New Insight into Aspartate Metabolic Pathways in Populus: Linking the Root Responsive Isoenzymes with Amino Acid Biosynthesis during Incompatible Interactions of Fusarium solani
Abstract
:1. Introduction
2. Results
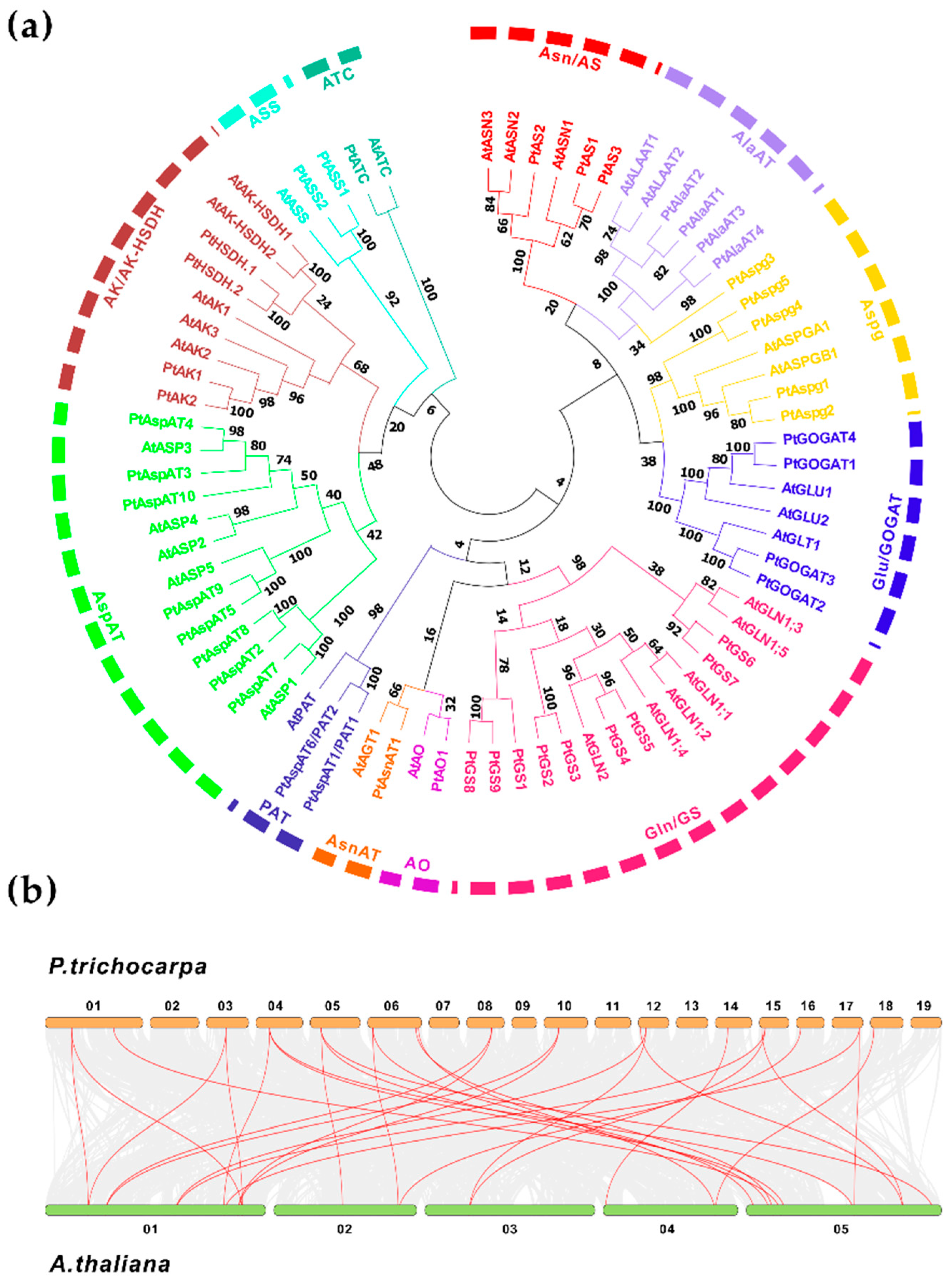
2.1. The Populus Candidate Isoenzymes Involved in Asp Metabolic Pathway
2.1.1. GS, AsnAT, AO, and PAT
2.1.2. GS, AsnAT, AO, and PAT
2.1.3. AspAT, AK/AK-HSDH, ASS, and ATC
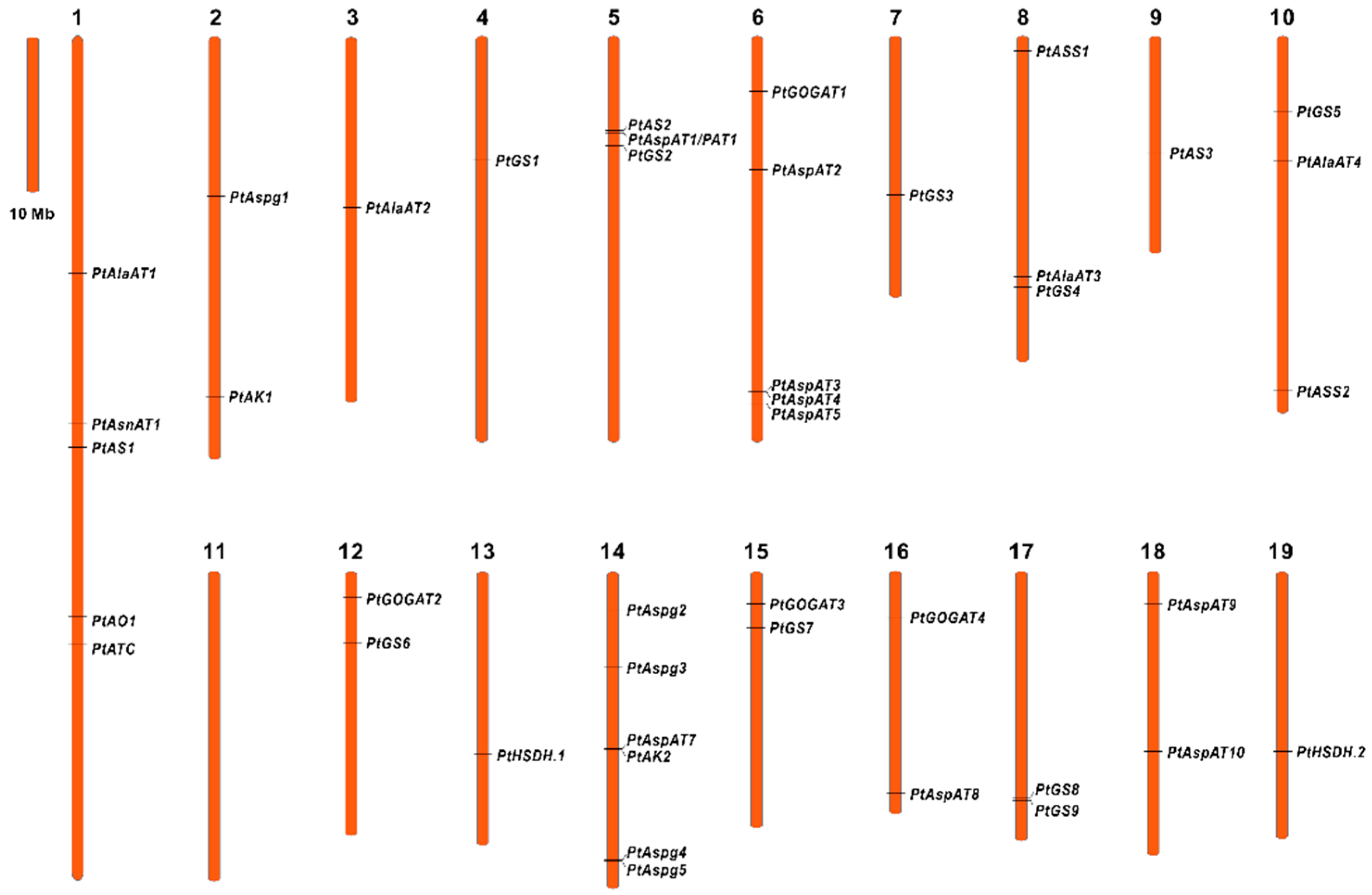
2.2. Identification of Enzyme Isogenes with Root-Specific Expression Patterns
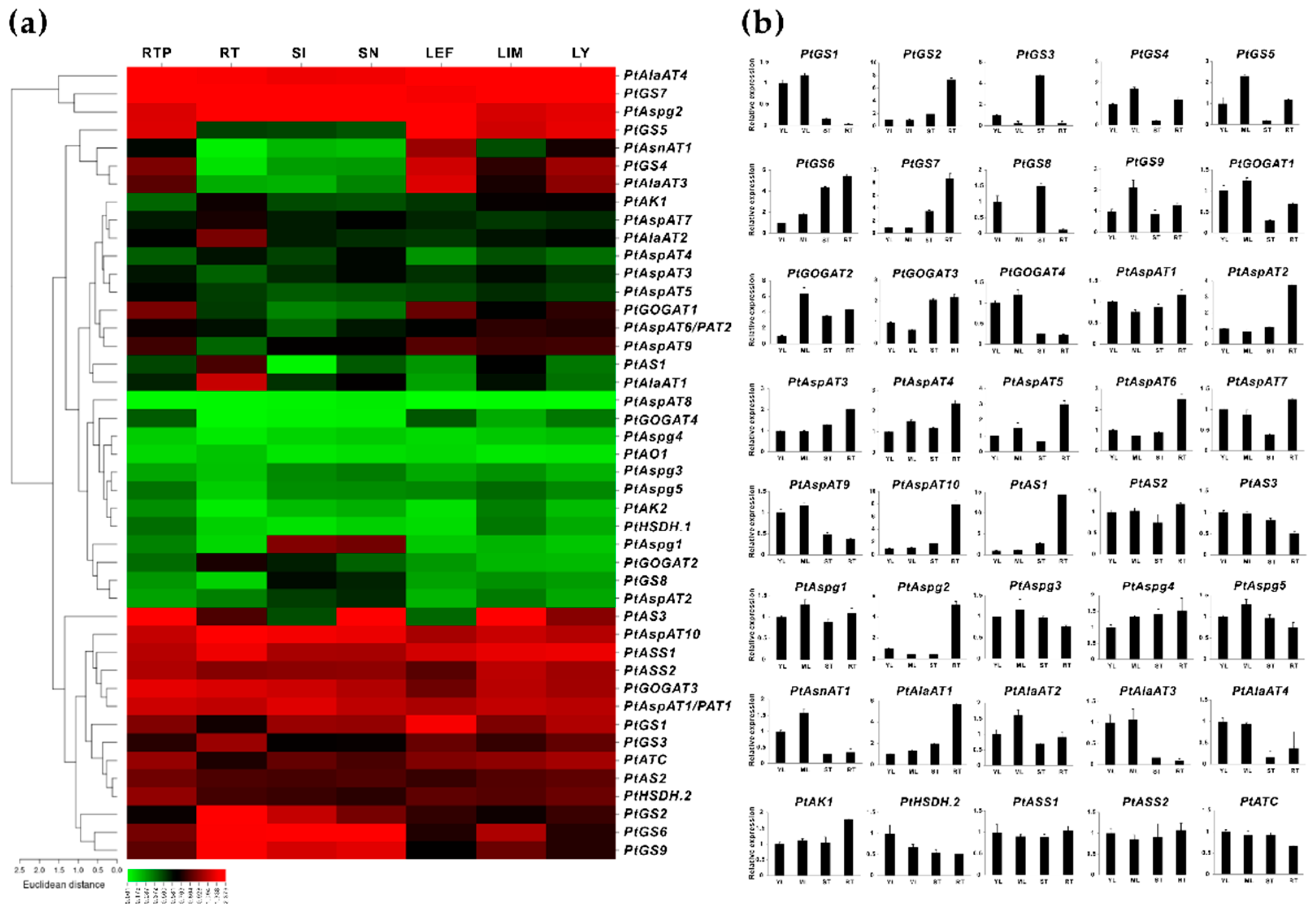
2.3. Transcriptomic Survey of the Fs Responsive Isogenes in Populus Roots
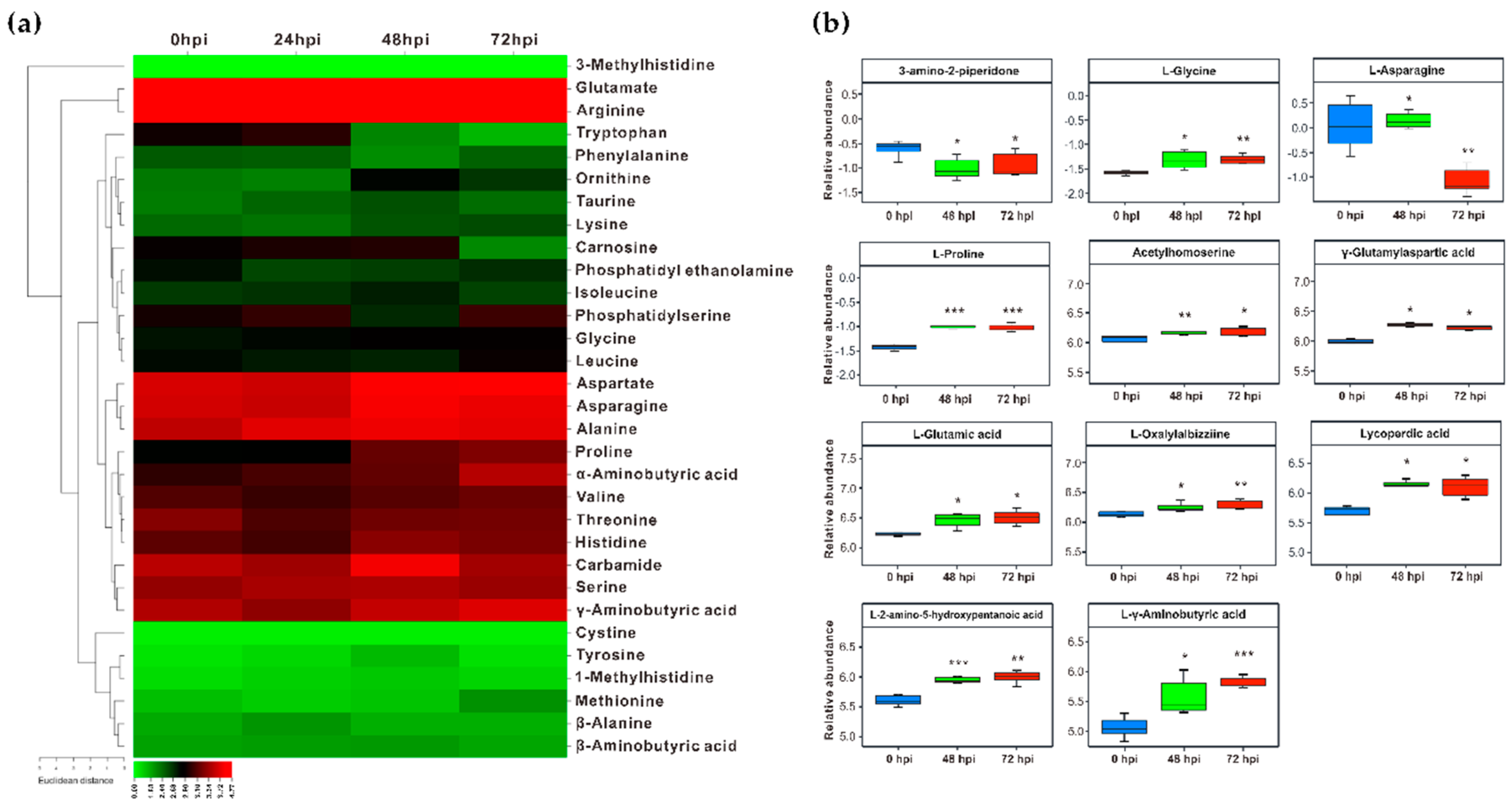
2.4. Perturbation of the Amino Acids and Derivatives in Fs Infected Roots
3. Discussion
3.1. The Functional Roles of Isoenzymes in the Asp Metabolic Pathways
3.2. Asp and the Asp-Derived Branches of Amino Acids Involved in Plant Defense and Immunity
4. Materials and Methods
4.1. Plant Materials, Growth Conditions, and Fungal Infection
4.2. Sequence Mining, Phylogeny, and Genomic Analyses
4.3. Transcriptome and Expression Validation by qRT-PCR
4.4. Qualitative and Quantitative Analyses of HPLC and Non-Targeted Metabolomics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Volaire, F. A unified framework of plant adaptive strategies to drought: Crossing scales and disciplines. Glob. Chang. Biol. 2018, 24, 2929–2938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Velásquez, A.C.; Castroverde, C.D.M.; He, S.Y. Plant–Pathogen Warfare under Changing Climate Conditions. Curr. Biol. 2018, 28, R619–R634. [Google Scholar] [CrossRef] [Green Version]
- Xin, X.-F.; Kvitko, B.; He, S.Y. Pseudomonas syringae: What it takes to be a pathogen. Nat. Rev. Microbiol. 2018, 16, 316–328. [Google Scholar] [CrossRef]
- Maitra, S.; Brestic, M.; Bhadra, P.; Shankar, T.; Praharaj, S.; Palai, J.B.; Shah, M.M.R.; Barek, V.; Ondrisik, P.; Skalický, M.; et al. Bioinoculants—Natural Biological Resources for Sustainable Plant Production. Microorganisms 2021, 10, 51. [Google Scholar] [CrossRef]
- Garcia-Bermudez, J.; Baudrier, L.; La, K.; Zhu, X.G.; Fidelin, J.; Sviderskiy, V.O.; Papagiannakopoulos, T.; Molina, H.; Snuderl, M.; Lewis, C.A.; et al. Aspartate is a limiting metabolite for cancer cell proliferation under hypoxia and in tumours. Nat. Cell Biol. 2018, 20, 775–781. [Google Scholar] [CrossRef]
- Jander, G.; Joshi, V. Recent Progress in Deciphering the Biosynthesis of Aspartate-Derived Amino Acids in Plants. Mol. Plant 2010, 3, 54–65. [Google Scholar] [CrossRef]
- Bellin, L.; Del Caño-Ochoa, F.; Velázquez-Campoy, A.; Möhlmann, T.; Ramón-Maiques, S. Mechanisms of feedback inhibition and sequential firing of active sites in plant aspartate transcarbamoylase. Nat. Commun. 2021, 12, 947. [Google Scholar] [CrossRef]
- Kirma, M.; Araujo, W.L.; Fernie, A.R.; Galili, G. The multifaceted role of aspartate-family amino acids in plant metabolism. J. Exp. Bot. 2012, 63, 4995–5001. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, K.; Arai, K.; Aoi, Y.; Tanaka, Y.; Hira, H.; Guo, R.; Hu, Y.; Ge, C.; Zhao, Y.; Kasahara, H.; et al. The main oxidative inactivation pathway of the plant hormone auxin. Nat. Commun. 2021, 12, 6752. [Google Scholar] [CrossRef]
- Wang, W.; Xu, M.; Wang, G.; Galili, G. New insights into the metabolism of aspartate-family amino acids in plant seeds. Plant Reprod. 2018, 31, 203–211. [Google Scholar] [CrossRef]
- Tegeder, M.; Masclaux-Daubresse, C. Source and sink mechanisms of nitrogen transport and use. New Phytol. 2018, 217, 35–53. [Google Scholar] [CrossRef] [Green Version]
- Graindorge, M.; Giustini, C.; Kraut, A.; Moyet, L.; Curien, G.; Matringe, M. Three Different Classes of Aminotransferases Evolved Prephenate Aminotransferase Functionality in Arogenate-competent Microorganisms. J. Biol. Chem. 2014, 289, 3198–3208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ritterhoff, J.; Young, S.; Villet, O.; Shao, D.; Neto, F.C.; Bettcher, L.F.; Hsu, Y.-W.A.; Kolwicz, S.C.; Raftery, D.; Tian, R. Metabolic Remodeling Promotes Cardiac Hypertrophy by Directing Glucose to Aspartate Biosynthesis. Circ. Res. 2020, 126, 182–196. [Google Scholar] [CrossRef]
