Differences in the Autophagy Response to Hypoxia in the Hippocampus and Neocortex of Rats
Abstract
:1. Introduction
2. Results
2.1. Assessment of Autophagic Flux in the Hippocampus and Neocortex of Rats under Normoxia
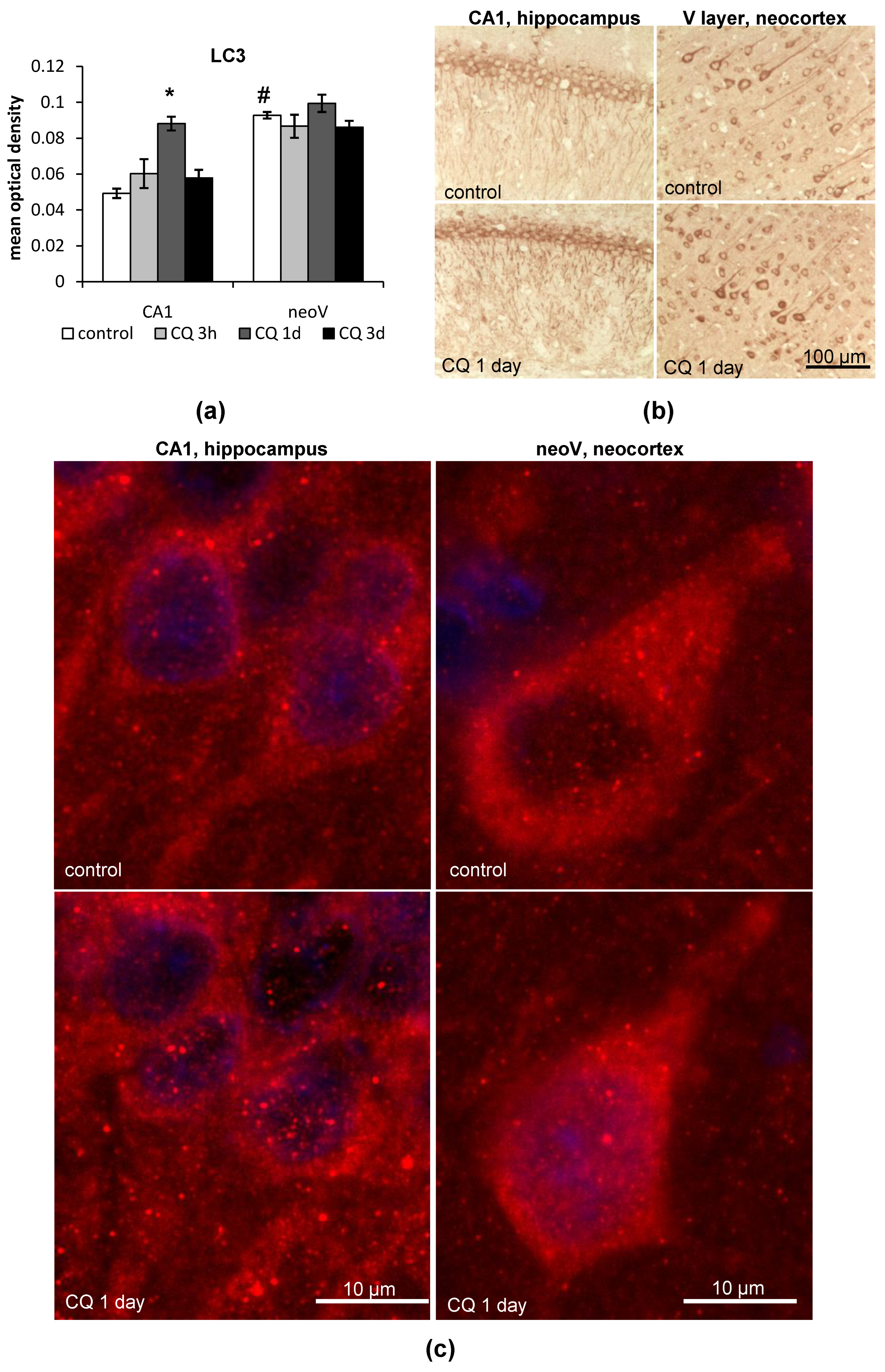
2.1.1. LC3
2.1.2. p62
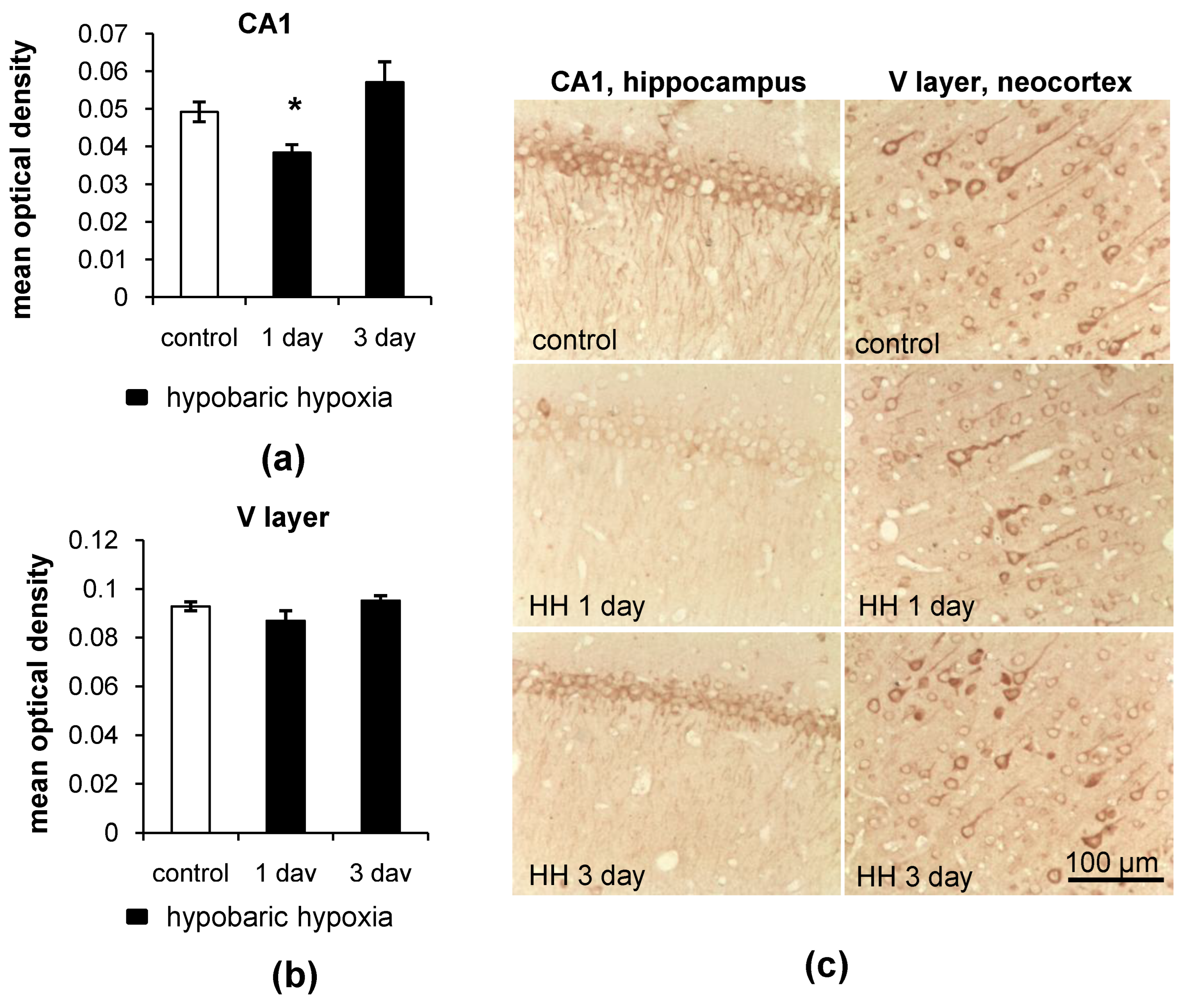
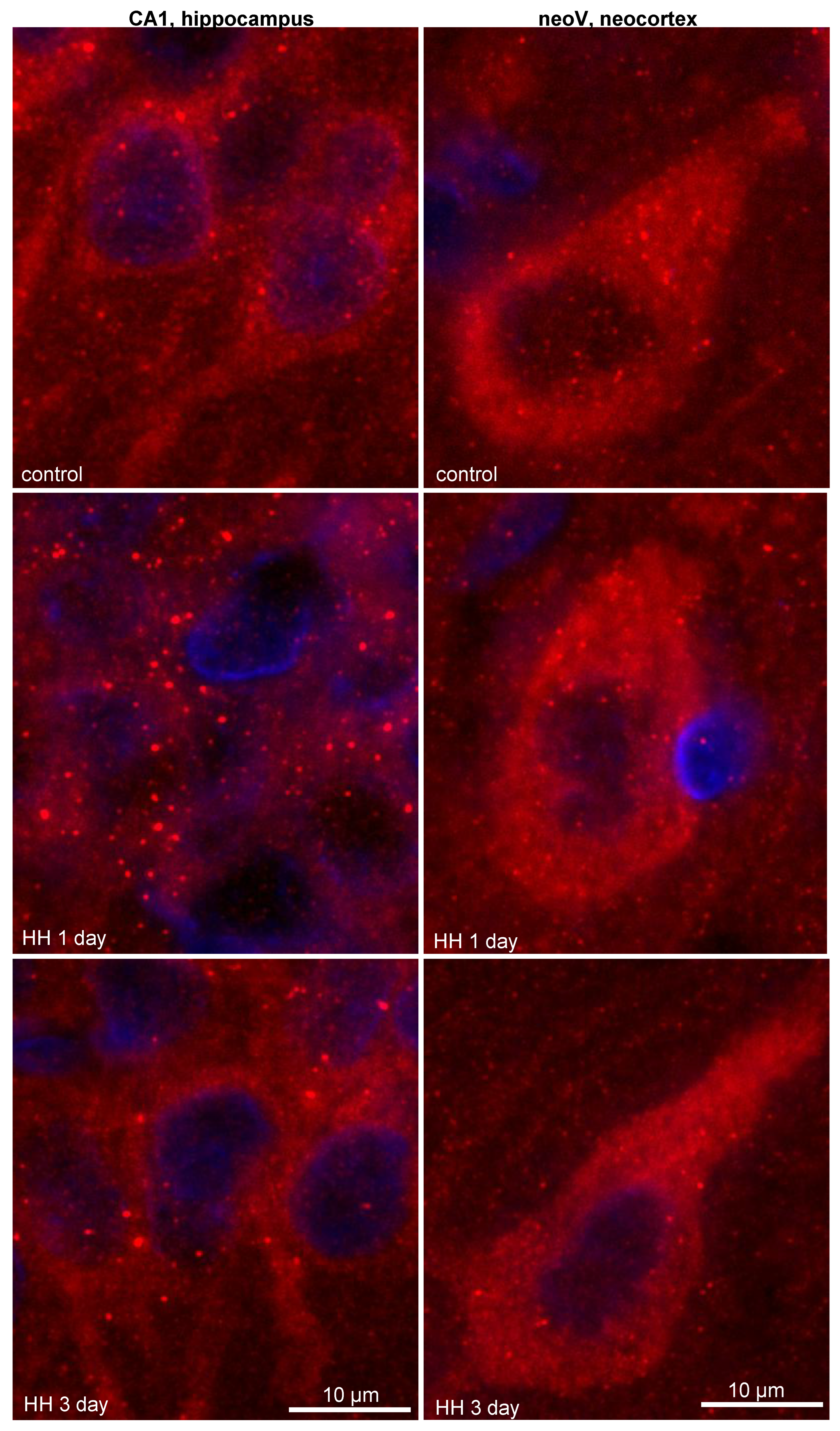
2.2. Changes in the LC3 and p62 Levels in the Hippocampus and Neocortex of Rats after Hypobaric Hypoxia
2.2.1. Immunohistochemistry
- LC3
- p62
2.2.2. Dot Analysis
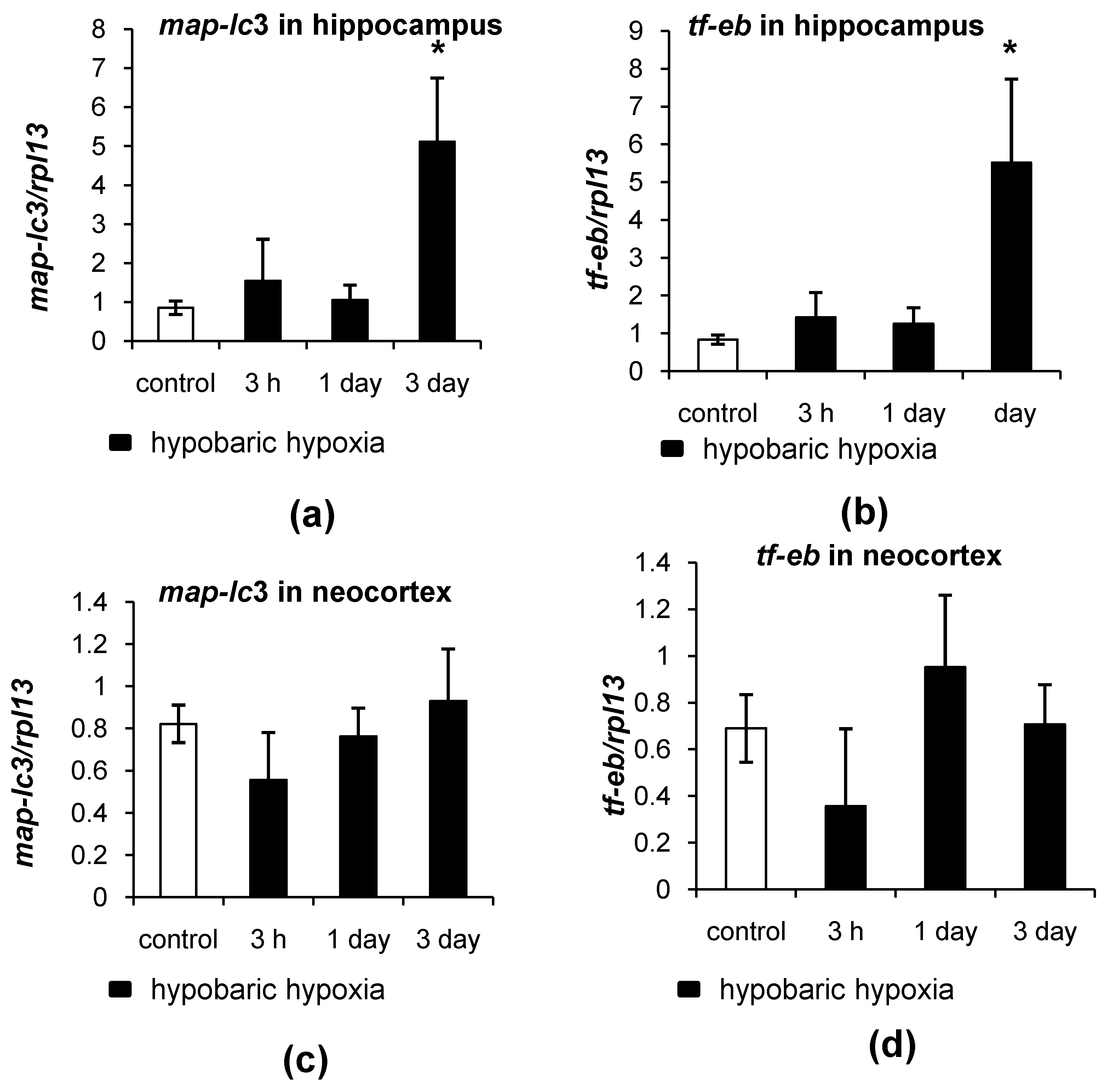
2.3. Different Patterns of Autophagy-Related Gene Expression in the Hippocampus and Neocortex of Rats in Response to Hypobaric Hypoxia
3. Discussion
Limitations of the Study
4. Materials and Methods
4.1. Experimental Groups and Hypoxia Model
4.2. Immunohistochemistry
4.3. Protein Extraction and RNA Isolation Procedures
4.4. Western Blot and Dot Analysis
4.5. qRT-PCR
4.6. Statistics
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

References
- Klionsky, D.J.; Emr, S.D. Autophagy as a regulated pathway of cellular degradation. Science 2000, 290, 1717–1721. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Komatsu, M. Autophagy: Renovation of cells and tissues. Cell 2011, 147, 728–741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, L.; Chen, Y.; Tooze, S.A. Autophagy pathway: Cellular and molecular mechanisms. Autophagy 2018, 14, 207–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stavoe, A.K.H.; Holzbaur, E.L.F. Autophagy in neurons. Annu. Rev. Cell Dev. Biol. 2019, 35, 477–500. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, A.; Yue, Z. Autophagy and its normal and pathogenic states in the brain. Annu. Rev. Neurosci. 2014, 37, 55–78. [Google Scholar] [CrossRef] [Green Version]
