Structural and Functional Insights into CP2c Transcription Factor Complexes
Abstract
:1. Introduction
2. Results
2.1. Differential Binding of tCP2c and CBP to the Erythroid Gene Regulatory Regions in MEL Cells
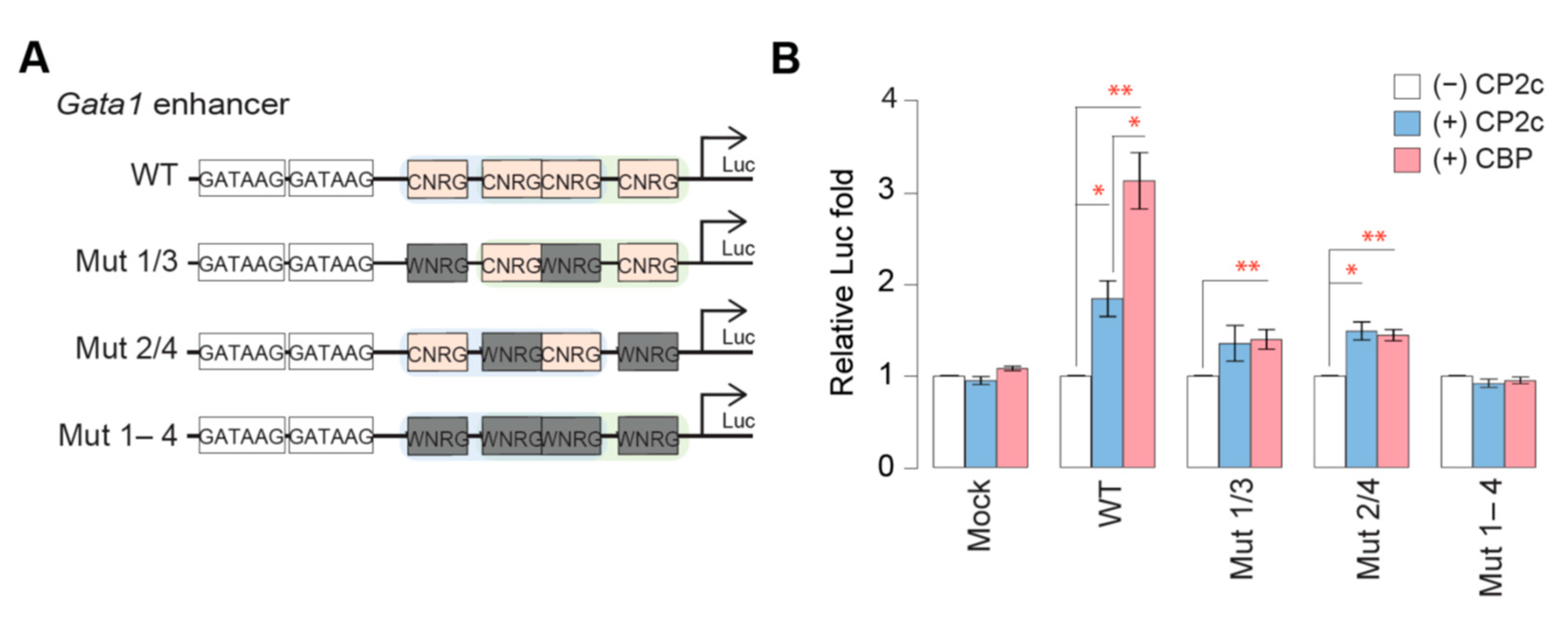
2.2. The tCP2c Binds to Sequences Containing Two Consecutive CP2c Half-Sites, Whereas the CBP Binds to Sequences Containing Three Consecutive CP2c Half-Sites or Two Staggered CP2c Binding Motifs
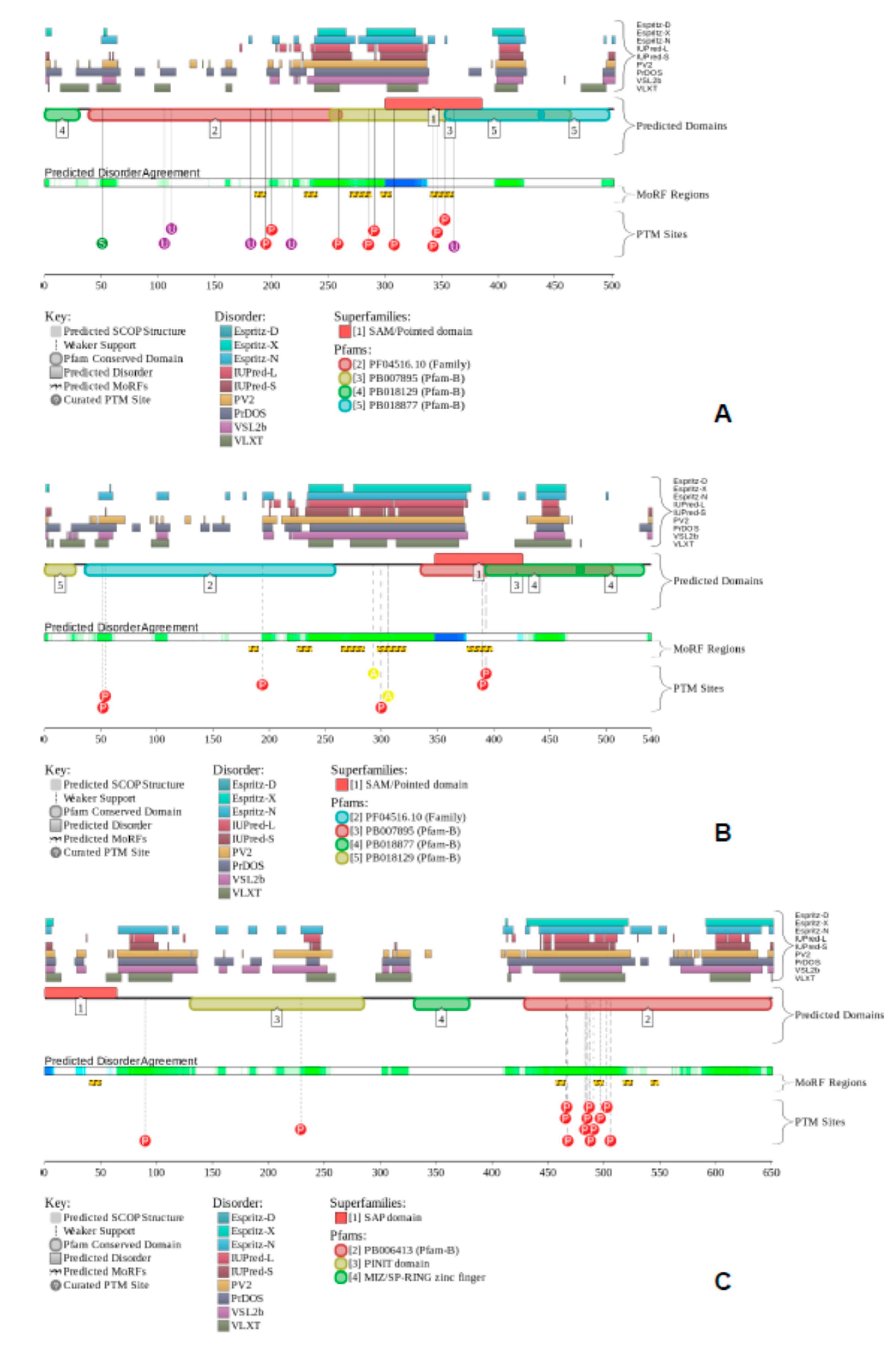
2.3. Stable DNA Binding Occurs Either by a Monomeric tCP2c or by a Dimeric CBP

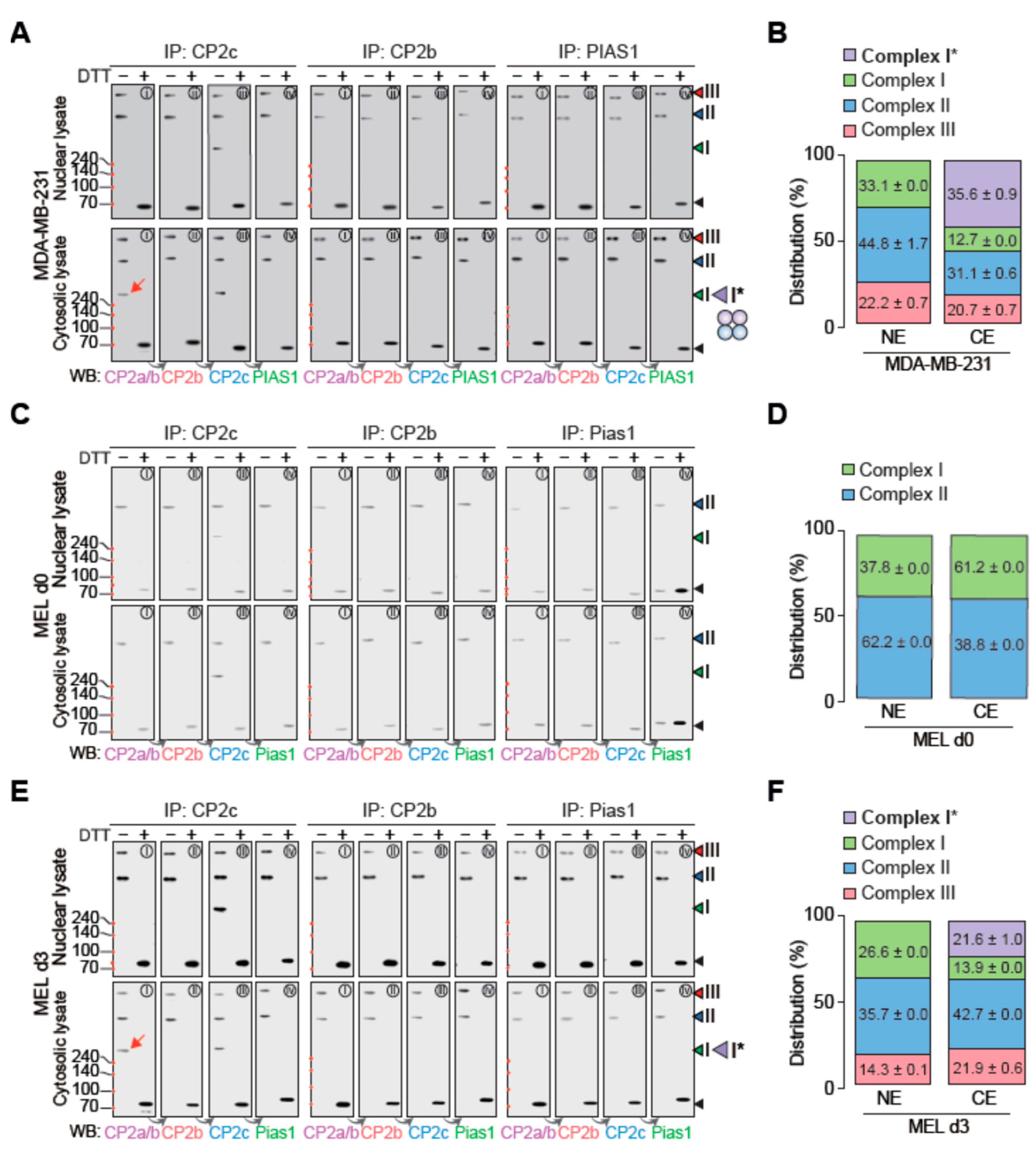
2.4. CP2c Exists in the Nucleus as a Complex of Either Monomeric tCP2c or Multimeric CBPs
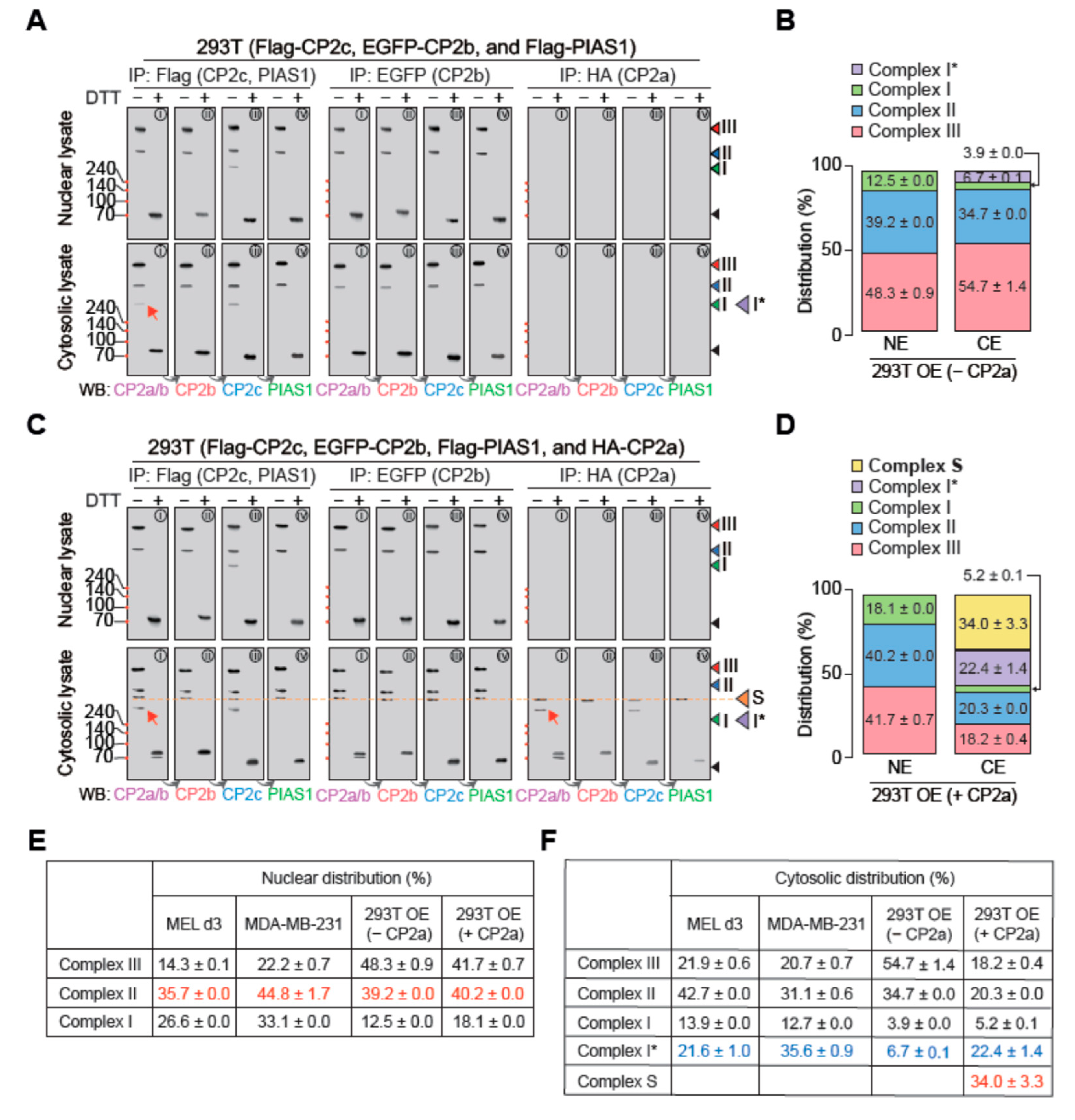
2.5. Cytosolic CP2a Regulates Subcellular Distribution and Dynamics of Various CP2c Complexes
2.6. tCP2c Exerts a Pioneering Function for Recruiting [C2B2P2]2 to the CP2c Binding Sites with Three or More CP2c Half-Sites
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Plasmid Construction
4.3. Chromatin Immunoprecipitation-Quantitative PCR (ChIP-qPCR)
4.4. Dual Luciferase Assay
4.5. Cell Extract Preparation
4.6. DNA Immunoprecipitation (DIP) Assay
4.7. Western Blot
4.8. Probe Titration Assay
4.9. DSP [Dithiobis(Succinimidyl Propionate)] Crosslinking and Western Blot (DSP XL-WB)
4.10. Quantification and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CBP | Heterohexameric CP2c, CP2b, and PIAS1 complex |
| ChIP-qPCR | Chromatin immunoprecipitation-quantitative PCR |
| DIP | DNA co-immunoprecipitation |
| DSP | Dithiobis (succinimidyl propionate) |
| DSP XL-WB | DSP crosslinking and Western blot |
| EMT | Epithelial–mesenchymal transition |
| FBS | Fetal bovine serum |
| HCC | Hepatocellular carcinoma |
| HIV/AIDS | Human immunodeficiency virus infection and acquired immunodeficiency syndrome |
| IDRs | Intrinsically disordered regions |
| MoRFs | Molecular recognition features |
| PAGE | Polyacrylamide gel electrophoresis |
| SDS | Sodium dodecyl sulfate |
| tCP2c | Homotetrameric CP2c complex |
| TFs | Transcription factors |