- Zhang, Q.; Lee, J.; Pandurangan, S.; Clarke, M.; Pajak, A.; Marsolais, F. Characterization of Arabidopsis serine: Glyoxylate aminotransferase, AGT1, as an asparagine aminotransferase. Phytochemistry 2013, 85, 30–35. [Google Scholar] [CrossRef]
- Gaufichon, L.; Rothstein, S.J.; Suzuki, A. Asparagine Metabolic Pathways in Arabidopsis. Plant Cell Physiol. 2016, 57, 675–689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clark, T.J.; Lu, Y. Analysis of Loss-of-Function Mutants in Aspartate Kinase and Homoserine Dehydrogenase Genes Points to Complexity in the Regulation of Aspartate-Derived Amino Acid Contents. Plant Physiol. 2015, 168, 1512–1526. [Google Scholar] [CrossRef] [Green Version]
- Winter, G.; Todd, C.D.; Trovato, M.; Forlani, G.; Funck, D. Physiological implications of arginine metabolism in plants. Front. Plant Sci. 2015, 6, 534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macho, A.P.; Boutrot, F.; Rathjen, J.P.; Zipfel, C. ASPARTATE OXIDASE Plays an Important Role in Arabidopsis Stomatal Immunity. Plant Physiol. 2012, 159, 1845–1856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Barclay, H.; Roitberg, B.; Lalonde, R. Forest Productivity Enhancement and Compensatory Growth: A Review and Synthesis. Front. Plant Sci. 2020, 11, 1494. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, R.A.; Lancien, M.; Lea, P.J. The aspartic acid metabolic pathway, an exciting and essential pathway in plants. Amino Acids 2006, 30, 143–162. [Google Scholar] [CrossRef] [PubMed]
- Cánovas, F.M.; Cañas, R.A.; de la Torre, F.N.; Pascual, M.B.; Castro-Rodríguez, V.; Avila, C. Nitrogen Metabolism and Biomass Production in Forest Trees. Front. Plant Sci. 2018, 9, 1449. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.-J.; Harding, S.A.; Cooke, J.E.K. Branching out: A new era of investigating physiological processes in forest trees using genomic tools. Tree Physiol. 2018, 38, 303–310. [Google Scholar] [CrossRef]
- Yang, Q.; Yu, W.; Wu, H.; Zhang, C.; Sun, S.S.; Liu, Q. Lysine biofortification in rice by modulating feedback inhibition of aspartate kinase and dihydrodipicolinate synthase. Plant Biotechnol. J. 2020, 19, 490–501. [Google Scholar] [CrossRef]
- Han, M.; Zhang, C.; Suglo, P.; Sun, S.; Wang, M.; Su, T. L-Aspartate: An Essential Metabolite for Plant Growth and Stress Acclimation. Molecules 2021, 26, 1887. [Google Scholar] [CrossRef]
- Castro-Rodríguez, V.; García-Gutiérrez, A.; Canales, J.; Avila, C.; Kirby, E.G.; Cánovas, F.M. The glutamine synthetase gene family in Populus. BMC Plant Biol. 2011, 11, 119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liepman, A.H.; Olsen, L.J. Genomic Analysis of Aminotransferases in Arabidopsis thaliana. CRC Crit. Rev. Plant Sci. 2004, 23, 73–89. [Google Scholar] [CrossRef]
- Xu, Z.; Ma, J.; Qu, C.; Hu, Y.; Hao, B.; Sun, Y.; Liu, Z.; Yang, H.; Yang, C.; Wang, H.; et al. Identification and expression analyses of the alanine aminotransferase (AlaAT) gene family in poplar seedlings. Sci. Rep. 2017, 7, 45933. [Google Scholar] [CrossRef] [Green Version]
- Michalska, K.; Hernandez-Santoyo, A.; Jaskolski, M. The Mechanism of Autocatalytic Activation of Plant-type L-Asparaginases. J. Biol. Chem. 2008, 283, 13388–13397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, T.; Han, M.; Min, J.; Cao, D.; Zhai, G.; Zhou, H.; Li, N.; Li, M. Genome-Wide Characterization of AspATs in Populus: Gene Expression Variation and Enzyme Activities in Response to Nitrogen Perturbations. Forests 2019, 10, 449. [Google Scholar] [CrossRef] [Green Version]
- Su, T.; Zhou, B.; Cao, D.; Pan, Y.; Hu, M.; Zhang, M.; Wei, H.; Han, M. Transcriptomic Profiling of Populus Roots Challenged with Fusarium Reveals Differential Responsive Patterns of Invertase and Invertase Inhibitor-Like Families within Carbohydrate Metabolism. J. Fungi 2021, 7, 89. [Google Scholar] [CrossRef] [PubMed]
- Guan, M.; de Bang, T.C.; Pedersen, C.; Schjoerring, J.K. Cytosolic Glutamine Synthetase Gln1;2 Is the Main Isozyme Contributing to GS1 Activity and Can Be Up-Regulated to Relieve Ammonium Toxicity. Plant Physiol. 2016, 171, 1921–1933. [Google Scholar] [CrossRef] [Green Version]
- Moison, M.; Marmagne, A.; Dinant, S.; Soulay, F.; Azzopardi, M.; Lothier, J.; Citerne, S.; Morin, H.; Legay, N.; Chardon, F.; et al. Three cytosolic glutamine synthetase isoforms localized in different-order veins act together for N remobilization and seed filling in Arabidopsis. J. Exp. Bot. 2018, 69, 4379–4393. [Google Scholar] [CrossRef] [PubMed]
