 335. [Google Scholar] [CrossRef] [Green Version]
- Ruegger, M.; Chapple, C. Mutations That Reduce Sinapoylmalate Accumulation in Arabidopsis thaliana Define Loci with Diverse Roles in Phenylpropanoid Metabolism. Genetics 2001, 159, 1741–1749. [Google Scholar] [CrossRef] [PubMed]
- Stout, J.; Romero-Severson, E.; Ruegger, M.O.; Chapple, C. Semidominant Mutations in Reduced Epidermal Fluorescence 4 Reduce Phenylpropanoid Content in Arabidopsis. Genetics 2008, 178, 2237–2251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonawitz, N.D.; Soltau, W.L.; Blatchley, M.R.; Powers, B.L.; Hurlock, A.K.; Seals, L.A.; Weng, J.-K.; Stout, J.; Chapple, C. REF4 and RFR1, Subunits of the Transcriptional Coregulatory Complex Mediator, Are Required for Phenylpropanoid Homeostasis in Arabidopsis. J. Biol. Chem. 2012, 287, 5434–5445. [Google Scholar] [CrossRef] [Green Version]
- Bonawitz, N.D.; Kim, J.I.; Tobimatsu, Y.; Ciesielski, P.N.; Anderson, N.A.; Ximenes, E.; Maeda, J.; Ralph, J.; Donohoe, B.S.; Ladisch, M.; et al. Disruption of Mediator rescues the stunted growth of a lignin-deficient Arabidopsis mutant. Nature 2014, 509, 376–380. [Google Scholar] [CrossRef]
- Mottiar, Y.; Vanholme, R.; Boerjan, W.; Ralph, J.; Mansfield, S. Designer lignins: Harnessing the plasticity of lignification. Curr. Opin. Biotechnol. 2016, 37, 190–200. [Google Scholar] [CrossRef] [Green Version]
- Anderson, N.A.; Bonawitz, N.D.; Nyffeler, K.; Chapple, C. Loss of ferulate 5-hydroxylase leads to Mediator-dependent inhibition of soluble phenylpropanoid biosynthesis in Arabidopsis. Plant Physiol. 2015, 169, 1557–1567. [Google Scholar] [CrossRef] [Green Version]
- Humphreys, J.M.; Hemm, M.R.; Chapple, C. New routes for lignin biosynthesis defined by biochemical characterization of recombinant ferulate 5-hydroxylase, a multifunctional cytochrome P450-dependent monooxygenase. Proc. Natl. Acad. Sci. USA 1999, 96, 10045–10050. [Google Scholar] [CrossRef] [Green Version]
- Dolan, W.L.; Dilkes, B.P.; Stout, J.M.; Bonawitz, N.D.; Chapple, C. Mediator Complex Subunits MED2, MED5, MED16, and MED23 Genetically Interact in the Regulation of Phenylpropanoid Biosynthesis. Plant Cell 2017, 29, 3269–3285. [Google Scholar] [CrossRef]
- Mao, X.; Kim, J.I.; Wheeler, M.T.; Heintzelman, A.K.; Weake, V.M.; Chapple, C. Mutation of Mediator subunit CDK 8 counteracts the stunted growth and salicylic acid hyperaccumulation phenotypes of an Arabidopsis MED 5 mutant. New Phytol. 2019, 223, 233–245. [Google Scholar] [CrossRef]
- Davoine, C.; Abreu, I.N.; Khajeh, K.; Blomberg, J.; Kidd, B.N.; Kazan, K.; Schenk, P.M.; Gerber, L.; Nilsson, O.; Moritz, T.; et al. Functional metabolomics as a tool to analyze Mediator function and structure in plants. PLoS ONE 2017, 12, e0179640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dolan, W.L.; Chapple, C. Transcriptome Analysis of Four Arabidopsis thaliana Mediator Tail Mutants Reveals Overlapping and Unique Functions in Gene Regulation. G3 Genes Genomes Genet. 2018, 8, 3093–3108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, X.; Weake, V.M.; Chapple, C. Mediator function in plant metabolism revealed by large-scale biology. J. Exp. Bot. 2019, 70, 5995–6003. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Du, X.; Mou, Z. The Mediator Complex Subunits MED14, MED15, and MED16 Are Involved in Defense Signaling Crosstalk in Arabidopsis. Front. Plant Sci. 2016, 7, 1947. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Wang, C.; Zhang, Y.; Sun, Y.; Mou, Z. The Arabidopsis Mediator Complex Subunit16 Positively Regulates Salicylate-Mediated Systemic Acquired Resistance and Jasmonate/Ethylene-Induced Defense Pathways. Plant Cell 2012, 24, 4294–4309. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Yao, J.; Zhang, Y.; Sun, Y.; Mou, Z. The Arabidopsis Mediator complex subunits MED14/SWP and MED16/SFR6/IEN1 differentially regulate defense gene expression in plant immune responses. Plant J. 2013, 75, 484–497. [Google Scholar] [CrossRef] [Green Version]


| Moule | Subunit | Gene Identifier | Homolog 1 | Roles in Plant Adaptive Responses 2 | ||
|---|---|---|---|---|---|---|
| Yeast | Human | Interacting Partners | Regulated Processes | |||
| Head | MED6 | At3g21350 | + | + | ||