| WT | Wild type |
References
- Yao, L.; Shen, H.; Laird, P.W.; Farnham, P.J.; Berman, B.P. Inferring regulatory element landscapes and transcription factor networks from cancer methylomes. Genome Biol. 2015, 16, 105. [Google Scholar] [CrossRef] [Green Version]
- Banerji, J.; Rusconi, S.; Schaffner, W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell 1981, 27, 299–308. [Google Scholar] [CrossRef]
- Latchman, D.S. Transcription factors: An overview. Int. J. Biochem. Cell Biol. 1997, 29, 1305–1312. [Google Scholar] [CrossRef] [Green Version]
- Karin, M. Too many transcription factors: Positive and negative interactions. New Biol. 1990, 2, 126–131. [Google Scholar]
- Venkatesan, K.; McManus, H.R.; Mello, C.C.; Smith, T.F.; Hansen, U. Functional conservation between members of an ancient duplicated transcription factor family, LSF/Grainyhead. Nucleic Acids Res. 2003, 31, 4304–4316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Traylor-Knowles, N.; Hansen, U.; Dubuc, T.Q.; Martindale, M.Q.; Kaufman, L.; Finnerty, J.R. The evolutionary diversification of LSF and Grainyhead transcription factors preceded the radiation of basal animal lineages. BMC Evol. Biol. 2010, 10, 101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, C.G.; Barnhart, K.M.; Sheffery, M. Purification of multiple erythroid cell proteins that bind the promoter of the alpha-globin gene. Mol. Cell. Biol. 1988, 8, 4270–4281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, H.C.; Chae, J.H.; Lee, Y.H.; Park, M.A.; Shin, J.H.; Kim, S.H.; Ye, S.K.; Cho, Y.S.; Fiering, S.; Kim, C.G. Erythroid cell-specific alpha-globin gene regulation by the CP2 transcription factor family. Mol. Cell. Biol. 2005, 25, 6005–6020. [Google Scholar] [CrossRef] [Green Version]
- Kim, C.H.; Heath, C.; Bertuch, A.; Hansen, U. Specific stimulation of simian virus 40 late transcription in vitro by a cellular factor binding the simian virus 40 21-base-pair repeat promoter element. Proc. Natl. Acad. Sci. USA 1987, 84, 6025–6029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, J.B.; Li, G.; Roeder, R.G. Characterization of a family of related cellular transcription factors which can modulate human immunodeficiency virus type 1 transcription in vitro. Mol. Cell. Biol. 1994, 14, 1776–1785. [Google Scholar] [CrossRef] [Green Version]
