6-Bromoindirubin-3′-Oxime Regulates Colony Formation, Apoptosis, and Odonto/Osteogenic Differentiation in Human Dental Pulp Stem Cells
Abstract
:1. Introduction
2. Results
2.1. Cell Characterisation
2.2. BIO Treatment Activated Wnt Signalling in hDPSCs
2.3. BIO Attenuated hDPSC Colony-Forming Unit Ability and Cell Migration by Inducing Early Apoptosis
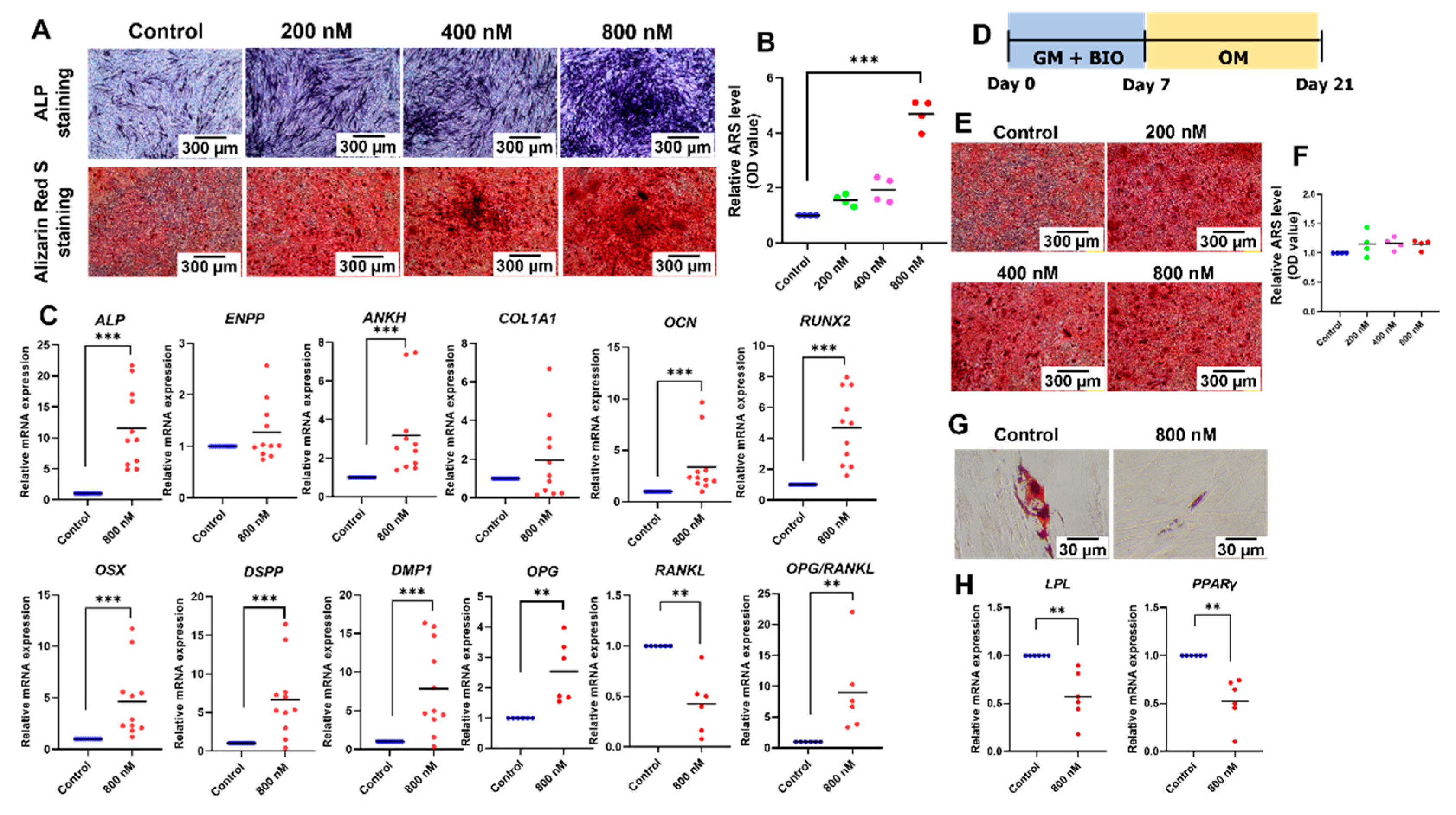
2.4. BIO Promoted Odonto/Osteogenic Differentiation but Attenuated Adipogenic Differentiation of hDPSCs
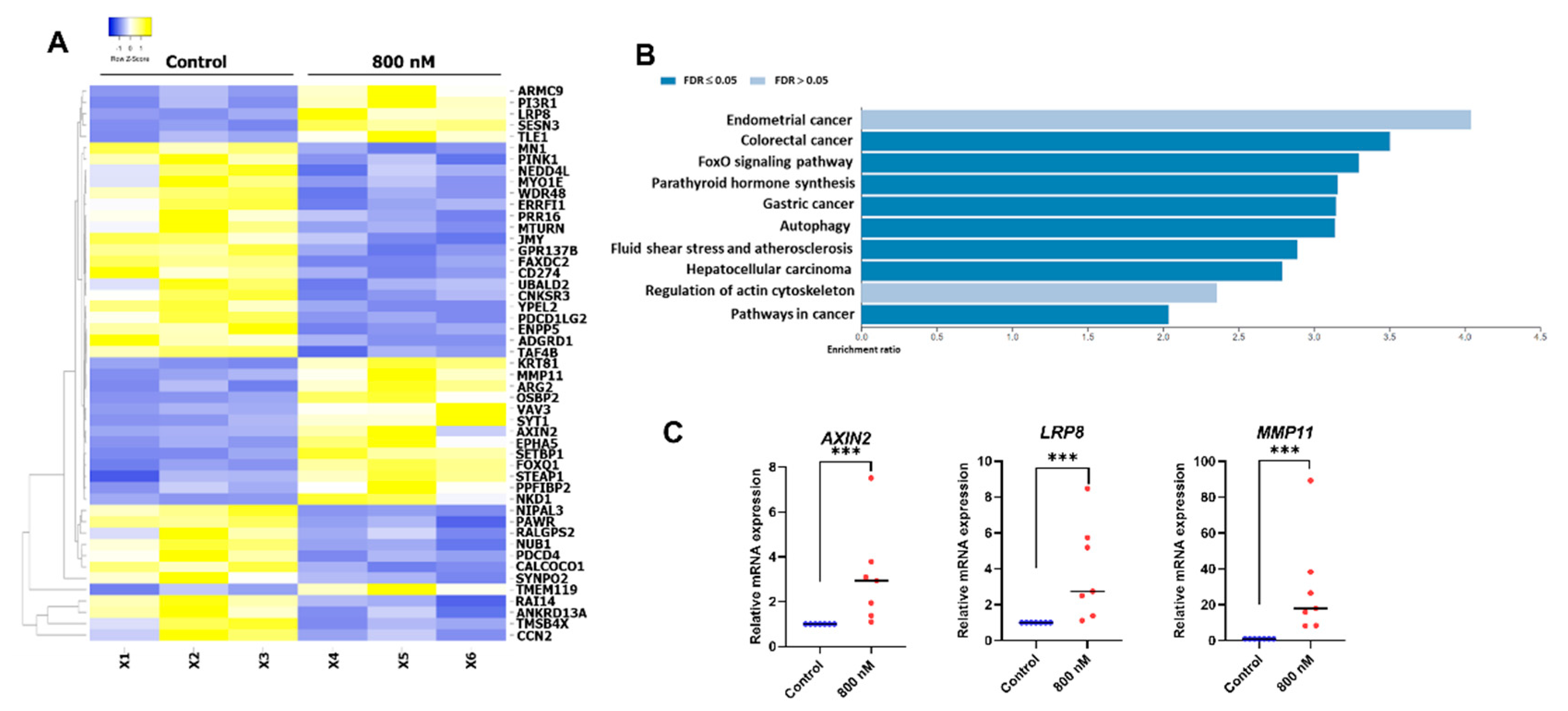
2.5. Gene Expression Profile of the BIO-Treated hDPSCs
3. Discussion
4. Materials and Methods
4.1. Cell Isolation and Culture
4.2. Differentiation Assays
4.3. Immunofluorescence Staining
4.4. Colony-Forming Unit Assay
4.5. In Vitro Scratch Assay
4.6. Apoptosis Assay
4.7. Cell Cycle Analysis
4.8. Alkaline Phosphatase Staining
4.9. Alizarin Red S Staining
4.10. Oil Red O Staining
4.11. Polymerase Chain Reaction
4.12. RNA Sequencing and Bioinformatic Analysis
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhu, L.; Yang, J.; Zhang, J.; Peng, B. A comparative study of BioAggregate and ProRoot MTA on adhesion, migration, and attachment of human dental pulp cells. J. Endod. 2014, 40, 1118–1123. [Google Scholar] [CrossRef] [PubMed]
- Komabayashi, T.; Zhu, Q.; Eberhart, R.; Imai, Y. Current status of direct pulp-capping materials for permanent teeth. Dent. Mater. J. 2016, 35, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuurs, A.H.; Gruythuysen, R.J.; Wesselink, P.R. Pulp capping with adhesive resin-based composite vs. calcium hydroxide: A review. Endod. Dent. Traumatol. 2000, 16, 240–250. [Google Scholar] [CrossRef] [PubMed]
- Dammaschke, T.; Stratmann, U.; Fischer, R.J.; Sagheri, D.; Schäfer, E. A histologic investigation of direct pulp capping in rodents with dentin adhesives and calcium hydroxide. Quintessence Int. 2010, 41, e62–e71. [Google Scholar] [PubMed]
- Huang, G.T.; Gronthos, S.; Shi, S. Mesenchymal stem cells derived from dental tissues vs. those from other sources: Their biology and role in regenerative medicine. J. Dent. Res. 2009, 88, 792–806. [Google Scholar] [CrossRef]
- Huang, G.T. Dental pulp and dentin tissue engineering and regeneration: Advancement and challenge. Front. Biosci. (Elite Ed.) 2011, 3, 788–800. [Google Scholar] [CrossRef] [Green Version]
- Huang, G.T.; Shagramanova, K.; Chan, S.W. Formation of odontoblast-like cells from cultured human dental pulp cells on dentin in vitro. J. Endod. 2006, 32, 1066–1073. [Google Scholar] [CrossRef]
- Goldberg, M.; Six, N.; Decup, F.; Lasfargues, J.J.; Salih, E.; Tompkins, K.; Veis, A. Bioactive molecules and the future of pulp therapy. Am. J. Dent. 2003, 16, 66–76. [Google Scholar]
- Imura, K.; Hashimoto, Y.; Okada, M.; Yoshikawa, K.; Yamamoto, K. Application of hydroxyapatite nanoparticle-assembled powder using basic fibroblast growth factor as a pulp-capping agent. Dent. Mater. J. 2019, 38, 713–720. [Google Scholar] [CrossRef] [Green Version]
- Iohara, K.; Nakashima, M.; Ito, M.; Ishikawa, M.; Nakasima, A.; Akamine, A. Dentin regeneration by dental pulp stem cell therapy with recombinant human bone morphogenetic protein 2. J. Dent. Res. 2004, 83, 590–595. [Google Scholar] [CrossRef]
