Oligonucleotides Isolation and Separation—A Review on Adsorbent Selection
Abstract
:1. Introduction
2. Types of Supports Used in OGNs Purification and Separation
2.1. Silica
2.2. Polymers
2.3. Magnetic Nanoparticles
3. Application of Adsorbents with Different Modifications for Separation of OGNs
3.1. RP Adsorbents
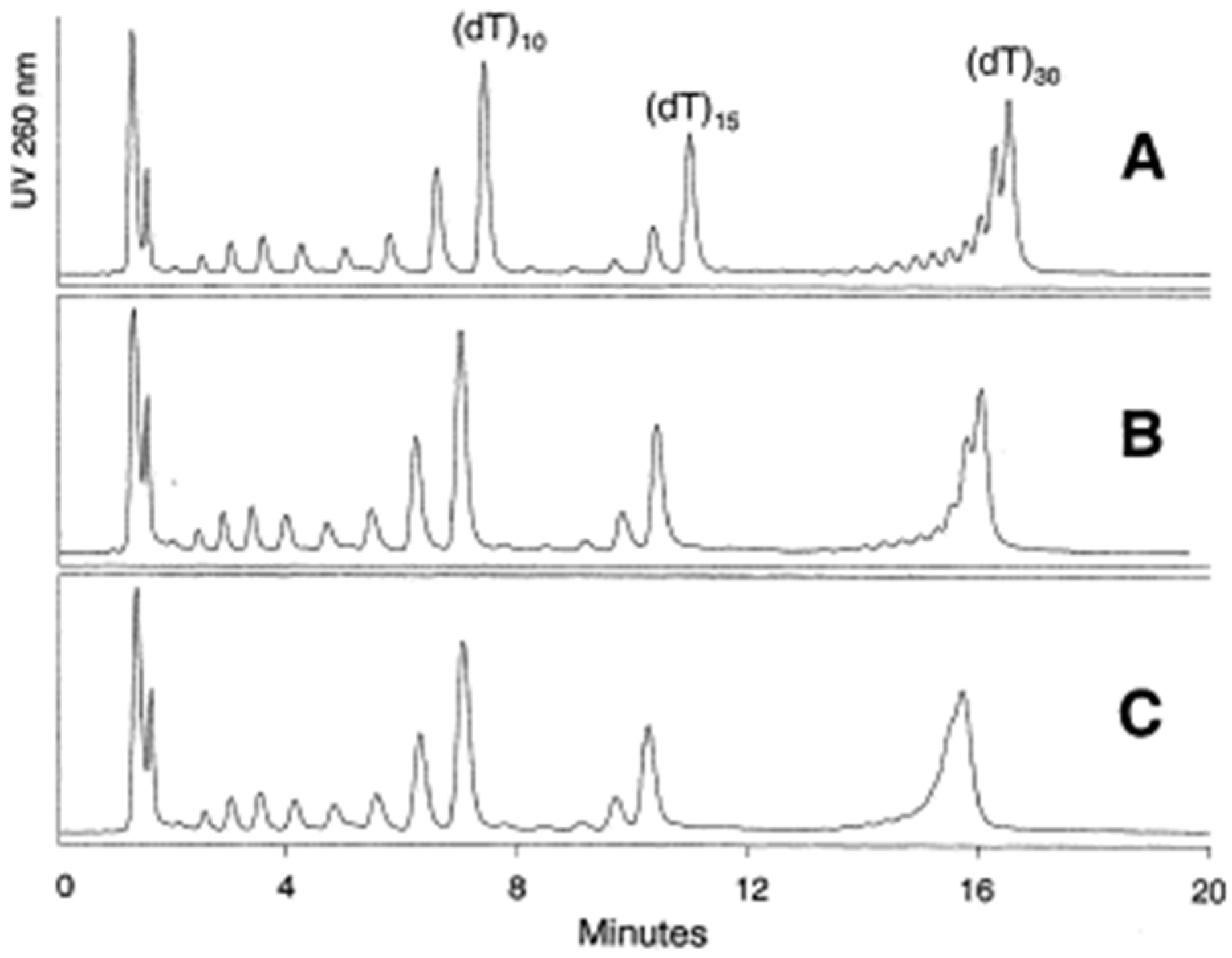
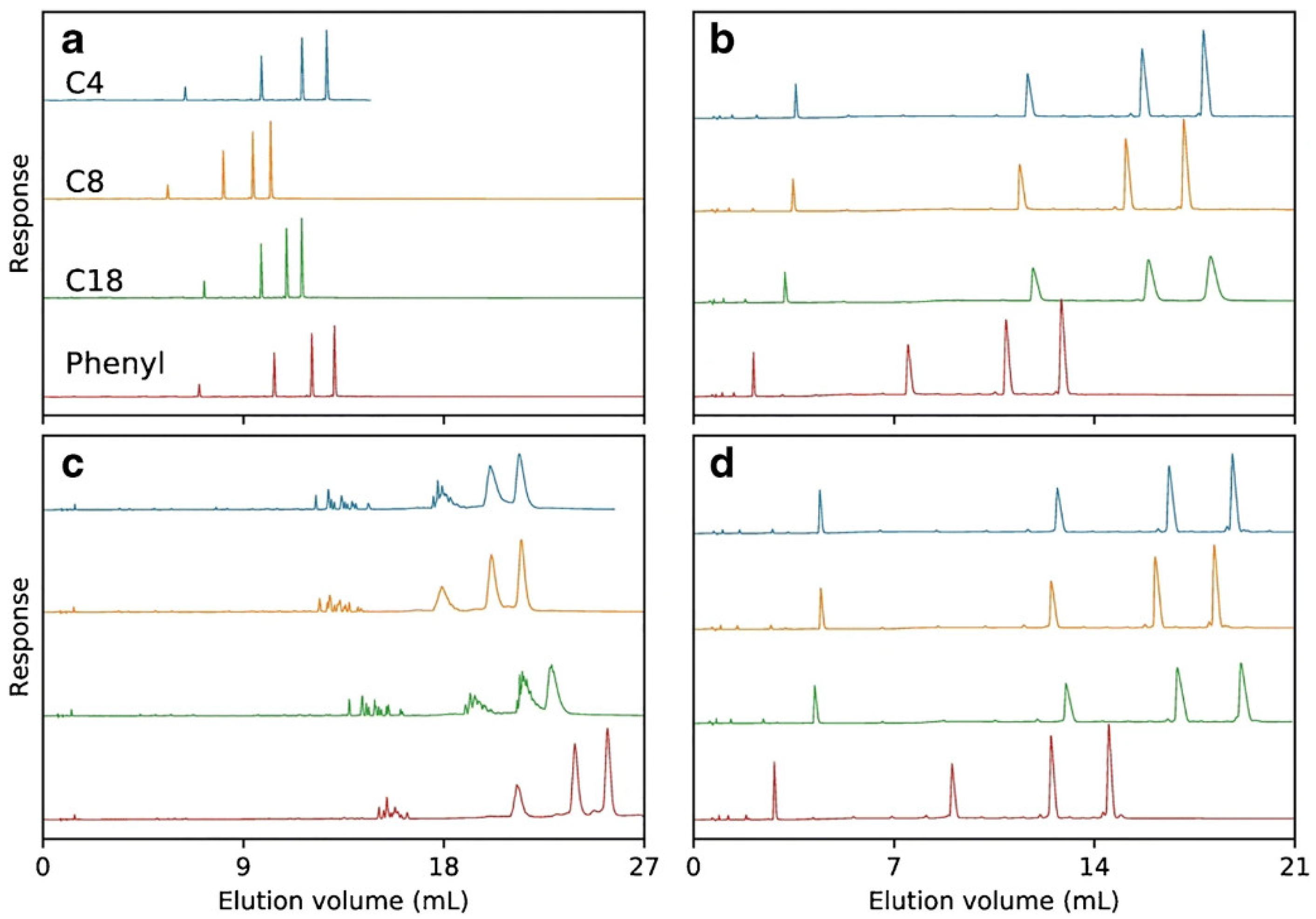
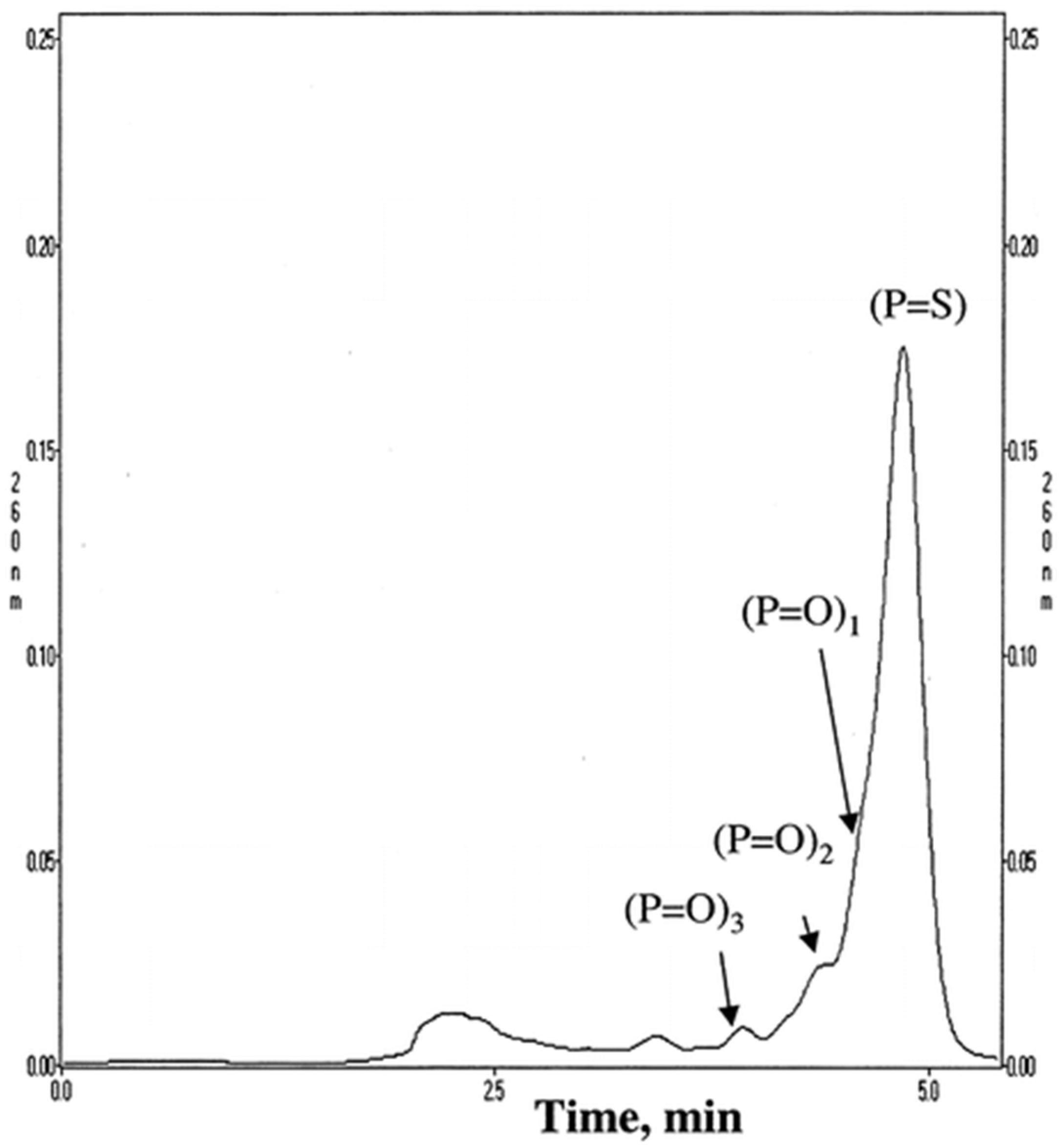
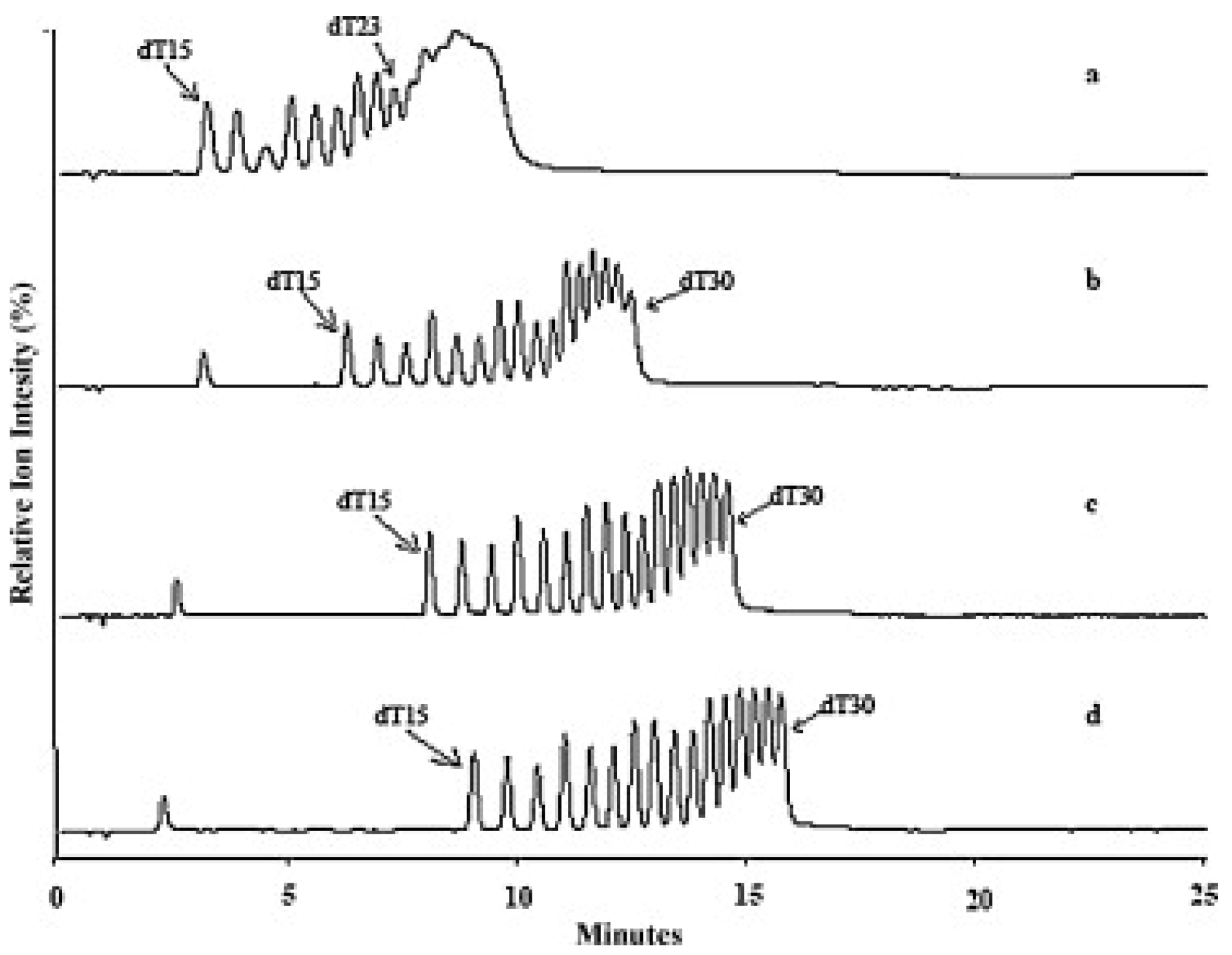
3.1.1. Adsorbents with Alkyl Ligands
3.1.2. Adsorbents with Aromatic Groups
3.1.3. Adsorbents with a Cholesterol Molecule
3.1.4. Adsorbents with Polar Groups Incorporated in Non-Polar Ligands
3.2. Adsorbents with Anion Exchange Groups
3.3. Polar ADSORBENTS
3.3.1. Adsorbents with Hydroxyl Ligands
3.3.2. Amide Adsorbents
3.3.3. Zwitterionic Adsorbents
3.4. OGNs-Modified Adsorbents
4. Application of Adsorbents with Different Modifications for Extraction of OGNs
5. Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wang, J.; Chen, J.; Sen, S. MicroRNA as Biomarkers and Diagnostics. J. Cell. Physiol. 2016, 231, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Huang, W. MicroRNAs: Biomarkers, diagnostics, and therapeutics. Methods Mol. Biol. 2017, 1617, 57–67. [Google Scholar] [PubMed]
- Dias, N.; Stein, C.A. Antisense oligonucleotides: Basic concepts and mechanisms. Mol. Cancer Ther. 2002, 1, 347–355. [Google Scholar] [PubMed]
- Urban, E.; Noe, C.R. Structural modifications of antisense oligonucleotides. Il Farm. 2003, 58, 243–258. [Google Scholar] [CrossRef]
- Rüger, J.; Ioannou, S.; Castanotto, D.; Stein, C.A. Oligonucleotides to the (Gene) Rescue: FDA Approvals 2017–2019. Trends Pharmacol. Sci. 2020, 41, 27–41. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.K.; Sharma, R.K.; Singh, S.K. Antisense oligonucleotides: Modifications and clinical trials. Med. Chem. Comm. 2014, 5, 1454–1471. [Google Scholar] [CrossRef]
- Bonilla, J.V.; Srivatsa, G.S. Handbook of Analysis of Oligonucleotides and Related Products; CRC Press: Boca Raton, FL, USA, 2011. [Google Scholar]
- Peng, J.; Tang, F.; Zhou, R.; Xie, X.; Li, S.; Xie, F.; Yu, P.; Mu, L. New techniques of on-line biological sample processing and their application in the field of biopharmaceutical analysis. Acta Pharm. Sin. B 2016, 6, 540–551. [Google Scholar] [CrossRef]
- Nuckowski, Ł.; Kaczmarkiewicz, A.; Studzińska, S. Review on sample preparation methods for oligonucleotides analysis by liquid chromatography. J. Chromatogr. B 2018, 1090, 90–100. [Google Scholar] [CrossRef]
- Biba, M.; Welch, C.J.; Foley, J.P.; Mao, B.; Vazquez, E.; Arvary, R.A. Evaluation of core-shell particle columns for ion-pair reversed-phase liquid chromatography analysis of oligonucleotides. J. Pharm. Biomed. Anal. 2013, 72, 25–32. [Google Scholar] [CrossRef]
- Gilar, M.; Bouvier, E.S.P. Purification of crude DNA oligonucleotides by solid-phase extraction and reversed-phase high-performance liquid chromatography. J. Chromatogr. A 2000, 890, 167–177. [Google Scholar] [CrossRef]
- Gilar, M.; Fountain, K.J.; Budman, Y.; Neue, U.D.; Yardley, K.R.; Rainville, P.D.; Russell, R.J.; Gebler, J.C. Ion-pair reversed-phase high-performance liquid chromatography analysis of oligonucleotides: Retention prediction. J. Chromatogr. A 2002, 958, 167–182. [Google Scholar] [CrossRef]
- Cen, Y.; Li, X.; Liu, D.; Pan, F.; Cai, Y.; Li, B.; Peng, W.; Wu, C.; Jiang, W.; Zhou, H. Development and validation of LC–MS/MS method for the detection and quantification of CpG oligonucleotides 107 (CpG ODN107) and its metabolites in mice plasma. J. Pharm. Biomed. Anal. 2012, 70, 447–455. [Google Scholar] [CrossRef] [PubMed]
- Qiao, J.-Q.; Liang, C.; Zhu, Z.-Y.; Cao, Z.-M.; Zheng, W.-J.; Lian, H.-Z. Monolithic alkylsilane column: A promising separation medium for oligonucleotides by ion-pair reversed-phase liquid chromatography. J. Chromatogr. A 2018, 1569, 168–177. [Google Scholar] [CrossRef] [PubMed]