- Kusano, M.; Fukushima, A.; Tabuchi-Kobayashi, M.; Funayama, K.; Kojima, S.; Maruyama, K.; Yamamoto, Y.Y.; Nishizawa, T.; Kobayashi, M.; Wakazaki, M.; et al. Cytosolic GLUTAMINE SYNTHETASE1;1 Modulates Metabolism and Chloroplast Development in Roots. Plant Physiol. 2020, 182, 1894–1909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohashi, M.; Ishiyama, K.; Kusano, M.; Fukushima, A.; Kojima, S.; Hayakawa, T.; Yamaya, T. Reduction in sucrose contents by downregulation of fructose-1,6-bisphosphatase 2 causes tiller outgrowth cessation in rice mutants lacking glutamine synthetase1;2. Rice 2018, 11, 65. [Google Scholar] [CrossRef]
- Ji, Y.; Li, Q.; Liu, G.; Selvaraj, G.; Zheng, Z.; Zou, J.; Wei, Y. Roles of Cytosolic Glutamine Synthetases in Arabidopsis Development and Stress Responses. Plant Cell Physiol. 2019, 60, 657–671. [Google Scholar] [CrossRef] [PubMed]
- James, D.; Borphukan, B.; Fartyal, D.; Ram, B.; Singh, J.; Manna, M.; Sheri, V.; Panditi, V.; Yadav, R.; Achary, V.M.M.; et al. Concurrent Overexpression of OsGS1;1 and OsGS2 Genes in Transgenic Rice (Oryza sativa L.): Impact on Tolerance to Abiotic Stresses. Front. Plant Sci. 2018, 9, 786. [Google Scholar] [CrossRef]
- Gao, Y.; Bang, T.C.; Schjoerring, J.K. Cisgenic overexpression of cytosolic glutamine synthetase improves nitrogen utilization efficiency in barley and prevents grain protein decline under elevated CO2. Plant Biotechnol. J. 2019, 17, 1209–1221. [Google Scholar] [CrossRef] [Green Version]
- Pascual, M.B.; Jing, Z.P.; Kirby, E.G.; Cánovas, F.M.; Gallardo, F. Response of transgenic poplar overexpressing cytosolic glutamine synthetase to phosphinothricin. Phytochemistry 2008, 69, 382–389. [Google Scholar] [CrossRef]
- Molina-Rueda, J.J.; Kirby, E.G. Transgenic poplar expressing the pine GS1a show alterations in nitrogen homeostasis during drought. Plant Physiol. Biochem. 2015, 94, 181–190. [Google Scholar] [CrossRef]
- Castro-Rodríguez, V.; García-Gutiérrez, A.; Canales, J.; Cañas, R.A.; Kirby, E.G.; Avila, C.; Cánovas, F.M. Poplar trees for phytoremediation of high levels of nitrate and applications in bioenergy. Plant Biotechnol. J. 2016, 14, 299–312. [Google Scholar] [CrossRef]
- Lu, T.; Liu, L.; Wei, M.; Liu, Y.; Qu, Z.; Yang, C.; Wei, H.; Wei, Z. The Effect of Poplar PsnGS1.2 Overexpression on Growth, Secondary Cell Wall, and Fiber Characteristics in Tobacco. Front. Plant Sci. 2018, 9, 1–13. [Google Scholar] [CrossRef]
- Wojciechowska, N.; Marzec-Schmidt, K.; Kalemba, E.M.; Ludwików, A.; Bagniewska-Zadworna, A. Seasonal senescence of leaves and roots of Populus trichocarpa—is the scenario the same or different? Tree Physiol. 2020, 40, 987–1000. [Google Scholar] [CrossRef]
- Konishi, N.; Ishiyama, K.; Matsuoka, K.; Maru, I.; Hayakawa, T.; Yamaya, T.; Kojima, S. NADH-dependent glutamate synthase plays a crucial role in assimilating ammonium in the Arabidopsis root. Physiol. Plant. 2014, 152, 138–151. [Google Scholar] [CrossRef]
- Coschigano, K.T.; Melo-Oliveira, R.; Lim, J.; Coruzzi, G.M. Arabidopsis gls Mutants and Distinct Fd-GOGAT Genes: Implications for Photorespiration and Primary Nitrogen Assimilation. Plant Cell 1998, 10, 741–752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lancien, M.; Martin, M.; Hsieh, M.-H.; Leustek, T.; Goodman, H.; Coruzzi, G.M. Arabidopsis glt1 -T mutant defines a role for NADH-GOGAT in the non-photorespiratory ammonium assimilatory pathway. Plant J. 2002, 29, 347–358. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Marmagne, A.; Park, J.; Fabien, C.; Yim, Y.; Kim, S.; Kim, T.; Lim, P.O.; Masclaux-Daubresse, C.; Nam, H.G. Concurrent activation of OsAMT1;2 and OsGOGAT1 in rice leads to enhanced nitrogen use efficiency under nitrogen limitation. Plant J. 2020, 103, 7–20. [Google Scholar] [CrossRef]
- Muñoz-Nortes, T.; Pérez-Pérez, J.M.; Sarmiento-Mañús, R.; Candela, H.; Micol, J.L. Deficient glutamate biosynthesis triggers a concerted upregulation of ribosomal protein genes in Arabidopsis. Sci. Rep. 2017, 7, 6164. [Google Scholar] [CrossRef] [Green Version]