- Chisini, L.A.; Conde, M.C.; Alcazar, J.C.; Silva, A.F.; Nor, J.E.; Tarquinio, S.B.; Demarco, F.F. Immunohistochemical Expression of TGF-beta1 and Osteonectin in engineered and Ca(OH)2-repaired human pulp tissues. Braz. Oral Res. 2016, 30, e93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steinhart, Z.; Angers, S. Wnt signaling in development and tissue homeostasis. Development 2018, 145, dev146589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Angelova Volponi, A.; Zaugg, L.K.; Neves, V.; Liu, Y.; Sharpe, P.T. Tooth Repair and Regeneration. Curr. Oral Health Rep. 2018, 5, 295–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uribe-Etxebarria, V.; García-Gallastegui, P.; Pérez-Garrastachu, M.; Casado-Andrés, M.; Irastorza, I.; Unda, F.; Ibarretxe, G.; Subirán, N. Wnt-3a Induces Epigenetic Remodeling in Human Dental Pulp Stem Cells. Cells 2020, 9, 652. [Google Scholar] [CrossRef] [Green Version]
- Uribe-Etxebarria, V.; Agliano, A.; Unda, F.; Ibarretxe, G. Wnt signaling reprograms metabolism in dental pulp stem cells. J. Cell Physiol. 2019, 234, 13068–13082. [Google Scholar] [CrossRef] [Green Version]
- Rolph, D.N.; Deb, M.; Kanji, S.; Greene, C.J.; Das, M.; Joseph, M.; Aggarwal, R.; Leblebicioglu, B.; Das, H. Ferutinin directs dental pulp-derived stem cells towards the osteogenic lineage by epigenetically regulating canonical Wnt signaling. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165314. [Google Scholar] [CrossRef]
- Hunter, D.J.; Bardet, C.; Mouraret, S.; Liu, B.; Singh, G.; Sadoine, J.; Dhamdhere, G.; Smith, A.; Tran, X.V.; Joy, A.; et al. Wnt Acts as a Prosurvival Signal to Enhance Dentin Regeneration. J. Bone Miner. Res. 2015, 30, 1150–1159. [Google Scholar] [CrossRef]
- Babb, R.; Chandrasekaran, D.; Carvalho Moreno Neves, V.; Sharpe, P.T. Axin2-expressing cells differentiate into reparative odontoblasts via autocrine Wnt/β-catenin signaling in response to tooth damage. Sci Rep. 2017, 7, 3102. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Yuan, X.; Liu, B.; Tulu, U.S.; Helms, J.A. Wnt-Responsive Odontoblasts Secrete New Dentin after Superficial Tooth Injury. J. Dent. Res. 2018, 97, 1047–1054. [Google Scholar] [CrossRef]
- Li, G.; Liu, J.; Wang, Y.; Yang, K.; Zhao, M.; Xiao, Y.; Wen, X.; Liu, L. LNGFR targets the Wnt/β-catenin pathway and promotes the osteogenic differentiation in rat ectomesenchymal stem cells. Sci Rep. 2017, 7, 11021. [Google Scholar] [CrossRef] [Green Version]
- Gong, Y.; Yuan, S.; Sun, J.; Wang, Y.; Liu, S.; Guo, R.; Dong, W.; Li, R. R-Spondin 2 Induces Odontogenic Differentiation of Dental Pulp Stem/Progenitor Cells via Regulation of Wnt/β-Catenin Signaling. Front. Physiol. 2020, 11, 918. [Google Scholar] [CrossRef] [PubMed]
- Yaemkleebbua, K.; Osathanon, T.; Nowwarote, N.; Limjeerajarus, C.N.; Sukarawan, W. Analysis of hard tissue regeneration and Wnt signalling in dental pulp tissues after direct pulp capping with different materials. Int. Endod. J. 2019, 52, 1605–1616. [Google Scholar] [CrossRef] [PubMed]
- Han, N.; Zheng, Y.; Li, R.; Li, X.; Zhou, M.; Niu, Y.; Zhang, Q. β-catenin enhances odontoblastic differentiation of dental pulp cells through activation of Runx2. PLoS ONE 2014, 9, e88890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mihara, E.; Hirai, H.; Yamamoto, H.; Tamura-Kawakami, K.; Matano, M.; Kikuchi, A.; Sato, T.; Takagi, J. Active and water-soluble form of lipidated Wnt protein is maintained by a serum glycoprotein afamin/α-albumin. Elife 2016, 5, e11621. [Google Scholar] [CrossRef] [PubMed]