- Kirkland, J.J.; Truszkowski, F.A.; Dilks, C.H.; Engel, G.S. Superficially porous silica microspheres for fast high-performance liquid chromatography of macromolecules. J. Chromatogr. A 2000, 890, 3–13. [Google Scholar] [CrossRef]
- Weng, G.; Liu, Z.; Chen, J.; Wang, F.; Pan, Y.; Zhang, Y. Enhancing the Mass Spectrometry Sensitivity for Oligonucleotide Detection by Organic Vapor Assisted Electrospray. Anal. Chem. 2017, 89, 10256–10263. [Google Scholar] [CrossRef]
- Deshmukh, R.R.; Miller, J.E.; De Leon, P.; Leitch, W.E.; Cole, D.L.; Sanghvi, Y.S. Process Development for Purification of Therapeutic Antisense Oligonucleotides by Anion-Exchange Chromatography. Org. Process Res. Dev. 2000, 4, 205–213. [Google Scholar] [CrossRef]
- Bunček, M.; Bačkovská, V.; Holasová, Š.; Radilová, H.; Šafářová, M.; Kunc, F.; Haluza, R. Retention behavior of oligonucleotides on a glycidyl methacrylate-based DEAE-modified sorbent. Chromatographia 2005, 62, 263–269. [Google Scholar] [CrossRef]
- Thayer, J.R.; Barreto, V.; Rao, S.; Pohl, C. Control of oligonucleotide retention on a pH-stabilized strong anion exchange column. Anal. Biochem. 2005, 338, 39–47. [Google Scholar] [CrossRef]
- Thayer, J.; Puri, N.; Burnett, C.; Hail, M.; Rao, S. Identification of RNA linkage isomers by anion exchange purification with electrospray ionization mass spectrometry of automatically desalted phosphodiesterase-II digests. Anal. Biochem. 2010, 399, 110–117. [Google Scholar] [CrossRef]
- Thayer, J.R.; Wu, Y.; Hansen, E.; Angelino, M.D.; Rao, S. Separation of oligonucleotide phosphorothioate diastereoisomers by pellicular anion-exchange chromatography. J. Chromatogr. A 2011, 1218, 802–808. [Google Scholar] [CrossRef]
- Lobue, P.A.; Jora, M.; Addepalli, B.; Limbach, P.A. Oligonucleotide analysis by hydrophilic interaction liquid chromatography-mass spectrometry in the absence of ion-pair reagents. J. Chromatogr. A 2019, 1595, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Holdšvendová, P.; Suchánková, J.; Bunček, M.; Bačkovská, V.; Coufal, P. Hydroxymethyl methacrylate-based monolithic columns designed for separation of oligonucleotides in hydrophilic-interaction capillary liquid chromatography. J. Biochem. Biophys. Methods 2007, 70, 23–29. [Google Scholar] [CrossRef]
- Oefner, P.J.; Huber, C.G. A decade of high-resolution liquid chromatography of nucleic acids on styrene–divinylbenzene copolymers. J. Chromatogr. B 2002, 782, 27–55. [Google Scholar] [CrossRef]
- Yamauchi, Y.; Taoka, M.; Nobe, Y.; Izumikawa, K.; Takahashi, N.; Nakayama, H.; Isobe, T. Denaturing reversed phase liquid chromatographic separation of non-coding ribonucleic acids on macro-porous polystyrene-divinylbenzene resins. J. Chromatogr. A 2013, 1312, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Kwaśniewska, K.; Gadzała-Kopciuch, R.; Buszewski, B. Magnetic molecular imprinted polymers as a tool for isolation and purification of biological samples. Open Chem. 2015, 13, 1228–1235. [Google Scholar] [CrossRef]
- Beveridge, J.S.; Stephens, J.R.; Williams, M.E. The Use of Magnetic Nanoparticles in Analytical Chemistry. Annu. Rev. Anal. Chem. 2011, 4, 251–273. [Google Scholar] [CrossRef] [PubMed]
- Dadfar, S.M.; Roemhild, K.; Drude, N.I.; von Stillfried, S.; Knüchel, R.; Kiessling, F.; Lammers, T. Iron oxide nanoparticles: Diagnostic, therapeutic and theranostic applications. Adv. Drug Deliv. Rev. 2019, 138, 302–325. [Google Scholar] [CrossRef]
- Anik, M.I.; Hossain, M.K.; Hossain, I.; Mahfuz, A.M.U.B.; Rahman, M.T.; Ahmed, I. Recent progress of magnetic nanoparticles in biomedical applications: A review. Nano Sel. 2021, 2, 1146–1186. [Google Scholar] [CrossRef]