- Cunningham, J.M.; Vanin, E.F.; Tran, N.; Valentine, M.; Jane, S.M. The human transcription factor CP2 (TFCP2), a component of the human gamma-globin stage selector protein, maps to chromosome region 12q13 and is within 250 kb of the NF-E2 gene. Genomics 1995, 30, 398–399. [Google Scholar]
- Taracha, A.; Kotarba, G.; Wilanowski, T. Neglected Functions of TFCP2/TFCP2L1/UBP1 Transcription Factors May Offer Valuable Insights into Their Mechanisms of Action. Int. J. Mol. Sci. 2018, 19, 2852. [Google Scholar] [CrossRef] [Green Version]
- Cheon, Y.P.; Choi, D.; Lee, S.H.; Kim, C.G. YY1 and CP2c in Unidirectional Spermatogenesis and Stemness. Dev. Reprod. 2020, 24, 249–262. [Google Scholar] [CrossRef]
- Veljkovic, J.; Hansen, U. Lineage-specific and ubiquitous biological roles of the mammalian transcription factor LSF. Gene 2004, 343, 23–40. [Google Scholar] [CrossRef] [Green Version]
- Kim, C.G.; Swendeman, S.L.; Barnhart, K.M.; Sheffery, M. Promoter elements and erythroid cell nuclear factors that regulate alpha-globin gene transcription in vitro. Mol. Cell. Biol. 1990, 10, 5958–5966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chae, J.H.; Kang, H.C.; Kim, C.G. The relative cellular levels of CP2a and CP2b potentiates erythroid cell-specific expression of the alpha-globin gene by regulating the nuclear localization of CP2c. Biochem. Biophys. Res. Commun. 2009, 380, 813–817. [Google Scholar] [CrossRef] [PubMed]
- Chae, J.H.; Kim, C.G. CP2 binding to the promoter is essential for the enhanced transcription of globin genes in erythroid cells. Mol. Cells 2003, 15, 40–47. [Google Scholar]
- Santhekadur, P.K.; Rajasekaran, D.; Siddiq, A.; Gredler, R.; Chen, D.; Schaus, S.E.; Hansen, U.; Fisher, P.B.; Sarkar, D. The transcription factor LSF: A novel oncogene for hepatocellular carcinoma. Am. J. Cancer Res. 2012, 2, 269–285. [Google Scholar]