- Kan, C.-C.; Chung, T.-Y.; Wu, H.-Y.; Juo, Y.-A.; Hsieh, M.-H. Exogenous glutamate rapidly induces the expression of genes involved in metabolism and defense responses in rice roots. BMC Genom. 2017, 18, 186. [Google Scholar] [CrossRef] [Green Version]
- Qiu, X.-M.; Sun, Y.-Y.; Ye, X.-Y.; Li, Z.-G. Signaling Role of Glutamate in Plants. Front. Plant Sci. 2020, 10, 1743. [Google Scholar] [CrossRef] [Green Version]
- Cui, M.; Gu, M.; Lu, Y.; Zhang, Y.; Chen, C.; Ling, H.; Wu, H. Glutamate synthase 1 is involved in iron-deficiency response and long-distance transportation in Arabidopsis. J. Integr. Plant Biol. 2020, 62, 1925–1941. [Google Scholar] [CrossRef]
- Chen, H.; Li, C.; Liu, L.; Zhao, J.; Cheng, X.; Jiang, G.; Zhai, W. The Fd-GOGAT1 mutant gene lc7 confers resistance to Xanthomonas oryzae pv. Oryzae in rice. Sci. Rep. 2016, 6, 26411. [Google Scholar] [CrossRef] [Green Version]
- Zhou, W.; Shi, W.; Xu, X.-W.; Li, Z.-G.; Yin, C.-F.; Peng, J.-B.; Pan, S.; Chen, X.-L.; Zhao, W.-S.; Zhang, Y.; et al. Glutamate synthase MoGlt1-mediated glutamate homeostasis is important for autophagy, virulence and conidiation in the rice blast fungus. Mol. Plant Pathol. 2018, 19, 564–578. [Google Scholar] [CrossRef] [Green Version]
- Schultz, C.J.; Hsu, M.; Miesak, B.; Coruzzi, G.M. Arabidopsis mutants define an in vivo role for isoenzymes of aspartate aminotransferase in plant nitrogen assimilation. Genetics 1998, 149, 491–499. [Google Scholar] [CrossRef]
- Murooka, Y.; Mori, Y.; Hayashi, M. Variation of the Amino Acid Content of Arabidopsis Seeds by Expressing Soybean Aspartate Aminotransferase Gene. J. Biosci. Bioeng. 2002, 94, 225–230. [Google Scholar] [CrossRef]
- Zhou, Y.; Cai, H.; Xiao, J.; Li, X.; Zhang, Q.; Lian, X. Over-expression of aspartate aminotransferase genes in rice resulted in altered nitrogen metabolism and increased amino acid content in seeds. Theor. Appl. Genet. 2009, 118, 1381–1390. [Google Scholar] [CrossRef]
- de la Torre, F.; Canas, R.A.; Pascual, M.B.; Avila, C.; Canovas, F.M. Plastidic aspartate aminotransferases and the biosynthesis of essential amino acids in plants. J. Exp. Bot. 2014, 65, 5527–5534. [Google Scholar] [CrossRef] [Green Version]
- Brauc, S.; De Vooght, E.; Claeys, M.; Höfte, M.; Angenon, G. Influence of over-expression of cytosolic aspartate aminotransferase on amino acid metabolism and defence responses against Botrytis cinerea infection in Arabidopsis thaliana. J. Plant Physiol. 2011, 168, 1813–1819. [Google Scholar] [CrossRef]
- Gaufichon, L.; Marmagne, A.; Belcram, K.; Yoneyama, T.; Sakakibara, Y.; Hase, T.; Grandjean, O.; Clément, G.; Citerne, S.; Boutet-Mercey, S.; et al. ASN1 -encoded asparagine synthetase in floral organs contributes to nitrogen filling in Arabidopsis seeds. Plant J. 2017, 91, 371–393. [Google Scholar] [CrossRef] [Green Version]
- Lam, H.-M.; Wong, P.; Chan, H.-K.; Yam, K.-M.; Chen, L.; Chow, C.-M.; Coruzzi, G.M. Overexpression of the ASN1 Gene Enhances Nitrogen Status in Seeds of Arabidopsis. Plant Physiol. 2003, 132, 926–935. [Google Scholar] [CrossRef] [Green Version]
- Gaufichon, L.; Masclaux-Daubresse, C.; Tcherkez, G.; Reisdorf-Cren, M.; Sakakibara, Y.; Hase, T.; Clément, G.; Avice, J.-C.; Grandjean, O.; Marmagne, A.; et al. Arabidopsis thaliana ASN2 encoding asparagine synthetase is involved in the control of nitrogen assimilation and export during vegetative growth. Plant. Cell Environ. 2013, 36, 328–342. [Google Scholar] [CrossRef]
- Maaroufi-Dguimi, H.; Debouba, M.; Gaufichon, L.; Clément, G.; Gouia, H.; Hajjaji, A.; Suzuki, A. An Arabidopsis mutant disrupted in ASN2 encoding asparagine synthetase 2 exhibits low salt stress tolerance. Plant Physiol. Biochem. 2011, 49, 623–628. [Google Scholar] [CrossRef]
- Lee, S.; Park, J.; Lee, J.; Shin, D.; Marmagne, A.; Lim, P.O.; Masclaux-Daubresse, C.; An, G.; Nam, H.G. OsASN1 Overexpression in Rice Increases Grain Protein Content and Yield under Nitrogen-Limiting Conditions. Plant Cell Physiol. 2020, 61, 1309–1320. [Google Scholar] [CrossRef]
- Ohashi, M.; Ishiyama, K.; Kojima, S.; Konishi, N.; Nakano, K.; Kanno, K.; Hayakawa, T.; Yamaya, T. Asparagine Synthetase1, but not Asparagine Synthetase2, is Responsible for the Biosynthesis of Asparagine Following the Supply of Ammonium to Rice Roots. Plant Cell Physiol. 2015, 56, 769–778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, L.; Qin, R.; Liu, T.; Yu, M.; Yang, T.; Xu, G. OsASN1 Plays a Critical Role in Asparagine-Dependent Rice Development. Int. J. Mol. Sci. 2018, 20, 130. [Google Scholar] [CrossRef] [Green Version]