- Tiwari, A.P.; Satvekar, R.K.; Rohiwal, S.S.; Karande, V.A.; Raut, A.V.; Patil, P.G.; Shete, P.B.; Ghosh, S.J.; Pawar, S.H. Magneto-separation of genomic deoxyribose nucleic acid using pH responsive Fe3O4@silica@chitosan nanoparticles in biological samples. RSC Adv. 2015, 5, 8463–8470. [Google Scholar] [CrossRef]
- Wang, J.; Ali, Z.; Wang, N.; Liang, W.; Liu, H.; Li, F.; Yang, H.; He, L.; Nie, L.; He, N.; et al. Simultaneous extraction of DNA and RNA from Escherichia coli BL 21 based on silica-coated magnetic nanoparticles. Sci. China Ser. B Chem. 2015, 58, 1774–1778. [Google Scholar] [CrossRef]
- Tang, C.; He, Z.; Liu, H.; Huang, H.; Yang, G.; Xiao, Z.; Li, S.; Liu, H.; Deng, Y.; Chen, Z.; et al. Application of magnetic nanoparticles in nucleic acid detection. J. Nanobiotechnol. 2020, 18, 62. [Google Scholar] [CrossRef] [PubMed]
- Ye, G.; Beverly, M. The use of strong anion-exchange (SAX) magnetic particles for the extraction of therapeutic siRNA and their analysis by liquid chromatography/mass spectrometry. Rapid Commun. Mass Spectrom. 2011, 25, 3207–3215. [Google Scholar] [CrossRef]
- Basiri, B.; Sutton, J.M.; Hooshfar, S.; Byrnes, C.C.; Murph, M.M.; Bartlett, M.G. Direct identification of microribonucleic acid miR-451 from plasma using liquid chromatography mass spectrometry. J. Chromatogr. A 2019, 1584, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Sips, L.; Ediage, E.N.; Ingelse, B.; Verhaeghe, T.; Dillen, L. LC–MS quantification of oligonucleotides in biological matrices with SPE or hybridization extraction. Bioanalysis 2019, 11, 1941–1954. [Google Scholar] [CrossRef] [PubMed]
- Dillen, L.; Sips, L.; Greway, T.; Verhaeghe, T. Quantitative analysis of imetelstat in plasma with LC–MS/MS using solid-phase or hybridization extraction. Bioanalysis 2017, 9, 1859–1872. [Google Scholar] [CrossRef] [PubMed]
- Neue, U.D. Chromatography: Liquid|Mechanisms: Reversed Phases; Elsevier: Amsterdam, The Netherlands, 2007; pp. 1–7. [Google Scholar] [CrossRef]
- Cecchi, T. Ion pairing chromatography. Crit. Rev. Anal. Chem. 2008, 38, 161–213. [Google Scholar] [CrossRef] [PubMed]
- Dai, G.; Wei, X.; Liu, Z.; Liu, S.; Marcucci, G.; Chan, K.K. Characterization and quantification of Bcl-2 antisense G3139 and metabolites in plasma and urine by ion-pair reversed phase HPLC coupled with electrospray ion-trap mass spectrometry. J. Chromatogr. B 2005, 825, 201–213. [Google Scholar] [CrossRef]
- Zhang, W.; Leighl, N.; Zawisza, D.; Moore, M.J.; Chen, E.X. Determination of GTI-2040, a novel antisense oligonucleotide, in human plasma by using HPLC combined with solid phase and liquid-liquid extractions. J. Chromatogr. B 2005, 829, 45–49. [Google Scholar] [CrossRef]
- Deng, P.; Chen, X.; Zhang, G.; Zhong, D. Bioanalysis of an oligonucleotide and its metabolites by liquid chromatography–tandem mass spectrometry. J. Pharm. Biomed. Anal. 2010, 52, 571–579. [Google Scholar] [CrossRef]
- Zhang, G.; Lin, J.; Srinivasan, K.; Kavetskaia, O.; Duncan, J.N. Strategies for Bioanalysis of an Oligonucleotide Class Macromolecule from Rat Plasma Using Liquid Chromatography−Tandem Mass Spectrometry. Anal. Chem. 2007, 79, 3416–3424. [Google Scholar] [CrossRef]
- Studzińska, S.; Rola, R.; Buszewski, B. Development of a method based on ultra high performance liquid chromatography coupled with quadrupole time-of-flight mass spectrometry for studying the in vitro metabolism of phosphorothioate oligonucleotides. Anal. Bioanal. Chem. 2016, 408, 1585–1595. [Google Scholar] [CrossRef] [PubMed]