- Yoo, B.K.; Emdad, L.; Gredler, R.; Fuller, C.; Dumur, C.I.; Jones, K.H.; Jackson-Cook, C.; Su, Z.Z.; Chen, D.; Saxena, U.H.; et al. Transcription factor Late SV40 Factor (LSF) functions as an oncogene in hepatocellular carcinoma. Proc. Natl. Acad. Sci. USA 2010, 107, 8357–8362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Kaushik, N.; Kang, J.H.; Kaushik, N.K.; Son, S.H.; Uddin, N.; Kim, M.J.; Kim, C.G.; Lee, S.J. A Feedback Loop Comprising EGF/TGFalpha Sustains TFCP2-Mediated Breast Cancer Progression. Cancer Res. 2020, 80, 2217–2229. [Google Scholar] [CrossRef] [Green Version]
- Kotarba, G.; Krzywinska, E.; Grabowska, A.I.; Taracha, A.; Wilanowski, T. TFCP2/TFCP2L1/UBP1 transcription factors in cancer. Cancer Lett. 2018, 420, 72–79. [Google Scholar] [CrossRef]
- Shirra, M.K.; Hansen, U. LSF and NTF-1 share a conserved DNA recognition motif yet require different oligomerization states to form a stable protein-DNA complex. J. Biol. Chem. 1998, 273, 19260–19268. [Google Scholar] [CrossRef] [Green Version]
- Kang, H.C.; Chae, J.H.; Jeon, J.; Kim, W.; Ha, D.H.; Shin, J.H.; Kim, C.G.; Kim, C.G. PIAS1 regulates CP2c localization and active promoter complex formation in erythroid cell-specific alpha-globin expression. Nucleic Acids Res. 2010, 38, 5456–5471. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.Y.; Kim, J.S.; Son, S.H.; Lim, C.S.; Eum, H.Y.; Ha, D.H.; Park, M.A.; Baek, E.J.; Ryu, B.Y.; Kang, H.C.; et al. Mbd2-CP2c loop drives adult-type globin gene expression and definitive erythropoiesis. Nucleic Acids Res. 2018, 46, 4933–4949. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.Y.; Chae, J.H.; Oh, C.H.; Kim, C.G. A DNA immunoprecipitation assay used in quantitative detection of in vitro DNA-protein complex binding. Anal. Biochem. 2013, 441, 147–151. [Google Scholar] [CrossRef]
- Friedman, P.N.; Chen, X.; Bargonetti, J.; Prives, C. The p53 protein is an unusually shaped tetramer that binds directly to DNA. Proc. Natl. Acad. Sci. USA 1993, 90, 3319–3323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elbaum-Garfinkle, S.; Kim, Y.; Szczepaniak, K.; Chen, C.C.; Eckmann, C.R.; Myong, S.; Brangwynne, C.P. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl. Acad. Sci. USA 2015, 112, 7189–7194. [Google Scholar] [CrossRef] [Green Version]