- Hara, T.; Nakamura, K.; Matsui, M.; Yamamoto, A.; Nakahara, Y.; Suzuki-Migishima, R.; Yokoyama, M.; Mishima, K.; Saito, I.; Okano, H.; et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 2006, 441, 885–889. [Google Scholar] [CrossRef]
- Komatsu, M.; Waguri, S.; Chiba, T.; Murata, S.; Iwata, J.-I.; Tanida, I.; Ueno, T.; Koike, M.; Uchiyama, Y.; Kominami, E.; et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 2006, 441, 880–884. [Google Scholar] [CrossRef]
- Nishiyama, J.; Miura, E.; Mizushima, N.; Watanabe, M.; Yuzaki, M. Aberrant membranes and double-membrane structures accumulate in the axons of Atg5-null Purkinje cells before neuronal death. Autophagy 2007, 3, 591–596. [Google Scholar] [CrossRef] [Green Version]
- Boland, B.; Yu, W.H.; Corti, O.; Mollereau, B.; Henriques, A.; Bezard, E.; Pastores, G.M.; Rubinsztein, D.C.; Nixon, R.A.; Duchen, M.R.; et al. Promoting the clearance of neurotoxic proteins in neurodegenerative disorders of ageing. Nat. Rev. Drug Discov. 2018, 17, 660–688. [Google Scholar] [CrossRef]
- Chu, C.T. Mechanisms of selective autophagy and mitophagy: Implications for neurodegenerative diseases. Neurobiol. Dis. 2019, 122, 23–34. [Google Scholar] [CrossRef]
- Frake, R.A.; Ricketts, T.; Menzies, F.M.; Rubinsztein, D.C. Autophagy and neurodegeneration. J. Clin. Investig. 2015, 125, 65–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghavami, S.; Shojaei, S.; Yeganeh, B.; Andee, S.R.; Jangamreddy, J.R.; Mehrpour, M.; Christoffersson, J.; Chaabane, W.; Moghadam, A.R.; Kashani, H.H.; et al. Autophagy and apoptosis dysfunction in neurodegenerative disorders. Prog. Neurobiol. 2014, 112, 24–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, B.H.; Jeziorek, M.; Kanal, H.D.; Contu, V.R.; Dobrowolski, R.; Levison, S.W. Moderately inducing autophagy reduces tertiary brain injury after perinatal hypoxia-ischemia. Cells 2021, 10, 898. [Google Scholar] [CrossRef] [PubMed]