- Ewles, M.; Goodwin, L.; Schneider, A.; Rothhammer-Hampl, T. Quantification of oligonucleotides by LC–MS/MS: The challenges of quantifying a phosphorothioate oligonucleotide and multiple metabolites. Bioanalysis 2014, 6, 447–464. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Dai, G.; Liu, Z.; Cheng, H.; Xie, Z.; Marcucci, G.; Chan, K.K. Metabolism of GTI-2040, a phosphorothioate oligonucleotide antisense, using ion-pair reversed phase high performance liquid chromatography (HPLC) coupled with electrospray ion-trap mass spectrometry. AAPS J. 2006, 8, E743–E755. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Dai, G.; Liu, Z.; Cheng, H.; Xie, Z.; Klisovic, R.; Marcucci, G.; Chan, K.K. Enzyme Kinetics of GTI-2040, a Phosphorothioate Oligonucleotide Targeting Ribonucleotide Reductase. Drug Metab. Dispos. 2008, 36, 2227–2233. [Google Scholar] [CrossRef]
- Husser, C.; Brink, A.; Zell, M.; Müller, M.B.; Koller, E.; Schadt, S. Identification of GalNAc-conjugated antisense oligonucleotide metabolites using an untargeted and generic approach based on high resolution mass spectrometry. Anal. Chem. 2017, 89, 6821–6826. [Google Scholar] [CrossRef]
- Zou, Y.; Tiller, P.; Chen, L.-W.; Beverly, M.; Hochman, J. Metabolite identification of small interfering RNA duplex by high-resolution accurate mass spectrometry. Rapid Commun. Mass Spectrom. 2008, 22, 1871–1881. [Google Scholar] [CrossRef]
- Shimizu, R.; Kitade, M.; Kobayashi, T.; Hori, S.-I.; Watanabe, A. Pharmacokinetic–pharmacodynamic modeling for reduction of hepatic apolipoprotein B mRNA and plasma total cholesterol after administration of antisense oligonucleotide in mice. J. Pharmacokinet. Pharmacodyn. 2015, 42, 67–77. [Google Scholar] [CrossRef]
- Hemsley, M.; Ewles, M.; Goodwin, L. Development of a bioanalytical method for quantification of a 15-mer oligonucleotide at sub-ng/ml concentrations using LC–MS/MS. Bioanalysis 2012, 4, 1457–1469. [Google Scholar] [CrossRef]
- Jiao, K.; Rashid, A.; Basu, S.K.; Zhu, S.; Brown, B.D.; Guerciolini, R.; Fambrough, U.M. Quantitative Analysis of Dicer Substrate Oligonucleotides in Mouse Liver by Ultra-High-Performance Liquid Chromatography—Electrospray Ionization Tandem Mass Spectrometry. ASSAY Drug Dev. Technol. 2012, 10, 278–288. [Google Scholar] [CrossRef]
- Ramanathan, L.; Shen, H. LC-TOF-MS methods to quantify siRNAs and major metabolite in plasma, urine and tissues. Bioanalysis 2019, 11, 1983–1992. [Google Scholar] [CrossRef]
- Kaczmarkiewicz, A.; Nuckowski, Ł.; Studzińska, S. Analysis of the first and second generation of antisense oligonucleotides in serum samples with the use of ultra high performance liquid chromatography coupled with tandem mass spectrometry. Talanta 2019, 196, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Wheller, R.; Summerfield, S.; Barfield, M. Comparison of accurate mass LC–MS and MRM LC–MS/MS for the quantification of a therapeutic small interfering RNA. Int. J. Mass Spectrom. 2013, 345–347, 45–53. [Google Scholar] [CrossRef]
- Wysoczynski, C.L.; Roemer, S.C.; Dostal, V.; Barkley, R.M.; Churchill, M.E.A.; Malarkey, C.S. Reversed-phase ion-pair liquid chromatography method for purification of duplex DNA with single base pair resolution. Nucleic Acids Res. 2013, 41, e194. [Google Scholar] [CrossRef] [PubMed]
- Studzińska, S.; Buszewski, B. Evaluation of ultrahigh-performance liquid chromatography columns for the analysis of unmodified and antisense oligonucleotides. Anal. Bioanal. Chem. 2014, 406, 7127–7136. [Google Scholar] [CrossRef] [PubMed]
- Studzińska, S.; Pietrzak, L.; Buszewski, B. The Effects of Stationary Phases on Retention and Selectivity of Oligonucleotides in IP-RP-HPLC. Chromatographia 2014, 77, 1589–1596. [Google Scholar] [CrossRef] [PubMed]
- Qiao, J.-Q.; Liang, C.; Wei, L.-C.; Cao, Z.-M.; Lian, H.-Z. Retention of nucleic acids in ion-pair reversed-phase high-performance liquid chromatography depends not only on base composition but also on base sequence. J. Sep. Sci. 2016, 39, 4502–4511. [Google Scholar] [CrossRef] [PubMed]