- Oddy, J.; Alarcón-Reverte, R.; Wilkinson, M.; Ravet, K.; Raffan, S.; Minter, A.; Mead, A.; Elmore, J.S.; de Almeida, I.M.; Cryer, N.C.; et al. Reduced free asparagine in wheat grain resulting from a natural deletion of TaASN-B2: Investigating and exploiting diversity in the asparagine synthetase gene family to improve wheat quality. BMC Plant Biol. 2021, 21, 302. [Google Scholar] [CrossRef]
- Jiang, J.; Batra, S.; Zhang, J. Asparagine: A Metabolite to Be Targeted in Cancers. Metabolites 2021, 11, 402. [Google Scholar] [CrossRef]
- Qu, C.; Hao, B.; Xu, X.; Wang, Y.; Yang, C.; Xu, Z.; Liu, G. Functional Research on Three Presumed Asparagine Synthetase Family Members in Poplar. Genes 2019, 10, 326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivanov, A.; Kameka, A.; Pajak, A.; Bruneau, L.; Beyaert, R.; Hernández-Sebastià, C.; Marsolais, F. Arabidopsis mutants lacking asparaginases develop normally but exhibit enhanced root inhibition by exogenous asparagine. Amino Acids 2012, 42, 2307–2318. [Google Scholar] [CrossRef] [PubMed]
- Pandurangan, S.; Pajak, A.; Rintoul, T.; Beyaert, R.; Hernández-Sebastià, C.; Brown, D.C.W.; Marsolais, F. Soybean seeds overexpressing asparaginase exhibit reduced nitrogen concentration. Physiol. Plant. 2015, 155, 126–137. [Google Scholar] [CrossRef]
- Yabuki, Y.; Ohashi, M.; Imagawa, F.; Ishiyama, K.; Beier, M.P.; Konishi, N.; Umetsu-Ohashi, T.; Hayakawa, T.; Yamaya, T.; Kojima, S. A temporal and spatial contribution of asparaginase to asparagine catabolism during development of rice grains. Rice 2017, 10, 3. [Google Scholar] [CrossRef] [Green Version]
- Sato, K.; Yamane, M.; Yamaji, N.; Kanamori, H.; Tagiri, A.; Schwerdt, J.G.; Fincher, G.B.; Matsumoto, T.; Takeda, K.; Komatsuda, T. Alanine aminotransferase controls seed dormancy in barley. Nat. Commun. 2016, 7, 11625. [Google Scholar] [CrossRef]
- Parthasarathy, A.; Borrego, E.J.; Savka, M.A.; Dobson, R.C.J.; Hudson, A.O. Amino acid–derived defense metabolites from plants: A potential source to facilitate novel antimicrobial development. J. Biol. Chem. 2021, 296, 100438. [Google Scholar] [CrossRef]
- Miyashita, Y.; Dolferus, R.; Ismond, K.P.; Good, A.G. Alanine aminotransferase catalyses the breakdown of alanine after hypoxia in Arabidopsis thaliana. Plant J. 2007, 49, 1108–1121. [Google Scholar] [CrossRef]
- Niessen, M.; Krause, K.; Horst, I.; Staebler, N.; Klaus, S.; Gaertner, S.; Kebeish, R.; Araujo, W.L.; Fernie, A.R.; Peterhansel, C. Two alanine aminotranferases link mitochondrial glycolate oxidation to the major photorespiratory pathway in Arabidopsis and rice. J. Exp. Bot. 2012, 63, 2705–2716. [Google Scholar] [CrossRef]
- Igarashi, D.; Tsuchida, H.; Miyao, M.; Ohsumi, C. Glutamate: Glyoxylate Aminotransferase Modulates Amino Acid Content during Photorespiration. Plant Physiol. 2006, 142, 901–910. [Google Scholar] [CrossRef] [Green Version]
- Shrawat, A.K.; Carroll, R.T.; DePauw, M.; Taylor, G.J.; Good, A.G. Genetic engineering of improved nitrogen use efficiency in rice by the tissue-specific expression of alanine aminotransferase. Plant Biotechnol. J. 2008, 6, 722–732. [Google Scholar] [CrossRef]
- Tiong, J.; Sharma, N.; Sampath, R.; MacKenzie, N.; Watanabe, S.; Metot, C.; Lu, Z.; Skinner, W.; Lu, Y.; Kridl, J.; et al. Improving Nitrogen Use Efficiency Through Overexpression of Alanine Aminotransferase in Rice, Wheat, and Barley. Front. Plant Sci. 2021, 12, 29. [Google Scholar] [CrossRef]
- Snyman, S.J.; Hajari, E.; Watt, M.P.; Lu, Y.; Kridl, J.C. Improved nitrogen use efficiency in transgenic sugarcane: Phenotypic assessment in a pot trial under low nitrogen conditions. Plant Cell Rep. 2015, 34, 667–669. [Google Scholar] [CrossRef] [Green Version]
- Sisharmini, A.; Apriana, A.; Khumaida, N.; Trijatmiko, K.R.; Purwoko, B.S. Expression of a cucumber alanine aminotransferase2 gene improves nitrogen use efficiency in transgenic rice. J. Genet. Eng. Biotechnol. 2019, 17, 9. [Google Scholar] [CrossRef]