- Willert, K.; Nusse, R. Wnt proteins. Cold Spring Harb. Perspect. Biol. 2012, 4, a007864. [Google Scholar] [CrossRef]
- Meijer, L.; Skaltsounis, A.L.; Magiatis, P.; Polychronopoulos, P.; Knockaert, M.; Leost, M.; Ryan, X.P.; Vonica, C.A.; Brivanlou, A.; Dajani, R.; et al. GSK-3-selective inhibitors derived from Tyrian purple indirubins. Chem. Biol. 2003, 10, 1255–1266. [Google Scholar] [CrossRef] [Green Version]
- Clough, B.H.; Zeitouni, S.; Krause, U.; Chaput, C.D.; Cross, L.M.; Gaharwar, A.K.; Gregory, C.A. Rapid Osteogenic Enhancement of Stem Cells in Human Bone Marrow Using a Glycogen-Synthease-Kinase-3-Beta Inhibitor Improves Osteogenic Efficacy In Vitro and In Vivo. Stem Cells Transl. Med. 2018, 7, 342–353. [Google Scholar] [CrossRef] [Green Version]
- Baghaban Eslaminejad, M.; Fallah, N. Small Molecule-BIO Accelerates and Enhances Marrow-Derived Mesenchymal Stem Cell in Vitro Chondrogenesis. Iran. J. Med. Sci. 2014, 39, 107–116. [Google Scholar]
- Neves, V.C.; Babb, R.; Chandrasekaran, D.; Sharpe, P.T. Promotion of natural tooth repair by small molecule GSK3 antagonists. Sci. Rep. 2017, 7, 39654. [Google Scholar] [CrossRef] [Green Version]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.; Krause, D.; Deans, R.; Keating, A.; Prockop, D.; Horwitz, E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef]
- Shah, D.; Lynd, T.; Ho, D.; Chen, J.; Vines, J.; Jung, H.D.; Kim, J.H.; Zhang, P.; Wu, H.; Jun, H.W.; et al. Pulp-Dentin Tissue Healing Response: A Discussion of Current Biomedical Approaches. J. Clin. Med. 2020, 9, 434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El Backly, R.M.; Marei, M.K. Dental pulp stem cells in tissue engineering and regenerative medicine: Opportunities for translational research. In Advances in Stem Cell Therapy: Bench to Bedside; El-Badri, N., Ed.; Springer International Publishing: Cham, Switzerland, 2017; pp. 171–196. [Google Scholar]
- Yoshida, S.; Tomokiyo, A.; Hasegawa, D.; Hamano, S.; Sugii, H.; Maeda, H. Insight into the Role of Dental Pulp Stem Cells in Regenerative Therapy. Biology 2020, 9, 160. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Liu, X.; Wang, J.; Chen, X.; Zhang, H.; Kim, S.H.; Cui, J.; Li, R.; Zhang, W.; Kong, Y.; et al. Wnt signaling in bone formation and its therapeutic potential for bone diseases. Ther. Adv. Musculoskelet. Dis. 2013, 5, 13–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komiya, Y.; Habas, R. Wnt signal transduction pathways. Organogenesis 2008, 4, 68–75. [Google Scholar] [CrossRef] [Green Version]
- Logan, C.Y.; Nusse, R. The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 2004, 20, 781–810. [Google Scholar] [CrossRef] [Green Version]
- Liu, K.; Li, J.; Wu, X.; Chen, M.; Luo, F.; Li, J. GSK-3β inhibitor 6-bromo-indirubin-3′-oxime promotes both adhesive activity and drug resistance in colorectal cancer cells. Int. J. Oncol. 2017, 51, 1821–1830. [Google Scholar] [CrossRef] [Green Version]
- Krause, U.; Harris, S.; Green, A.; Ylostalo, J.; Zeitouni, S.; Lee, N.; Gregory, C.A. Pharmaceutical modulation of canonical Wnt signaling in multipotent stromal cells for improved osteoinductive therapy. Proc. Natl. Acad. Sci. USA 2010, 107, 4147–4152. [Google Scholar] [CrossRef] [Green Version]