- Uv, A.E.; Thompson, C.R.; Bray, S.J. The Drosophila tissue-specific factor Grainyhead contains novel DNA-binding and dimerization domains which are conserved in the human protein CP2. Mol. Cell. Biol. 1994, 14, 4020–4031. [Google Scholar] [CrossRef] [Green Version]
- Berg, O.G.; Ehrenberg, M. Association kinetics with coupled three- and one-dimensional diffusion. Chain-length dependence of the association rate of specific DNA sites. Biophys. Chem. 1982, 15, 41–51. [Google Scholar] [CrossRef]
- Lomholt, M.A.; van den Broek, B.; Kalisch, S.M.; Wuite, G.J.; Metzler, R. Facilitated diffusion with DNA coiling. Proc. Natl. Acad. Sci. USA 2009, 106, 8204–8208. [Google Scholar] [CrossRef] [Green Version]
- Elf, J.; Li, G.W.; Xie, X.S. Probing transcription factor dynamics at the single-molecule level in a living cell. Science 2007, 316, 1191–1194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lieberman-Aiden, E.; van Berkum, N.L.; Williams, L.; Imakaev, M.; Ragoczy, T.; Telling, A.; Amit, I.; Lajoie, B.R.; Sabo, P.J.; Dorschner, M.O.; et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009, 326, 289–293. [Google Scholar] [CrossRef] [Green Version]
- Kokoszynska, K.; Ostrowski, J.; Rychlewski, L.; Wyrwicz, L.S. The fold recognition of CP2 transcription factors gives new insights into the function and evolution of tumor suppressor protein p53. Cell Cycle 2008, 7, 2907–2915. [Google Scholar] [CrossRef] [Green Version]
- Ming, Q.; Roske, Y.; Schuetz, A.; Walentin, K.; Ibraimi, I.; Schmidt-Ott, K.M.; Heinemann, U. Structural basis of gene regulation by the Grainyhead/CP2 transcription factor family. Nucleic Acids Res. 2018, 46, 2082–2095. [Google Scholar] [CrossRef] [Green Version]
- Frith, M.C.; Hansen, U.; Weng, Z. Detection of cis-element clusters in higher eukaryotic DNA. Bioinformatics 2001, 17, 878–889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- el-Deiry, W.S.; Kern, S.E.; Pietenpol, J.A.; Kinzler, K.W.; Vogelstein, B. Definition of a consensus binding site for p53. Nat. Genet. 1992, 1, 45–49. [Google Scholar] [CrossRef]