- Dohi, E.; Tanaka, S.; Seki, T.; Miyagi, T.; Hide, I.; Takahashi, T.; Matsumoto, M.; Sakai, N. Hypoxic stress activates chaperone-mediated autophagy and modulates neuronal cell survival. Neurochem. Int. 2012, 60, 431–442. [Google Scholar] [CrossRef] [PubMed]
- Giordano, S.; Darley-Usmar, V.; Zhang, J. Autophagy as an essential cellular antioxidant pathway in neurodegenerative disease. Redox Biol. 2013, 2, 82–90. [Google Scholar] [CrossRef] [Green Version]
- Hubbi, M.E.; Hu, H.; Kshitiz; Ahmed, I.; Levchenko, A.; Semenza, G.L. Chaperone-mediated autophagy targets hypoxia-inducible factor-1α (HIF-1α) for lysosomal degradation. J. Biol. Chem. 2013, 288, 10703–10714. [Google Scholar] [CrossRef] [Green Version]
- Tooze, S.A.; Codogno, P. Compartmentalized regulation of autophagy regulators: Fine-tuning AMBRA1 by Bcl-2. EMBO J. 2011, 30, 1185–1186. [Google Scholar] [CrossRef]
- Adhami, F.; Liao, G.; Morozov, Y.M.; Schloemer, A.; Schmithorst, V.J.; Lorenz, J.N.; Dunn, R.S.; Vorhees, C.V.; Wills-Karp, M.; Degen, J.L.; et al. Cerebral ischemia-hypoxia induces intravascular coagulation and autophagy. Am. J. Pathol. 2006, 169, 566–583. [Google Scholar] [CrossRef] [Green Version]
- Koike, M.; Shibata, M.; Tadakoshi, M.; Gotoh, K.; Komatsu, M.; Waguri, S.; Kawahara, N.; Kuida, K.; Nagata, S.; Kominami, E.; et al. Inhibition of Autophagy Prevents Hippocampal Pyramidal Neuron Death after Hypoxic-Ischemic Injury. Am. J. Pathol. 2008, 172, 454–469. [Google Scholar] [CrossRef] [Green Version]
- Puyal, J.; Clarke, P.G. Targeting autophagy to prevent neonatal stroke damage. Autophagy 2009, 5, 1060–1061. [Google Scholar] [CrossRef] [Green Version]