- Sankaranarayanan, K.; Ramachandran, R.P.; Sundararajan, R. 6—Electrically-enhanced proliferation control of cancer-stem-cells-like adult human mesenchymal stem cells—A novel modality of treatment. In Electroporation-Based Therapies for Cancer; Sundararajan, R., Ed.; Woodhead Publishing: Sawston, UK, 2014; pp. 127–159. [Google Scholar]
- Yin, J.Y.; Luo, X.H.; Feng, W.Q.; Miao, S.H.; Ning, T.T.; Lei, Q.; Jiang, T.; Ma, D.D. Multidifferentiation potential of dental-derived stem cells. World J. Stem Cells 2021, 13, 342–365. [Google Scholar] [CrossRef]
- Baghaban Eslaminejad, M.; Salami, F.; Soleimani Mehranjani, M.; Abnoosi, M.-H.; Eftekhari-yazdi, P. BIO treatment enhances rat marrow-derived mesenchymal stem cell in vitro proliferation and viability. Physiol. Pharmacol. 2009, 13, 57–67. [Google Scholar]
- Tseng, A.S.; Engel, F.B.; Keating, M.T. The GSK-3 inhibitor BIO promotes proliferation in mammalian cardiomyocytes. Chem. Biol. 2006, 13, 957–963. [Google Scholar] [CrossRef] [Green Version]
- Al-Habib, M.; Yu, Z.; Huang, G.T.J. Small molecules affect human dental pulp stem cell properties via multiple signaling pathways. Stem Cells Dev. 2013, 22, 2402–2413. [Google Scholar] [CrossRef] [Green Version]
- Yu, A.S.; Zhao, L. Effects of the GSK-3β inhibitor (2Z,3E)-6-bromoindirubin-3′-oxime upon ovarian cancer cells. Tumour Biol. 2016, 37, 4857–4864. [Google Scholar] [CrossRef] [PubMed]
- Ragozzino, E.; Brancaccio, M.; Di Costanzo, A.; Scalabrì, F.; Andolfi, G.; Wanderlingh, L.G.; Patriarca, E.J.; Minchiotti, G.; Altamura, S.; Summa, V.; et al. 6-Bromoindirubin-3’-oxime intercepts GSK3 signaling to promote and enhance skeletal muscle differentiation affecting miR-206 expression in mice. Sci. Rep. 2019, 9, 18091. [Google Scholar] [CrossRef]
- Zhao, X.E.; Yang, Z.; Gao, Z.; Ge, J.; Wei, Q.; Ma, B. 6-Bromoindirubin-3′-oxime promotes osteogenic differentiation of canine BMSCs through inhibition of GSK3β activity and activation of the Wnt/β-catenin signaling pathway. An. Acad. Bras. Cienc. 2019, 91, e20180459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chon, E.; Flanagan, B.; de Sá Rodrigues, L.C.; Piskun, C.; Stein, T.J. 6-Bromoindirubin-3′oxime (BIO) decreases proliferation and migration of canine melanoma cell lines. Vet. J. 2015, 205, 305–312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Zou, D.; Wang, N.; Tan, T.; Liu, Y.; Zhao, Q.; Pu, Y.; Thapa, R.J.; Chen, J. SFRP5 inhibits the migration and invasion of melanoma cells through Wnt signaling pathway. OncoTargets Ther. 2018, 11, 8761–8772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yi, Y.; Liao, B.; Zheng, Z.; Yang, X.; Yang, Y.; Zhou, Y.; Tan, B.; Yang, X. Downregulation of DEC1 inhibits proliferation, migration and invasion, and induces apoptosis in ovarian cancer cells via regulation of Wnt/β-catenin signaling pathway. Exp. Ther. Med. 2021, 21, 372. [Google Scholar] [CrossRef]
- De Jaime-Soguero, A.; Aulicino, F.; Ertaylan, G.; Griego, A.; Cerrato, A.; Tallam, A.; Del Sol, A.; Cosma, M.P.; Lluis, F. Wnt/Tcf1 pathway restricts embryonic stem cell cycle through activation of the Ink4/Arf locus. PLoS Genet. 2017, 13, e1006682. [Google Scholar] [CrossRef] [Green Version]
- Lianguzova, M.S.; Chuykin, I.A.; Nordheim, A.; Pospelov, V.A. Phosphoinositide 3-kinase inhibitor LY294002 but not serum withdrawal suppresses proliferation of murine embryonic stem cells. Cell Biol. Int. 2007, 31, 330–337. [Google Scholar] [CrossRef]