- Hacham, Y.; Matityahu, I.; Schuster, G.; Amir, R. Overexpression of mutated forms of aspartate kinase and cystathionine γ-synthase in tobacco leaves resulted in the high accumulation of methionine and threonine. Plant J. 2008, 54, 260–271. [Google Scholar] [CrossRef]
- Van Bochaute, P.; Novoa, A.; Ballet, S.; Rognes, S.E.; Angenon, G. Regulatory mechanisms after short- and long-term perturbed lysine biosynthesis in the aspartate pathway: The need for isogenes in Arabidopsis thaliana. Physiol. Plant. 2013, 149, 449–460. [Google Scholar] [CrossRef]
- Stuttmann, J.; Hubberten, H.-M.; Rietz, S.; Kaur, J.; Muskett, P.; Guerois, R.; Bednarek, P.; Hoefgen, R.; Parker, J.E. Perturbation of Arabidopsis Amino Acid Metabolism Causes Incompatibility with the Adapted Biotrophic Pathogen Hyaloperonospora arabidopsidis. Plant Cell 2011, 23, 2788–2803. [Google Scholar] [CrossRef] [Green Version]
- Modde, K.; Timm, S.; Florian, A.; Michl, K.; Fernie, A.R.; Bauwe, H. High serine: Glyoxylate aminotransferase activity lowers leaf daytime serine levels, inducing the phosphoserine pathway in Arabidopsis. J. Exp. Bot. 2016, 68, 643–656. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.; Yang, L.; Han, X.; Zhao, Y.; Zhao, L.; Xiang, B.; Zhu, Y.; Bai, Y.; Wang, Y. Overexpression of AtAGT1 promoted root growth and development during seedling establishment. Plant Cell Rep. 2019, 38, 1165–1180. [Google Scholar] [CrossRef] [PubMed]
- Gakière, B.; Hao, J.; de Bont, L.; Pétriacq, P.; Nunes-Nesi, A.; Fernie, A.R. NAD + Biosynthesis and Signaling in Plants. CRC Crit. Rev. Plant Sci. 2018, 37, 259–307. [Google Scholar] [CrossRef]
- Hao, J.; Pétriacq, P.; de Bont, L.; Hodges, M.; Gakière, B. Characterization of l -aspartate oxidase from Arabidopsis thaliana. Plant Sci. 2018, 271, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Katoh, A.; Uenohara, K.; Akita, M.; Hashimoto, T. Early Steps in the Biosynthesis of NAD in Arabidopsis Start with Aspartate and Occur in the Plastid. Plant Physiol. 2006, 141, 851–857. [Google Scholar] [CrossRef] [Green Version]
- Pétriacq, P.; de Bont, L.; Hager, J.; Didierlaurent, L.; Mauve, C.; Guérard, F.; Noctor, G.; Pelletier, S.; Renou, J.-P.; Tcherkez, G.; et al. Inducible NAD overproduction in Arabidopsis alters metabolic pools and gene expression correlated with increased salicylate content and resistance to Pst-AvrRpm1. Plant J. 2012, 70, 650–665. [Google Scholar] [CrossRef]
- Anwar, S. Phenotypic analysis of Arabidopsis thaliana arginine-deficient mutants. Pure Appl. Biol. 2022, 11, 302–314. [Google Scholar] [CrossRef]
- Joshi, V.; Nimmakayala, P.; Song, Q.; Abburi, V.; Natarajan, P.; Levi, A.; Crosby, K.; Reddy, U.K. Genome-wide association study and population structure analysis of seed-bound amino acids and total protein in watermelon. PeerJ 2021, 9, e12343. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Liu, L.; Peng, S.; Yuan, L.; Li, H.; Wang, H. Heterologous Expression of Argininosuccinate Synthase from Oenococcus oeni Enhances the Acid Resistance of Lactobacillus plantarum. Front. Microbiol. 2019, 10, 1393. [Google Scholar] [CrossRef] [PubMed]
- Sagawa, C.H.D.; Assis, R.d.A.B.; Zaini, P.A.; Saxe, H.; Wilmarth, P.A.; Salemi, M.; Phinney, B.S.; Dandekar, A.M. De novo arginine synthesis is required for full virulence of Xanthomonas arboricola pv. juglandis during Walnut Bacterial Blight Disease. Phytopathology 2021, 1–55. [Google Scholar] [CrossRef]
- Mao, Y.; Shi, D.; Li, G.; Jiang, P. Citrulline depletion by ASS1 is required for proinflammatory macrophage activation and immune responses. Mol. Cell 2022, 82, 527–541.e7. [Google Scholar] [CrossRef]
- Luengo, A.; Gui, D.Y.; Vander Heiden, M.G. Targeting Metabolism for Cancer Therapy. Cell Chem. Biol. 2017, 24, 1161–1180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.T.; Slocum, R.D. Expression and functional analysis of aspartate transcarbamoylase and role of de novo pyrimidine synthesis in regulation of growth and development in Arabidopsis. Plant Physiol. Biochem. 2008, 46, 150–159. [Google Scholar] [CrossRef]
- Yon, R.J.; Grayson, J.E.; Chawda, A.; Butterworth, P.J. The quaternary structure of wheat-germ aspartate transcarbamoylase. Biochem. J. 1982, 203, 413–417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaynab, M.; Fatima, M.; Sharif, Y.; Zafar, M.H.; Ali, H.; Khan, K.A. Role of primary metabolites in plant defense against pathogens. Microb. Pathog. 2019, 137, 103728. [Google Scholar] [CrossRef]