- Rami, A.; Langhagen, A.; Steiger, S. Focal cerebral ischemia induces upregulation of Beclin 1 and autophagy-like cell death. Neurobiol. Dis. 2008, 29, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Rybnikova, E.; Vataeva, L.; Tyulkova, E.; Gluschenko, T.; Otellin, V.; Pelto-Huikko, M.; Samoilov, M.O. Mild hypoxia preconditioning prevents impairment of passive avoidance learning and suppression of brain NGFI-A expression induced by severe hypoxia. Behav. Brain Res. 2005, 160, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Rybnikova, E.; Sitnik, N.; Gluschenko, T.; Tyulkova, E.; Samoilov, M.O. The preconditioning modified neuronal expression of apoptosis-related proteins of Bcl-2 superfamily following severe hypobaric hypoxia in rats. Brain Res. 2006, 1089, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Rybnikova, E.; Nalivaeva, N. Glucocorticoid-dependent mechanisms of brain tolerance to hypoxia. Int. J. Mol. Sci. 2021, 22, 7982. [Google Scholar] [CrossRef]
- Rybnikova, E.; Glushchenko, T.; Churilova, A.; Pivina, S.; Samoilov, M. Expression of glucocorticoid and mineralocorticoid receptors in hippocampus of rats exposed to various modes of hypobaric hypoxia: Putative role in hypoxic preconditioning. Brain Res. 2011, 1381, 66–77. [Google Scholar] [CrossRef]
- Stroev, S.A.; Gluschenko, T.S.; Tjulkova, E.I.; Rybnikova, E.A.; Samoilov, M.O.; Pelto-Huikko, M. The effect of preconditioning on the Cu, Zn superoxide dismutase expression and enzyme activity in rat brain at the early period after severe hypobaric hypoxia. Neurosci. Res. 2005, 53, 39–47. [Google Scholar] [CrossRef]
- Samoilov, M.; Churilova, A.; Gluschenko, T.; Rybnikova, E. Neocortical pCREB and BDNF expression under different modes of hypobaric hypoxia: Role in brain hypoxic tolerance in rats. Acta Histochem. 2014, 116, 949–957. [Google Scholar] [CrossRef]