- Nikcevic, I.; Wyrzykiewicz, T.K.; Limbach, P.A. Detecting low-level synthesis impurities in modified phosphorothioate oligonucleotides using liquid chromatography–high resolution mass spectrometry. Int. J. Mass Spectrom. 2011, 304, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Enmark, M.; Rova, M.; Samuelsson, J.; Örnskov, E.; Schweikart, F.; Fornstedt, T. Investigation of factors influencing the separation of diastereomers of phosphorothioated oligonucleotides. Anal. Bioanal. Chem. 2019, 411, 3383–3394. [Google Scholar] [CrossRef]
- Nuckowski, Ł.; Kaczmarkiewicz, A.; Studzińska, S.; Buszewski, B. A new approach to preparation of antisense oligonucleotide samples with microextraction by packed sorbent. Analyst 2019, 144, 4622–4632. [Google Scholar] [CrossRef]
- Enmark, M.; Bagge, J.; Samuelsson, J.; Thunberg, L.; Örnskov, E.; Leek, H.; Limé, F.; Fornstedt, T. Analytical and preparative separation of phosphorothioated oligonucleotides: Columns and ion-pair reagents. Anal. Bioanal. Chem. 2020, 412, 299–309. [Google Scholar] [CrossRef]
- Studzińska, S.; Nuckowski, Ł.; Kilanowska, A. Ultra-High-Performance Reversed-Phase Liquid Chromatography Hyphenated with ESI-Q-TOF-MS for the Analysis of Unmodified and Antisense Oligonucleotides. Chromatographia 2020, 83, 349–360. [Google Scholar] [CrossRef]
- Studzińska, S.; Bocian, S.; Siecińska, L.; Buszewski, B. Application of phenyl-based stationary phases for the study of retention and separation of oligonucleotides. J. Chromatogr. B 2017, 1060, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Xiong, W.; Glick, J.; Lin, Y.; Vouros, P. Separation and Sequencing of Isomeric Oligonucleotide Adducts Using Monolithic Columns by Ion-Pair Reversed-Phase Nano-HPLC Coupled to Ion Trap Mass Spectrometry. Anal. Chem. 2007, 79, 5312–5321. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.L.; Guo, W.; Zang, J.; Khan, S.; Bardin, S.; Ahmad, A.; Duggan, J.X.; Ahmad, I. Quantification of raf antisense oligonucleotide (rafAON) in biological matrices by LC-MS/MS to support pharmacokinetics of a liposome-entrapped rafAON formulation. Biomed. Chromatogr. 2005, 19, 272–278. [Google Scholar] [CrossRef]
- Erb, R.; Leithner, K.; Bernkop-Schnürch, A.; Oberacher, H. Phosphorothioate Oligonucleotide Quantification by μ-Liquid Chromatography-Mass Spectrometry. AAPS J. 2012, 14, 728–737. [Google Scholar] [CrossRef] [PubMed]
- Andrews, C.L.; Harsch, A.; Vouros, P. Analysis of the in vitro digestion of modified DNA to oligonucleotides by LC–MS and LC–MS/MS. Int. J. Mass Spectrom. 2004, 231, 169–177. [Google Scholar] [CrossRef]
- Li, J.; Liu, J.; Enders, J.; Arciprete, M.; Tran, C.; Aluri, K.; Guan, L.H.; O’Shea, J.; Bisbe, A.; Charissé, K.; et al. Discovery of a novel deaminated metabolite of a single-stranded oligonucleotide in vivo by mass spectrometry. Bioanalysis 2019, 11, 1955–1965. [Google Scholar] [CrossRef]
- Liu, J.; Li, J.; Tran, C.; Aluri, K.; Zhang, X.; Clausen, V.; Zlatev, I.; Guan, L.; Chong, S.; Charisse, K.; et al. Oligonucleotide quantification and metabolite profiling by high-resolution and accurate mass spectrometry. Bioanalysis 2019, 11, 1967–1980. [Google Scholar] [CrossRef]
- Studzińska, S.; Krzemińska, K.; Szumski, M.; Buszewski, B. Application of a cholesterol stationary phase in the analysis of phosphorothioate oligonucleotides by means of ion pair chromatography coupled with tandem mass spectrometry. Talanta 2016, 154, 270–277. [Google Scholar] [CrossRef]
- Li, N.; El Zahar, N.M.; Saad, J.G.; van der Hage, E.R.E.; Bartlett, M.G. Alkylamine ion-pairing reagents and the chromatographic separation of oligonucleotides. J. Chromatogr. A 2018, 1580, 110–119. [Google Scholar] [CrossRef]
- Buszewski, B.; Safaei, Z.; Studzińska, S. Analysis of oligonucleotides by liquid chromatography with alkylamide stationary phase. Open Chem. 2015, 13, 1286–1292. [Google Scholar] [CrossRef]