- Fagard, M.; Launay, A.; Clement, G.; Courtial, J.; Dellagi, A.; Farjad, M.; Krapp, A.; Soulie, M.-C.; Masclaux-Daubresse, C. Nitrogen metabolism meets phytopathology. J. Exp. Bot. 2014, 65, 5643–5656. [Google Scholar] [CrossRef]
- Rojas, C.M.; Senthil-Kumar, M.; Tzin, V.; Mysore, K.S. Regulation of primary plant metabolism during plant-pathogen interactions and its contribution to plant defense. Front. Plant Sci. 2014, 5, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Less, H.; Angelovici, R.; Tzin, V.; Galili, G. Coordinated Gene Networks Regulating Arabidopsis Plant Metabolism in Response to Various Stresses and Nutritional Cues. Plant Cell 2011, 23, 1264–1271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeier, J. New insights into the regulation of plant immunity by amino acid metabolic pathways. Plant Cell Environ. 2013, 36, 2085–2103. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Gu, Z.; Wang, R.; Guo, J.; Ling, N.; Firbank, L.G.; Guo, S. Plant Primary Metabolism Regulated by Nitrogen Contributes to Plant–Pathogen Interactions. Plant Cell Physiol. 2019, 60, 329–342. [Google Scholar] [CrossRef] [Green Version]
- van Damme, M.; Zeilmaker, T.; Elberse, J.; Andel, A.; de Sain-van der Velden, M.; van den Ackerveken, G. Downy Mildew Resistance in Arabidopsis by Mutation of HOMOSERINE KINASE. Plant Cell 2009, 21, 2179–2189. [Google Scholar] [CrossRef] [Green Version]
- Hwang, I.S.; An, S.H.; Hwang, B.K. Pepper asparagine synthetase 1 (CaAS1) is required for plant nitrogen assimilation and defense responses to microbial pathogens. Plant J. 2011, 67, 749–762. [Google Scholar] [CrossRef]
- Hartmann, M.; Zeier, J. l-lysine metabolism to N-hydroxypipecolic acid: An integral immune-activating pathway in plants. Plant J. 2018, 96, 5–21. [Google Scholar] [CrossRef] [Green Version]
- Návarová, H.; Bernsdorff, F.; Döring, A.-C.; Zeier, J. Pipecolic Acid, an Endogenous Mediator of Defense Amplification and Priming, Is a Critical Regulator of Inducible Plant Immunity. Plant Cell 2013, 24, 5123–5141. [Google Scholar] [CrossRef] [Green Version]
- Ward, J.L.; Forcat, S.; Beckmann, M.; Bennett, M.; Miller, S.J.; Baker, J.M.; Hawkins, N.D.; Vermeer, C.P.; Lu, C.; Lin, W.; et al. The metabolic transition during disease following infection of Arabidopsis thaliana by Pseudomonas syringae pv. tomato. Plant J. 2010, 63, 443–457. [Google Scholar] [CrossRef]
- Jin, L.; Cai, Y.; Sun, C.; Huang, Y.; Yu, T. Exogenous l-glutamate treatment could induce resistance against Penicillium expansum in pear fruit by activating defense-related proteins and amino acids metabolism. Postharvest Biol. Technol. 2019, 150, 148–157. [Google Scholar] [CrossRef]
- Goto, Y.; Maki, N.; Ichihashi, Y.; Kitazawa, D.; Igarashi, D.; Kadota, Y.; Shirasu, K. Exogenous Treatment with Glutamate Induces Immune Responses in Arabidopsis. Mol. Plant-Microbe Interact. 2020, 33, 474–487. [Google Scholar] [CrossRef]
- Szabados, L.; Savouré, A. Proline: A multifunctional amino acid. Trends Plant Sci. 2010, 15, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Dickman, M.B. Proline suppresses apoptosis in the fungal pathogen Colletotrichum trifolii. Proc. Natl. Acad. Sci. USA 2005, 102, 3459–3464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- da Rocha, I.M.A.; Vitorello, V.A.; Silva, J.S.; Ferreira-Silva, S.L.; Viégas, R.A.; Silva, E.N.; Silveira, J.A.G. Exogenous ornithine is an effective precursor and the δ-ornithine amino transferase pathway contributes to proline accumulation under high N recycling in salt-stressed cashew leaves. J. Plant Physiol. 2012, 169, 41–49. [Google Scholar] [CrossRef] [PubMed]
- El-Beltagi, H.S.; Mohamed, H.I.; Sofy, M.R. Role of Ascorbic acid, Glutathione and Proline Applied as Singly or in Sequence Combination in Improving Chickpea Plant through Physiological Change and Antioxidant Defense under Different Levels of Irrigation Intervals. Molecules 2020, 25, 1702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haudecoeur, E.; Planamente, S.; Cirou, A.; Tannières, M.; Shelp, B.J.; Moréra, S.; Faure, D. Proline antagonizes GABA-induced quenching of quorum-sensing in Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. USA 2009, 106, 14587–14592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matysiak, K.; Kierzek, R.; Siatkowski, I.; Kowalska, J.; Krawczyk, R.; Miziniak, W. Effect of Exogenous Application of Amino Acids L-Arginine and Glycine on Maize under Temperature Stress. Agronomy 2020, 10, 769. [Google Scholar] [CrossRef]