- Boland, B.; Kumar, A.; Lee, S.; Platt, F.M.; Wegiel, J.; Yu, W.H.; Nixon, R.A.; Boland, B.; Kumar, A.; Lee, S.; et al. Autophagy Induction and Autophagosome Clearance in Neurons: Relationship to Autophagic Pathology in Alzheimer’s Disease. J. Neurosci. 2008, 28, 6926–6937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klionsky, D.J.; Abdel-Aziz, A.K.; Abdelfatah, S.; Abdellatif, M.; Abdoli, A.; Abel, S.; Abeliovich, H.; Abildgaard, M.H.; Abudu, Y.P.; Acevedo-Arozena, A.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition). Autophagy 2021, 17, 1–382. [Google Scholar] [CrossRef]
- Mizushima, N.; Yoshimori, T. How to Interpret LC3 Immunoblotting. Autophagy 2007, 3, 542–545. [Google Scholar] [CrossRef]
- Vodicka, P.; Lim, J.; Williams, D.T.; Kegel, K.B.; Chase, K.; Park, H.; Marchionini, D.; Wilkinson, S.; Mead, T.; Birch, H.; et al. Assessment of chloroquine treatment for modulating autophagy flux in brain of WT and HD mice. J. Huntington’s Dis. 2014, 3, 159–174. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Yoshimori, T.; Levine, B. Methods in mammalian autophagy research. Cell 2010, 140, 313–326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill, S.; Colón-Ramos, D.A. The journey of the synaptic autophagosome: A cell biological perspective. Neuron 2020, 105, 961–973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Birdsall, V.; Waites, C.L. Autophagy at the synapse. Neurosci. Lett. 2019, 697, 24–28. [Google Scholar] [CrossRef]
- Kuijpers, M.; Kochlamazashvili, G.; Stumpf, A.; Puchkov, D.; Swaminathan, A.; Lucht, M.T.; Krause, E.; Maritzen, T.; Schmitz, D.; Haucke, V. Neuronal autophagy regulates presynaptic neurotransmission by controlling the axonal endoplasmic reticulum. Neuron 2021, 109, 299–313.e9. [Google Scholar] [CrossRef]
- Lieberman, O.J.; Sulzer, D. The synaptic autophagy cycle. J. Mol. Biol. 2020, 432, 2589–2604. [Google Scholar] [CrossRef]