- Studzińska, S.; Rola, R.; Buszewski, B. The impact of ion-pairing reagents on the selectivity and sensitivity in the analysis of modified oligonucleotides in serum samples by liquid chromatography coupled with tandem mass spectrometry. J. Pharm. Biomed. Anal. 2017, 138, 146–152. [Google Scholar] [CrossRef]
- Cook, K.; Thayer, J. Advantages of ion-exchange chromatography for oligonucleotide analysis. Bioanalysis 2011, 3, 1109–1120. [Google Scholar] [CrossRef] [PubMed]
- Cummins, P.M.; Rochfort, K.D.; O’Connor, B.F. Ion-Exchange chromatography: Basic principles and application. In Protein Chromatography; Walls, D., Loughran, S.T., Eds.; Springer: New York, NY, USA, 2017; pp. 209–223. [Google Scholar] [CrossRef]
- Arora, V.; Knapp, D.C.; Reddy, M.T.; Weller, D.D.; Iversen, P.L. Bioavailability and Efficacy of Antisense Morpholino Oligomers Targeted to c-myc and Cytochrome P-450 3A2 Following Oral Administration in Rats. J. Pharm. Sci. 2002, 91, 1009–1018. [Google Scholar] [CrossRef] [PubMed]
- Devi, G.R.; Beer, T.M.; Corless, C.L.; Arora, V.; Weller, D.L.; Iversen, P.L. In vivo bioavailability and pharmacokinetics of a c-MYC antisense phosphorodiamidate morpholino oligomer, AVI-4126, in solid tumors. Clin. Cancer Res. 2005, 11, 3930–3938. [Google Scholar] [CrossRef]
- Yang, X.; Hodge, R.P.; Luxon, B.A.; Shope, R.; Gorenstein, D.G. Separation of Synthetic Oligonucleotide Dithioates from Monothiophosphate Impurities by Anion-Exchange Chromatography on a Mono-Q Column. Anal. Biochem. 2002, 306, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Thayer, J.R.; Flook, K.J.; Woodruff, A.; Rao, S.; Pohl, C.A. New monolith technology for automated anion-exchange purification of nucleic acids. J. Chromatogr. B 2010, 878, 933–941. [Google Scholar] [CrossRef] [PubMed]
- Goyon, A.; Zhang, K. Characterization of antisense oligonucleotide impurities by ion-pairing reversed-phase and anion exchange chromatography coupled to hydrophilic interaction liquid chromatography/mass spectrometry using a versatile two-dimensional liquid chromatography set. Anal. Chem. 2020, 92, 5944–5951. [Google Scholar] [CrossRef]
- Crean, C.; Uvaydov, Y.; Geacintov, N.E.; Shafirovich, V. Oxidation of single-stranded oligonucleotides by carbonate radical anions: Generating intrastrand cross-links between guanine and thymine bases separated by cytosines. Nucleic Acids Res. 2008, 36, 742–755. [Google Scholar] [CrossRef]
- Grant, G.P.G.; Popova, A.; Qin, P.Z. Diastereomer characterizations of nitroxide-labeled nucleic acids. Biochem. Biophys. Res. Commun. 2008, 371, 451–455. [Google Scholar] [CrossRef]
- Totsingan, F.; Rossi, S.; Corradini, R.; Tedeschi, T.; Sforza, S.; Juris, A.; Scaravelli, E.; Marchelli, R. Label-free selective DNA detection with high mismatch recognition by PNA beacons and ion exchange HPLC. Org. Biomol. Chem. 2008, 6, 1232–1237. [Google Scholar] [CrossRef] [PubMed]
- Katevatis, C.; Fan, A.; Klapperich, C.M. Low concentration DNA extraction and recovery using a silica solid phase. PLoS ONE 2017, 12, e0176848. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Lynen, F.; Wang, J.; Li, H.; Xu, G.; Sandra, P. Comprehensive hydrophilic interaction and ion-pair reversed-phase liquid chromatography for analysis of di- to deca-oligonucleotides. J. Chromatogr. A 2012, 1255, 237–243. [Google Scholar] [CrossRef]
- Gong, L. Analysis of oligonucleotides by ion-pairing hydrophilic interaction liquid chromatography/electrospray ionization mass spectrometry. Rapid Commun. Mass Spectrom. 2017, 31, 2125–2134. [Google Scholar] [CrossRef] [PubMed]
- Easter, R.N.; Kröning, K.K.; Caruso, J.A.; Limbach, P.A. Separation and identification of oligonucleotides by hydrophilic interaction liquid chromatography (HILIC)—Inductively coupled plasma mass spectrometry (ICPMS). Analyst 2010, 135, 2560–2565. [Google Scholar] [CrossRef]