- Freitas, I.S.; Trennepohl, B.I.; Acioly, T.M.S.; Conceição, V.J.; Mello, S.C.; Dourado Neto, D.; Kluge, R.A.; Azevedo, R.A. Exogenous Application of L-Arginine Improves Protein Content and Increases Yield of Pereskia aculeata Mill. Grown in Soilless Media Container. Horticulturae 2022, 8, 142. [Google Scholar] [CrossRef]
- Ramadan, A.A.; Abd Elhamid, E.M.; Sadak, M.S. Comparative study for the effect of arginine and sodium nitroprusside on sunflower plants grown under salinity stress conditions. Bull. Natl. Res. Cent. 2019, 43, 118. [Google Scholar] [CrossRef]
- Silveira, N.M.; Ribeiro, R.V.; de Morais, S.F.N.; de Souza, S.C.R.; da Silva, S.F.; Seabra, A.B.; Hancock, J.T.; Machado, E.C. Leaf arginine spraying improves leaf gas exchange under water deficit and root antioxidant responses during the recovery period. Plant Physiol. Biochem. 2021, 162, 315–326. [Google Scholar] [CrossRef]
- Su, T.; Han, M.; Min, J.; Zhou, H.; Zhang, Q.; Zhao, J.; Fang, Y. Functional Characterization of Invertase Inhibitors PtC/VIF1 and 2 Revealed Their Involvements in the Defense Response to Fungal Pathogen in Populus trichocarpa. Front. Plant Sci. 2020, 10, 1654. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [Green Version]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Almagro Armenteros, J.J.; Sønderby, C.K.; Sønderby, S.K.; Nielsen, H.; Winther, O. DeepLoc: Prediction of protein subcellular localization using deep learning. Bioinformatics 2017, 33, 3387–3395. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Han, M.; Heppel, S.C.; Su, T.; Bogs, J.; Zu, Y.; An, Z.; Rausch, T. Enzyme Inhibitor Studies Reveal Complex Control of Methyl-D-Erythritol 4-Phosphate (MEP) Pathway Enzyme Expression in Catharanthus roseus. PLoS ONE 2013, 8, e62467. [Google Scholar] [CrossRef] [Green Version]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, M.; Xu, M.; Wang, S.; Wu, L.; Sun, S.; Su, T. Effects of exogenous L-Glutamine as a sole nitrogen source on physiological characteristics and nitrogen use efficiency of poplar. Plant Physiol. Biochem. 2022, 172, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Du, W.; Liu, X.; Zhao, L.; Xu, Y.; Yin, Y.; Wu, J.; Ji, R.; Sun, Y.; Guo, H. Response of cucumber (Cucumis sativus) to perfluorooctanoic acid in photosynthesis and metabolomics. Sci. Total Environ. 2020, 724, 138257. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhang, Y.; Lai, J.; Luo, X.; Han, M.; Zhao, S.; Zhu, Y. Analysis of the biodegradation and phytotoxicity mechanism of TNT, RDX, HMX in alfalfa (Medicago sativa). Chemosphere 2021, 281, 130842. [Google Scholar] [CrossRef] [PubMed]
- Tomé, D. Amino acid metabolism and signalling pathways: Potential targets in the control of infection and immunity. Nutr. Diabetes 2021, 11, 20. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, M.; Xu, X.; Li, X.; Xu, M.; Hu, M.; Xiong, Y.; Feng, J.; Wu, H.; Zhu, H.; Su, T. New Insight into Aspartate Metabolic Pathways in Populus: Linking the Root Responsive Isoenzymes with Amino Acid Biosynthesis during Incompatible Interactions of Fusarium solani. Int. J. Mol. Sci. 2022, 23, 6368. https://doi.org/10.3390/ijms23126368
Han M, Xu X, Li X, Xu M, Hu M, Xiong Y, Feng J, Wu H, Zhu H, Su T. New Insight into Aspartate Metabolic Pathways in Populus: Linking the Root Responsive Isoenzymes with Amino Acid Biosynthesis during Incompatible Interactions of Fusarium solani. International Journal of Molecular Sciences. 2022; 23(12):6368. https://doi.org/10.3390/ijms23126368
Chicago/Turabian StyleHan, Mei, Xianglei Xu, Xue Li, Mingyue Xu, Mei Hu, Yuan Xiong, Junhu Feng, Hao Wu, Hui Zhu, and Tao Su. 2022. "New Insight into Aspartate Metabolic Pathways in Populus: Linking the Root Responsive Isoenzymes with Amino Acid Biosynthesis during Incompatible Interactions of Fusarium solani" International Journal of Molecular Sciences 23, no. 12: 6368. https://doi.org/10.3390/ijms23126368