- Shehata, M.; Abdou, K.; Choko, K.; Matsuo, M.; Nishizono, H.; Inokuchi, K. Autophagy enhances memory erasure through synaptic destabilization. J. Neurosci. 2018, 38, 3809–3822. [Google Scholar] [CrossRef] [Green Version]
- Shen, W.; Ganetzky, B. Autophagy promotes synapse development in Drosophila. J. Cell Biol. 2009, 187, 71–79. [Google Scholar] [CrossRef] [Green Version]
- Stavoe, A.K.; Hill, S.E.; Hall, D.H.; Colón-Ramos, D.A. KIF1A/UNC-104 transports ATG-9 to regulate neurodevelopment and autophagy at synapses. Dev. Cell 2016, 38, 171–185. [Google Scholar] [CrossRef] [Green Version]
- Nikoletopoulou, V.; Sidiropoulou, K.; Kallergi, E.; Dalezios, Y.; Tavernarakis, N. Modulation of autophagy by BDNF underlies synaptic plasticity. Cell Metab. 2017, 26, 230–242.e5. [Google Scholar] [CrossRef] [Green Version]
- Spruston, N. Pyramidal neurons: Dendritic structure and synaptic integration. Nat. Rev. Neurosci. 2008, 9, 206–221. [Google Scholar] [CrossRef] [PubMed]
- Kessels, H.W.; Malinow, R. Synaptic AMPA receptor plasticity and behavior. Neuron 2009, 61, 340–350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cotman, C.W.; Monaghan, D.T. Anatomical organization of excitatory amino acid receptors and their properties. Adv. Exp. Med. 1986, 203, 237–252. [Google Scholar] [CrossRef]
- Shehata, M.; Matsumura, H.; Okubo-Suzuki, R.; Ohkawa, N.; Inokuchi, K. Neuronal stimulation induces autophagy in hippocampal neurons that is involved in AMPA receptor degradation after chemical long-term depression. J. Neurosci. 2012, 32, 10413–10422. [Google Scholar] [CrossRef] [Green Version]
- Glatigny, M.; Moriceau, S.; Rivagorda, M.; Ramos-Brossier, M.; Nascimbeni, A.C.; Lante, F.; Shanley, M.R.; Boudarene, N.; Rousseaud, A.; Friedman, A.K.; et al. Autophagy is required for memory formation and reverses age-related memory decline. Curr. Biol. 2019, 29, 435–448.e8. [Google Scholar] [CrossRef] [Green Version]