- Easter, R.; Barry, C.; Caruso, J.; Limbach, P. Separation and identification of phosphorothioate oligonucleotides by HILIC-ESIMS. Anal. Methods 2013, 5, 2657–2659. [Google Scholar] [CrossRef]
- Studzińska, S.; Łobodziński, F.; Buszewski, B. Application of hydrophilic interaction liquid chromatography coupled with mass spectrometry in the analysis of phosphorothioate oligonucleotides in serum. J. Chromatogr. B 2017, 1040, 282–288. [Google Scholar] [CrossRef] [PubMed]
- Kilanowska, A.; Buszewski, B.; Studzińska, S. Application of hydrophilic interaction liquid chromatography coupled with tandem mass spectrometry for the retention and sensitivity studies of antisense oligonucleotides. J. Chromatogr. A 2020, 1622, 461100. [Google Scholar] [CrossRef]
- Nuckowski, Ł.; Kilanowska, A.; Studzińska, S. Hydrophilic interaction in solid-phase extraction of antisense oligonucleotides. J. Chromatogr. Sci. 2020, 58, 383–387. [Google Scholar] [CrossRef]
- Gong, L.; McCullagh, J.S.O. Analysis of oligonucleotides by hydrophilic interaction liquid chromatography coupled to negative ion electrospray ionization mass spectrometry. J. Chromatogr. A 2011, 1218, 5480–5486. [Google Scholar] [CrossRef]
- MacNeill, R.; Hutchinson, T.; Acharya, V.; Stromeyer, R.; Ohorodnik, S. An oligonucleotide bioanalytical LC–SRM methodology entirely liberated from ion-pairing. Bioanalysis 2019, 11, 1155–1167. [Google Scholar] [CrossRef] [PubMed]
- Buszewski, B.; Noga, S. Hydrophilic interaction liquid chromatography (HILIC)—A powerful separation technique. Anal. Bioanal. Chem. 2012, 402, 231–247. [Google Scholar] [CrossRef] [PubMed]
- Martins, R.; Queiroz, J.A.; Sousa, F. Ribonucleic acid purification. J. Chromatogr. A 2014, 1355, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Chockalingam, P.S.; Jurado, L.A.; Jarret, H.W. DNA affinity chromatography. Mol. Biotechnol. 2001, 19, 189–199. [Google Scholar] [CrossRef]
- Sousa, F.; Prazeres, D.M.F.; Queiroz, J.A. Affinity chromatography approaches to overcome the challenges of purifying plasmid DNA. Trends Biotechnol. 2008, 26, 518–525. [Google Scholar] [CrossRef]
- Studzińska, S.; Zawadzka, E.; Bocian, S.; Szumski, M. Synthesis and application of stationary phase for DNA-affinity chromatographic analysis of unmodified and antisense oligonucleotide. Anal. Bioanal. Chem. 2021, 413, 5109–5119. [Google Scholar] [CrossRef]
- Studzińska, S.; Skoczylas, M.; Bocian, S.; Dembska, A.; Buszewski, B. Attachment of hybridizable oligonucleotides to a silica support and its application for selective extraction of unmodified and antisense oligonucleotides from serum samples. RSC Adv. 2020, 10, 16221–16230. [Google Scholar] [CrossRef]
- Nuckowski, Ł.; Kaczmarkiewicz, A.; Studzińska, S. Development of SPE method for the extraction of phosphorothioate oligonucleotides from serum samples. Bioanalysis 2018, 10, 1667–1677. [Google Scholar] [CrossRef]
- Qureshi, M.N.; Stecher, G.; Huck, C.; Bonn, G.K. Preparation of polymer based sorbents for solid phase extraction of polyphenolic compounds. Cent. Eur. J. Chem. 2011, 9, 206–212. [Google Scholar] [CrossRef]
- Chen, B.; Bartlett, M.G. A One-Step Solid Phase Extraction Method for Bioanalysis of a Phosphorothioate Oligonucleotide and Its 3′ n-1 Metabolite from Rat Plasma by uHPLC–MS/MS. AAPS J. 2012, 14, 772–780. [Google Scholar] [CrossRef]
- Lu, Z.; Diehl, D.; Mazzeo, J.; Oehrle, S.A.; Mallet, C.R.; Young, M.S.; Chambers, E. Methods for Separating and Analyzing Anionic Compounds. U.S. Patent US20080169242A1, 17 July 2008. [Google Scholar]
- Sutton, J.M.; Kim, J.; El Zahar, N.M.; Bartlett, M.G. Bioanalysis and biotransformation of oligonucleotide therapeutics by liquid chromatography-mass spectrometry. Mass Spectrom. Rev. 2021, 40, 334–358. [Google Scholar] [CrossRef] [PubMed]