- Wei, C.L.; Wu, Q.; Vega, V.B.; Chiu, K.P.; Ng, P.; Zhang, T.; Shahab, A.; Yong, H.C.; Fu, Y.; Weng, Z.; et al. A global map of p53 transcription-factor binding sites in the human genome. Cell 2006, 124, 207–219. [Google Scholar] [CrossRef] [Green Version]
- Rastogi, C.; Rube, H.T.; Kribelbauer, J.F.; Crocker, J.; Loker, R.E.; Martini, G.D.; Laptenko, O.; Freed-Pastor, W.A.; Prives, C.; Stern, D.L.; et al. Accurate and sensitive quantification of protein-DNA binding affinity. Proc. Natl. Acad. Sci. USA 2018, 115, E3692–E3701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jana, T.; Brodsky, S.; Barkai, N. Speed-Specificity Trade-Offs in the Transcription Factors Search for Their Genomic Binding Sites. Trends Genet. 2021, 37, 421–432. [Google Scholar] [CrossRef] [PubMed]
- Oates, M.E.; Romero, P.; Ishida, T.; Ghalwash, M.; Mizianty, M.J.; Xue, B.; Dosztanyi, Z.; Uversky, V.N.; Obradovic, Z.; Kurgan, L.; et al. D(2)P(2): Database of disordered protein predictions. Nucleic Acids Res. 2013, 41, D508–D516. [Google Scholar] [CrossRef] [Green Version]
- Dosztányi, Z.; Csizmok, V.; Tompa, P.; Simon, I. IUPred: Web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 2005, 21, 3433–3434. [Google Scholar] [CrossRef] [Green Version]
- Romero, P.; Obradovic, Z.; Li, X.; Garner, E.C.; Brown, C.J.; Dunker, A.K. Sequence complexity of disordered protein. Proteins 2001, 42, 38–48. [Google Scholar] [CrossRef]
- Ishida, T.; Kinoshita, K. PrDOS: Prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 2007, 35, W460–W464. [Google Scholar] [CrossRef]
- Obradovic, Z.; Peng, K.; Vucetic, S.; Radivojac, P.; Dunker, A.K. Exploiting heterogeneous sequence properties improves prediction of protein disorder. Proteins Struct. Funct. Bioinform. 2005, 61, 176–182. [Google Scholar] [CrossRef]
- Peng, K.; Radivojac, P.; Vucetic, S.; Dunker, A.K.; Obradovic, Z. Length-dependent prediction of protein intrinsic disorder. BMC Bioinform. 2006, 7, 208. [Google Scholar] [CrossRef] [Green Version]