- Niu, M.; Zheng, N.; Wang, Z.; Gao, Y.; Luo, X.; Chen, Z.; Fu, X.; Wang, Y.; Wang, T.; Liu, M.; et al. RAB39B Deficiency impairs learning and memory partially through compromising autophagy. Front. Cell Dev. Biol. 2020, 8, 598622. [Google Scholar] [CrossRef]
- Vetrovoy, O.; Sarieva, K.; Galkina, O.; Eschenko, N.; Lyanguzov, A.; Gluschenko, T.; Tyulkova, E.; Rybnikova, E. Neuroprotective mechanism of hypoxic post-conditioning involves HIF1-associated regulation of the pentose phosphate pathway in rat brain. Neurochem. Res. 2019, 44, 1425–1436. [Google Scholar] [CrossRef]
- Tremblay, R.; Lee, S.; Rudy, B. GABAergic interneurons in the neocortex: From cellular properties to circuits. Neuron 2016, 91, 260–292. [Google Scholar] [CrossRef] [Green Version]
- Schaaf, M.B.E.; Keulers, T.G.; Vooijs, M.A.; Rouschop, K.M.A. LC3/GABARAP family proteins: Autophagy-(un)related functions. FASEB J. 2016, 30, 3961–3978. [Google Scholar] [CrossRef] [Green Version]
- Hui, K.K.; Takashima, N.; Watanabe, A.; Chater, T.E.; Matsukawa, H.; Nekooki-Machida, Y.; Nilsson, P.; Endo, R.; Goda, Y.; Saido, T.C.; et al. GABARAPs dysfunction by autophagy deficiency in adolescent brain impairs GABAA receptor trafficking and social behavior. Sci. Adv. 2019, 5, eaau8237. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.K.; Kim, Y.S.; Lee, H.-M.; Jin, H.S.; Neupane, C.; Kim, S.; Lee, S.-H.; Min, J.-J.; Sasai, M.; Jeong, J.-H.; et al. GABAergic signaling linked to autophagy enhances host protection against intracellular bacterial infections. Nat. Commun. 2018, 9, 4184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lakhani, R.; Vogel, K.R.; Till, A.; Liu, J.; Burnett, S.F.; Gibson, K.M.; Subramani, S. Defects in GABA metabolism affect selective autophagy pathways and are alleviated by m TOR inhibition. EMBO Mol. Med. 2014, 6, 551–566. [Google Scholar] [CrossRef] [PubMed]