- Lin, X.; Zha, Y.; Zeng, X.Z.; Dong, R.; Wang, Q.H.; Wang, D.T. Role of the Wnt/β-catenin signaling pathway in inducing apoptosis and renal fibrosis in 5/6-nephrectomized rats. Mol. Med. Rep. 2017, 15, 3575–3582. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Nam, S.; Tian, Y.; Yang, F.; Wu, J.; Wang, Y.; Scuto, A.; Polychronopoulos, P.; Magiatis, P.; Skaltsounis, L.; et al. 6-Bromoindirubin-3′-oxime inhibits JAK/STAT3 signaling and induces apoptosis of human melanoma cells. Cancer Res. 2011, 71, 3972–3979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Ma, X.; Yan, S.; Shen, S.; Zhu, H.; Gu, Y.; Wang, H.; Qin, G.; Yu, Q. 17-hydroxy-jolkinolide B inhibits signal transducers and activators of transcription 3 signaling by covalently cross-linking Janus kinases and induces apoptosis of human cancer cells. Cancer Res. 2009, 69, 7302–7310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Epling-Burnette, P.K.; Liu, J.H.; Catlett-Falcone, R.; Turkson, J.; Oshiro, M.; Kothapalli, R.; Li, Y.; Wang, J.M.; Yang-Yen, H.F.; Karras, J.; et al. Inhibition of STAT3 signaling leads to apoptosis of leukemic large granular lymphocytes and decreased Mcl-1 expression. J. Clin. Investig. 2001, 107, 351–362. [Google Scholar] [CrossRef]
- Oshiro, M.M.; Landowski, T.H.; Catlett-Falcone, R.; Hazlehurst, L.A.; Huang, M.; Jove, R.; Dalton, W.S. Inhibition of JAK kinase activity enhances Fas-mediated apoptosis but reduces cytotoxic activity of topoisomerase II inhibitors in U266 myeloma cells. Clin. Cancer Res. 2001, 7, 4262–4271. [Google Scholar] [PubMed]
- Xiong, H.; Zhang, Z.-G.; Tian, X.-Q.; Sun, D.-F.; Liang, Q.-C.; Zhang, Y.-J.; Lu, R.; Chen, Y.-X.; Fang, J.-Y. Inhibition of JAK1, 2/STAT3 signaling induces apoptosis, cell cycle arrest, and reduces tumor cell invasion in colorectal cancer cells. Neoplasia 2008, 10, 287–297. [Google Scholar] [CrossRef] [Green Version]
- Amin, H.M.; Medeiros, L.J.; Ma, Y.; Feretzaki, M.; Das, P.; Leventaki, V.; Rassidakis, G.Z.; O’Connor, S.L.; McDonnell, T.J.; Lai, R. Inhibition of JAK3 induces apoptosis and decreases anaplastic lymphoma kinase activity in anaplastic large cell lymphoma. Oncogene 2003, 22, 5399–5407. [Google Scholar] [CrossRef] [Green Version]
- Heo, J.S.; Lee, S.Y.; Lee, J.C. Wnt/β-catenin signaling enhances osteoblastogenic differentiation from human periodontal ligament fibroblasts. Mol. Cells 2010, 30, 449–454. [Google Scholar] [CrossRef]
- Shen, S.; Zhang, Y.; Zhang, S.; Wang, B.; Shang, L.; Shao, J.; Lin, M.; Cui, Y.; Sun, S.; Ge, S. 6-Bromoindirubin-3′-oxime Promotes Osteogenic Differentiation of Periodontal Ligament Stem Cells and Facilitates Bone Regeneration in a Mouse Periodontitis Model. ACS Biomater. Sci. Eng. 2021, 7, 232–241. [Google Scholar] [CrossRef]
- Wang, J.; Liu, B.; Gu, S.; Liang, J. Effects of Wnt/β-catenin signalling on proliferation and differentiation of apical papilla stem cells. Cell Prolif. 2012, 45, 121–131. [Google Scholar] [CrossRef]
- Vijaykumar, A.; Root, S.H.; Mina, M. Wnt/β-Catenin Signaling Promotes the Formation of Preodontoblasts In Vitro. J. Dent. Res. 2021, 100, 387–396. [Google Scholar] [CrossRef]