- Walsh, I.; Martin, A.J.; Di Domenico, T.; Tosatto, S.C. ESpritz: Accurate and fast prediction of protein disorder. Bioinformatics 2012, 28, 503–509. [Google Scholar] [CrossRef] [Green Version]
- Hnisz, D.; Shrinivas, K.; Young, R.A.; Chakraborty, A.K.; Sharp, P.A. A Phase Separation Model for Transcriptional Control. Cell 2017, 169, 13–23. [Google Scholar] [CrossRef] [Green Version]
- Alberti, S.; Gladfelter, A.; Mittag, T. Considerations and Challenges in Studying Liquid-Liquid Phase Separation and Biomolecular Condensates. Cell 2019, 176, 419–434. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Perumal, N.B.; Oldfield, C.J.; Su, E.W.; Uversky, V.N.; Dunker, A.K. Intrinsic disorder in transcription factors. Biochemistry 2006, 45, 6873–6888. [Google Scholar] [CrossRef] [Green Version]
- Oldfield, C.J.; Ulrich, E.L.; Cheng, Y.; Dunker, A.K.; Markley, J.L. Addressing the intrinsic disorder bottleneck in structural proteomics. Proteins 2005, 59, 444–453. [Google Scholar] [CrossRef]
- van der Lee, R.; Lang, B.; Kruse, K.; Gsponer, J.; de Groot, N.S.; Huynen, M.A.; Matouschek, A.; Fuxreiter, M.; Babu, M.M. Intrinsically disordered segments affect protein half-life in the cell and during evolution. Cell Rep. 2014, 8, 1832–1844. [Google Scholar] [CrossRef] [Green Version]
- Tompa, P.; Schad, E.; Tantos, A.; Kalmar, L. Intrinsically disordered proteins: Emerging interaction specialists. Curr. Opin. Struct. Biol. 2015, 35, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Turoverov, K.K.; Kuznetsova, I.M.; Fonin, A.V.; Darling, A.L.; Zaslavsky, B.Y.; Uversky, V.N. Stochasticity of Biological Soft Matter: Emerging Concepts in Intrinsically Disordered Proteins and Biological Phase Separation. Trends Biochem. Sci. 2019, 44, 716–728. [Google Scholar] [CrossRef]
- Uversky, V.N. Intrinsically disordered proteins in overcrowded milieu: Membrane-less organelles, phase separation, and intrinsic disorder. Curr. Opin. Struct. Biol. 2017, 44, 18–30. [Google Scholar] [CrossRef]