| MED8 | At2g03070 | + | + | MED25, FAMA, NOT2 | Plant defense, ROS response | |
| MED11 | At3g01435 | + | + | |||
| MED17 | At5g20170 | + | + | HsfA1 | Heat response | |
| MED18 | At2g22370 | + | + | NRPD2a, YY1, HLS1, ABI4 | Plant defense, ABA signaling | |
| MED19a | At5g12230 | + | + | ELENA1, HaRxL44 | Plant–pathogen interactions | |
| MED19b | At5g19480 | + | + | Unknown | Unknown | |
| MED20a | At2g28230 | + | + | Unknown | Unknown | |
| MED20b | At4g09070 | + | + | Unknown | Unknown | |
| MED20c | At2g28020 | + | + | Unknown | Unknown | |
| MED22a | At1g16430 | + | + | Unknown | Unknown | |
| MED22b | At1g07950 | + | + | Unknown | Unknown | |
| MED28 | At3g52860 | − | + | Unknown | Unknown | |
| MED30 | At5g63480 | − | + | Unknown | Unknown | |
| Middle | MED1 | At2g15890 | + | + | Unknown | Unknown |
| MED4 | At5g02850 | + | + | Unknown | Unknown | |
| MED7a | At5g03220 | + | + | Unknown | Unknown | |
| MED7b | At5g03500 | + | + | Unknown | Unknown | |
| MED9 | At1g55080 | + | + | Unknown | Unknown | |
| MED10a | At5g41910 | + | + | TPL | JA signaling | |
| MED10b | At1g26665 | + | + | Unknown | Unknown | |
| MED14 | At3g04740 | + | + | MED2. MED16, HsfA1 | Cold response, heat response, plant defense | |
| MED21 | At4g04780 | + | + | TPL, HUB1 | Plant defense | |
| MED26a | At3g10820 | − | + | Unknown | Phenylpropanoid biosynthesis | |
| MED26b | At5g05140 | − | + | Unknown | Unknown | |
| MED26c | At5g09850 | − | + | Unknown | Unknown | |
| MED31 | At5g19910 | + | + | Unknown | Unknown | |
| Tail | MED2 | At1g11760 | + | + | CBFs | Cold response, phenylpropanoid biosynthesis |
| MED3 | At3g09180 | + | + | Unknown | Unknown | |
| MED5a | At3g23590 | + | + | Unknown | Phenylpropanoid biosynthesis | |
| MED5b | At2g48110 | + | + | Unknown | Phenylpropanoid biosynthesis | |
| MED15a | At1g15780 | + | + | Unknown | Plant defense | |
| MED15b | At1g15770 | + | + | Unknown | Plant defense | |
| MED15c | At2g10440 | + | + | Unknown | Plant defense | |
| MED16 | At4g04920 | + | + | MED25, ABI5, CBFs, FIT, STOP1 | ABA signaling, cold responses, response to Fe and Pi limitation, phenylpropanoid biosynthesis | |
| MED23 | At1g23230 | − | + | Unknown | Phenylpropanoid biosynthesis | |
| MED25 | At1g25540 | − | + | COI1, JAZ, ORA59, ERF1, MYC2, HAC1, PRP39a, PRP40a, ABI5, EIN3/EIL1, PIF4 | JA signaling, plant defense, ABA signaling, ethylene signaling, shade response | |
| CKM | MED12 | At4g00450 | + | + | WRKY6, WRKY18. TGAs | SAR |
| MED13 | At1g55325 | + | + | TPL | SAR | |
| CDK8 | At5g63610 | + | + | WIN1, RAP2.6 | SAR, plant defense, ABA signaling, phenylpropanoid biosynthesis | |
| CYCCa | At5g48630 | + | + | Unknown | Unknown | |
| CYCCb | At5g48640 | + | + | Unknown | Unknown | |
| Unknown | MED34 | At1g31360 | − | − | Unknown | Unknown |
| MED35a | At1g44910 | − | − | Unknown | Unknown | |
| MED35b | At3g19670 | − | − | Unknown | Unknown | |
| MED35c | At3g19840 | − | − | Unknown | Unknown | |
| MED36a | At4g25630 | − | − | ELENA1 | SAR | |
| MED36b | At5g52470 | − | − | Unknown | Unknown | |
| MED37a | At5g28540 | − | − | Unknown | Unknown | |
| MED37b | At1g09080 | − | − | Unknown | Unknown | |
| MED37c | At3g12580 | − | − | Unknown | Unknown | |
| MED37d | At5g02500 | − | − | Unknown | Unknown | |
| MED37e | At5g42020 | − | − | Unknown | Unknown | |
| HAC1 | At1g79000 | − | − | MED25 | JA signaling | |
| HAC5 | At3g12980 | − | − | MED25 | JA signaling | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, J.; Yang, S.; Fan, B.; Zhu, C.; Chen, Z. The Mediator Complex: A Central Coordinator of Plant Adaptive Responses to Environmental Stresses. Int. J. Mol. Sci. 2022, 23, 6170. https://doi.org/10.3390/ijms23116170
Chen J, Yang S, Fan B, Zhu C, Chen Z. The Mediator Complex: A Central Coordinator of Plant Adaptive Responses to Environmental Stresses. International Journal of Molecular Sciences. 2022; 23(11):6170. https://doi.org/10.3390/ijms23116170
Chicago/Turabian StyleChen, Jialuo, Su Yang, Baofang Fan, Cheng Zhu, and Zhixiang Chen. 2022. "The Mediator Complex: A Central Coordinator of Plant Adaptive Responses to Environmental Stresses" International Journal of Molecular Sciences 23, no. 11: 6170. https://doi.org/10.3390/ijms23116170