- Luscher, B.; Fuchs, T.; Kilpatrick, C.L. GABAA receptor trafficking-mediated plasticity of inhibitory synapses. Neuron 2011, 70, 385–409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du Sert, N.P.; Hurst, V.; Ahluwalia, A.; Alam, S.; Avey, M.T.; Baker, M.; Browne, W.J.; Clark, A.; Cuthill, I.C.; Dirnagl, U.; et al. The ARRIVE guidelines 2.0: Updated guidelines for reporting animal research. PLoS Biol. 2020, 18, e3000410. [Google Scholar] [CrossRef]
- Churilova, A.V.; Gluschenko, T.S.; Rybnikova, E.A.; Samoilov, M.O. The effect of histone deacetylase inhibitor on the expression level of glucococrticoid receptor in rat forebrain under hypoxia. Cell Tissue Biol. 2019, 13, 79–84. [Google Scholar] [CrossRef]
- Nurzynska, K.; Mikhalkin, A.; Piorkowski, A. CAS: Cell annotation software—Research on neuronal tissue has never been so transparent. Neuroinformatics 2017, 15, 365–382. [Google Scholar] [CrossRef] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Churilova, A.; Zachepilo, T.; Baranova, K.; Rybnikova, E. Differences in the Autophagy Response to Hypoxia in the Hippocampus and Neocortex of Rats. Int. J. Mol. Sci. 2022, 23, 8002. https://doi.org/10.3390/ijms23148002
Churilova A, Zachepilo T, Baranova K, Rybnikova E. Differences in the Autophagy Response to Hypoxia in the Hippocampus and Neocortex of Rats. International Journal of Molecular Sciences. 2022; 23(14):8002. https://doi.org/10.3390/ijms23148002
Chicago/Turabian StyleChurilova, Anna, Tatiana Zachepilo, Ksenia Baranova, and Elena Rybnikova. 2022. "Differences in the Autophagy Response to Hypoxia in the Hippocampus and Neocortex of Rats" International Journal of Molecular Sciences 23, no. 14: 8002. https://doi.org/10.3390/ijms23148002