- Gaur, T.; Lengner, C.J.; Hovhannisyan, H.; Bhat, R.A.; Bodine, P.V.; Komm, B.S.; Javed, A.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; et al. Canonical WNT signaling promotes osteogenesis by directly stimulating Runx2 gene expression. J. Biol. Chem. 2005, 280, 33132–33140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Khavandgar, Z.; Lin, S.H.; Murshed, M. Lithium chloride attenuates BMP-2 signaling and inhibits osteogenic differentiation through a novel WNT/GSK3- independent mechanism. Bone 2011, 48, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Rattanawarawipa, P.; Pavasant, P.; Osathanon, T.; Sukarawan, W. Effect of lithium chloride on cell proliferation and osteogenic differentiation in stem cells from human exfoliated deciduous teeth. Tissue Cell 2016, 48, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Ishimoto, K.; Hayano, S.; Yanagita, T.; Kurosaka, H.; Kawanabe, N.; Itoh, S.; Ono, M.; Kuboki, T.; Kamioka, H.; Yamashiro, T. Topical application of lithium chloride on the pulp induces dentin regeneration. PLoS ONE 2015, 10, e0121938. [Google Scholar] [CrossRef]
- Sustmann, C.; Flach, H.; Ebert, H.; Eastman, Q.; Grosschedl, R. Cell-type-specific function of BCL9 involves a transcriptional activation domain that synergizes with beta-catenin. Mol. Cell Biol. 2008, 28, 3526–3537. [Google Scholar] [CrossRef] [Green Version]
- Moussaieff, A.; Rouleau, M.; Kitsberg, D.; Cohen, M.; Levy, G.; Barasch, D.; Nemirovski, A.; Shen-Orr, S.; Laevsky, I.; Amit, M.; et al. Glycolysis-mediated changes in acetyl-CoA and histone acetylation control the early differentiation of embryonic stem cells. Cell Metab. 2015, 21, 392–402. [Google Scholar] [CrossRef] [Green Version]
- Nowwarote, N.; Sukarawan, W.; Pavasant, P.; Foster, B.L.; Osathanon, T. Basic fibroblast growth factor regulates phosphate/pyrophosphate regulatory genes in stem cells isolated from human exfoliated deciduous teeth. Stem Cell Res. Ther. 2018, 9, 345. [Google Scholar] [CrossRef]
- Foster, B.L.; Tompkins, K.A.; Rutherford, R.B.; Zhang, H.; Chu, E.Y.; Fong, H.; Somerman, M.J. Phosphate: Known and potential roles during development and regeneration of teeth and supporting structures. Birth Defects Res. C Embryo Today 2008, 84, 281–314. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.M.; Lee, E.H. Transcriptional regulatory cascades in Runx2-dependent bone development. Tissue Eng. Part. B Rev. 2013, 19, 254–263. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Rani, S.; Wu, Y.; Unterbrink, A.; Gu, T.T.; Gluhak-Heinrich, J.; Chuang, H.H.; Macdougall, M. Differential regulation of dentin sialophosphoprotein expression by Runx2 during odontoblast cytodifferentiation. J. Biol. Chem. 2005, 280, 29717–29727. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Gu, T.T.; Sreenath, T.; Kulkarni, A.B.; Karsenty, G.; MacDougall, M. Spatial expression of Cbfa1/Runx2 isoforms in teeth and characterization of binding sites in the DSPP gene. Connect. Tissue Res. 2002, 43, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Liu, M.; Ono, N.; Bringhurst, F.R.; Kronenberg, H.M.; Guo, J. Loss of wnt/β-catenin signaling causes cell fate shift of preosteoblasts from osteoblasts to adipocytes. J. Bone Miner. Res. 2012, 27, 2344–2358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McBeath, R.; Pirone, D.M.; Nelson, C.M.; Bhadriraju, K.; Chen, C.S. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev. Cell 2004, 6, 483–495. [Google Scholar] [CrossRef] [Green Version]
- Boyce, B.F.; Xing, L. Functions of RANKL/RANK/OPG in bone modeling and remodeling. Arch. Biochem. Biophys. 2008, 473, 139–146. [Google Scholar] [CrossRef] [Green Version]
- Palumbo, S.; Li, W.J. Osteoprotegerin enhances osteogenesis of human mesenchymal stem cells. Tissue Eng. Part. A 2013, 19, 2176–2187. [Google Scholar] [CrossRef] [PubMed]