- Uversky, V.N. Protein intrinsic disorder-based liquid-liquid phase transitions in biological systems: Complex coacervates and membrane-less organelles. Adv. Colloid Interface Sci. 2017, 239, 97–114. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Finkelstein, A.V. Life in Phases: Intra- and Inter-Molecular Phase Transitions in Protein Solutions. Biomolecules 2019, 9, 842. [Google Scholar] [CrossRef] [Green Version]
- Uversky, V.N.; Kuznetsova, I.M.; Turoverov, K.K.; Zaslavsky, B. Intrinsically disordered proteins as crucial constituents of cellular aqueous two phase systems and coacervates. FEBS Lett. 2015, 589, 15–22. [Google Scholar] [CrossRef] [Green Version]
- Darling, A.L.; Zaslavsky, B.Y.; Uversky, V.N. Intrinsic Disorder-Based Emergence in Cellular Biology: Physiological and Pathological Liquid-Liquid Phase Transitions in Cells. Polymers 2019, 11, 990. [Google Scholar] [CrossRef] [Green Version]
- Darling, A.L.; Liu, Y.; Oldfield, C.J.; Uversky, V.N. Intrinsically Disordered Proteome of Human Membrane-Less Organelles. Proteomics 2018, 18, e1700193. [Google Scholar] [CrossRef] [PubMed]
- Boija, A.; Klein, I.A.; Sabari, B.R.; Dall’Agnese, A.; Coffey, E.L.; Zamudio, A.V.; Li, C.H.; Shrinivas, K.; Manteiga, J.C.; Hannett, N.M.; et al. Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell 2018, 175, 1842–1855.e1816. [Google Scholar] [CrossRef] [Green Version]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Son, S.H.; Kim, M.Y.; Jo, E.; Uversky, V.N.; Kim, C.G. Structural and Functional Insights into CP2c Transcription Factor Complexes. Int. J. Mol. Sci. 2022, 23, 6369. https://doi.org/10.3390/ijms23126369
Son SH, Kim MY, Jo E, Uversky VN, Kim CG. Structural and Functional Insights into CP2c Transcription Factor Complexes. International Journal of Molecular Sciences. 2022; 23(12):6369. https://doi.org/10.3390/ijms23126369
Chicago/Turabian StyleSon, Seung Han, Min Young Kim, Eunbi Jo, Vladimir N. Uversky, and Chul Geun Kim. 2022. "Structural and Functional Insights into CP2c Transcription Factor Complexes" International Journal of Molecular Sciences 23, no. 12: 6369. https://doi.org/10.3390/ijms23126369