- Berensmeier, S. Magnetic particles for the separation and purification of nucleic acids. Appl. Microbiol. Biotechnol. 2006, 73, 495–504. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Basiri, B.; Hassan, C.; Punt, C.; van der Hage, E.; den Besten, C.; Bartlett, M.G. Metabolite Profiling of the Antisense Oligonucleotide Eluforsen Using Liquid Chromatography-Mass Spectrometry. Mol. Ther. Nucleic Acids 2019, 17, 714–725. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Gong, Y.; Kim, J.; Liu, X.; Gilbert, J.; Kerns, H.M.; Groth, R.; Rooney, M. Hybridization Liquid Chromatography–Tandem Mass Spectrometry: An Alternative Bioanalytical Method for Antisense Oligonucleotide Quantitation in Plasma and Tissue Samples. Anal. Chem. 2020, 92, 10548–10559. [Google Scholar] [CrossRef] [PubMed]



| Functional Groups at the Surface of the Support | Parameter | References | ||||
|---|---|---|---|---|---|---|
| LC | SPE | Types of Analyzed OGNs | ||||
| Resolution | Selectivity | Time | Recovery | |||
| Alkyl chains | high | high | short | very high | homoligonucleotides, shortmers, longmers, metabolites, sequence isomers, depurination products, ASO, phosphorothioate OGNs diastereoisomers | [10,11,13,16,59,36,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,60,61,62,66,101] |
| Alkyl chains with incorporated polar groups | medium | low | long | high for polymer-based adsorbents | sequence isomers, shortmers, metabolites | [13,39,40,41,42,44,45,46,47,48,49,53,56,57,63,64,73,74,101] |
| Aromatics | high | high | medium | high, but just for unmodified OGNs; low for ASO | sequence isomers, ASO, shortmers, longmers, metabolites | [9,10,35,43,53,56,62,63,64,65,66,67,68,69,70] |
| Pentafluorophenyl | medium | medium | short | - | sequence isomers, ASO shortmers, longmers, metabolites | [53,56,63] |
| Cholesterol | medium | low | short | - | sequence isomers, shortmers | [57,71] |
| Alkyl chain with quaternary nitrogen | high | medium | long | usually high, depend on OGNs modification | homooligonucleotides, shortmers, longmers, phosphorothioate OGNs diastereoisomers, ASO | [17,19,20,33,75,77,78,79,80,82,83] |
| Diethylaminoethyl | medium | medium | very high | sequence isomers, ASO, shortmers | [18,24,25,35,36,51,52,54,69,70,84,94,103] | |
| Silica | low or even irreversible adsorption | low | long | medium | homooligonucleotides, shortmers, longmers, | [22,23,25,30,87,88,89,90,91] |
| Amide | very high | very high | medium | - | homooligonucleotides, sequence isomers, shortmers, ASO | [81,88,89,90,93,94] |
| Zwitterionic | high | very high | short | - | homooligonucleotides, shortmers, metabolites, ASO | [91,93] |
| Diol | good | high but just for polymer-based supports | long | - | homooligonucleotides, sequence isomers, shortmers | [22,23,87,88,90,91] |
| Oligonucleotides | good | very high | short | very high | unmodified OGNs and ASO, which are complementary to the OGN strand at the surface | [33,35,36,96,97,98,99,100,105,106,107] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Studzińska, S.; Nuckowski, Ł.; Buszewski, B. Oligonucleotides Isolation and Separation—A Review on Adsorbent Selection. Int. J. Mol. Sci. 2022, 23, 9546. https://doi.org/10.3390/ijms23179546
Studzińska S, Nuckowski Ł, Buszewski B. Oligonucleotides Isolation and Separation—A Review on Adsorbent Selection. International Journal of Molecular Sciences. 2022; 23(17):9546. https://doi.org/10.3390/ijms23179546
Chicago/Turabian StyleStudzińska, Sylwia, Łukasz Nuckowski, and Bogusław Buszewski. 2022. "Oligonucleotides Isolation and Separation—A Review on Adsorbent Selection" International Journal of Molecular Sciences 23, no. 17: 9546. https://doi.org/10.3390/ijms23179546