- Bao, Q.; Chen, S.; Qin, H.; Feng, J.; Liu, H.; Liu, D.; Li, A.; Shen, Y.; Zhao, Y.; Li, J.; et al. An appropriate Wnt/β-catenin expression level during the remodeling phase is required for improved bone fracture healing in mice. Sci. Rep. 2017, 7, 2695. [Google Scholar] [CrossRef]
- Xu, X.J.; Shen, L.; Yang, Y.P.; Zhu, R.; Shuai, B.; Li, C.G.; Wu, M.X. Serum β-Catenin Levels Associated with the Ratio of RANKL/OPG in Patients with Postmenopausal Osteoporosis. Int. J. Endocrinol. 2013, 2013, 534352. [Google Scholar] [CrossRef] [Green Version]
- Huang, T.B.; Li, Y.Z.; Yu, K.; Yu, Z.; Wang, Y.; Jiang, Z.W.; Wang, H.M.; Yang, G.L. Effect of the Wnt signal-RANKL/OPG axis on the enhanced osteogenic integration of a lithium incorporated surface. Biomater. Sci. 2019, 7, 1101–1116. [Google Scholar] [CrossRef]
- Kramer, I.; Halleux, C.; Keller, H.; Pegurri, M.; Gooi, J.H.; Weber, P.B.; Feng, J.Q.; Bonewald, L.F.; Kneissel, M. Osteocyte Wnt/beta-catenin signaling is required for normal bone homeostasis. Mol. Cell Biol. 2010, 30, 3071–3085. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.-x.; Cheng, X.-y.; Wang, Y.; Yin, W. Toosendanin inhibits adipogenesis by activating Wnt/β-catenin signaling. Sci. Rep. 2018, 8, 4626. [Google Scholar] [CrossRef]
- Zaragosi, L.E.; Wdziekonski, B.; Fontaine, C.; Villageois, P.; Peraldi, P.; Dani, C. Effects of GSK3 inhibitors on in vitro expansion and differentiation of human adipose-derived stem cells into adipocytes. BMC Cell Biol. 2008, 9, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ross, S.E.; Hemati, N.; Longo, K.A.; Bennett, C.N.; Lucas, P.C.; Erickson, R.L.; MacDougald, O.A. Inhibition of adipogenesis by Wnt signaling. Science 2000, 289, 950–953. [Google Scholar] [CrossRef] [PubMed]
- Ma, E.B.; Sahar, N.E.; Jeong, M.; Huh, J.Y. Irisin Exerts Inhibitory Effect on Adipogenesis Through Regulation of Wnt Signaling. Front. Physiol. 2019, 10, 1085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, B.H.; Qiang, L.; Farmer, S.R. Phosphorylation of C/EBPbeta at a consensus extracellular signal-regulated kinase/glycogen synthase kinase 3 site is required for the induction of adiponectin gene expression during the differentiation of mouse fibroblasts into adipocytes. Mol. Cell Biol. 2004, 24, 8671–8680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prestwich, T.C.; Macdougald, O.A. Wnt/beta-catenin signaling in adipogenesis and metabolism. Curr. Opin. Cell Biol. 2007, 19, 612–617. [Google Scholar] [CrossRef] [Green Version]
- Bennett, C.N.; Ross, S.E.; Longo, K.A.; Bajnok, L.; Hemati, N.; Johnson, K.W.; Harrison, S.D.; MacDougald, O.A. Regulation of Wnt signaling during adipogenesis. J. Biol. Chem. 2002, 277, 30998–31004. [Google Scholar] [CrossRef] [Green Version]
- Xie, Y.Y.; Mo, C.L.; Cai, Y.H.; Wang, W.J.; Hong, X.X.; Zhang, K.K.; Liu, Q.F.; Liu, Y.J.; Hong, J.J.; He, T.; et al. Pygo2 Regulates Adiposity and Glucose Homeostasis via β-Catenin-Axin2-GSK3β Signaling Pathway. Diabetes 2018, 67, 2569–2584. [Google Scholar] [CrossRef] [Green Version]
- Uematsu, K.; He, B.; You, L.; Xu, Z.; McCormick, F.; Jablons, D.M. Activation of the Wnt pathway in non small cell lung cancer: Evidence of dishevelled overexpression. Oncogene 2003, 22, 7218–7221. [Google Scholar] [CrossRef] [Green Version]
- Coopes, A.; Henry, C.E.; Llamosas, E.; Ford, C.E. An update of Wnt signalling in endometrial cancer and its potential as a therapeutic target. Endocr. Relat. Cancer 2018, 25, R647–R662. [Google Scholar] [CrossRef]
- Schatoff, E.M.; Leach, B.I.; Dow, L.E. Wnt Signaling and Colorectal Cancer. Curr. Colorectal. Cancer Rep. 2017, 13, 101–110. [Google Scholar] [CrossRef] [Green Version]
- Su, N.; Wang, P.; Li, Y. Role of Wnt/β-catenin pathway in inducing autophagy and apoptosis in multiple myeloma cells. Oncol. Lett. 2016, 12, 4623–4629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ríos, J.A.; Godoy, J.A.; Inestrosa, N.C. Wnt3a ligand facilitates autophagy in hippocampal neurons by modulating a novel GSK-3β-AMPK axis. Cell Commun. Signal. 2018, 16, 15. [Google Scholar] [CrossRef] [PubMed]
- Yun, E.J.; Kim, S.; Hsieh, J.T.; Baek, S.T. Wnt/β-catenin signaling pathway induces autophagy-mediated temozolomide-resistance in human glioblastoma. Cell Death Dis. 2020, 11, 771. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; McLean, S.; Carter, D.E.; Leask, A. The gene expression profile induced by Wnt 3a in NIH 3T3 fibroblasts. J. Cell Commun. Signal. 2007, 1, 175–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salinas, P.C. Modulation of the microtubule cytoskeleton: A role for a divergent canonical Wnt pathway. Trends Cell Biol. 2007, 17, 333–342. [Google Scholar] [CrossRef]
- Lai, S.L.; Chien, A.J.; Moon, R.T. Wnt/Fz signaling and the cytoskeleton: Potential roles in tumorigenesis. Cell Res. 2009, 19, 532–545. [Google Scholar] [CrossRef]
- Gumber, D.; Do, M.; Suresh Kumar, N.; Sonavane, P.R.; Wu, C.C.N.; Cruz, L.S.; Grainger, S.; Carson, D.; Gaasterland, T.; Willert, K. Selective activation of FZD7 promotes mesendodermal differentiation of human pluripotent stem cells. eLife 2020, 9, e63060. [Google Scholar] [CrossRef]
- Sebastian, A.; Hum, N.R.; Murugesh, D.K.; Hatsell, S.; Economides, A.N.; Loots, G.G. Wnt co-receptors Lrp5 and Lrp6 differentially mediate Wnt3a signaling in osteoblasts. PLoS ONE 2017, 12, e0188264. [Google Scholar] [CrossRef] [Green Version]
- Chang, B.; Ma, C.; Liu, X. Nanofibrous Tubular Three-Dimensional Platform for Single Dental Pulp Stem Cell Polarization. ACS Appl. Mater. Interfaces 2020, 12, 54481–54488. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 1 March 2021).
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kornsuthisopon, C.; Rochanavibhata, S.; Nowwarote, N.; Tompkins, K.A.; Sukarawan, W.; Osathanon, T. 6-Bromoindirubin-3′-Oxime Regulates Colony Formation, Apoptosis, and Odonto/Osteogenic Differentiation in Human Dental Pulp Stem Cells. Int. J. Mol. Sci. 2022, 23, 8676. https://doi.org/10.3390/ijms23158676
Kornsuthisopon C, Rochanavibhata S, Nowwarote N, Tompkins KA, Sukarawan W, Osathanon T. 6-Bromoindirubin-3′-Oxime Regulates Colony Formation, Apoptosis, and Odonto/Osteogenic Differentiation in Human Dental Pulp Stem Cells. International Journal of Molecular Sciences. 2022; 23(15):8676. https://doi.org/10.3390/ijms23158676
Chicago/Turabian StyleKornsuthisopon, Chatvadee, Sunisa Rochanavibhata, Nunthawan Nowwarote, Kevin A. Tompkins, Waleerat Sukarawan, and Thanaphum Osathanon. 2022. "6-Bromoindirubin-3′-Oxime Regulates Colony Formation, Apoptosis, and Odonto/Osteogenic Differentiation in Human Dental Pulp Stem Cells" International Journal of Molecular Sciences 23, no. 15: 8676. https://doi.org/10.3390/ijms23158676





