Genome-Wide Identification and Characterization of Banana Ca2+-ATPase Genes and Expression Analysis under Different Concentrations of Ca2+ Treatments
Abstract
:1. Introduction
2. Results
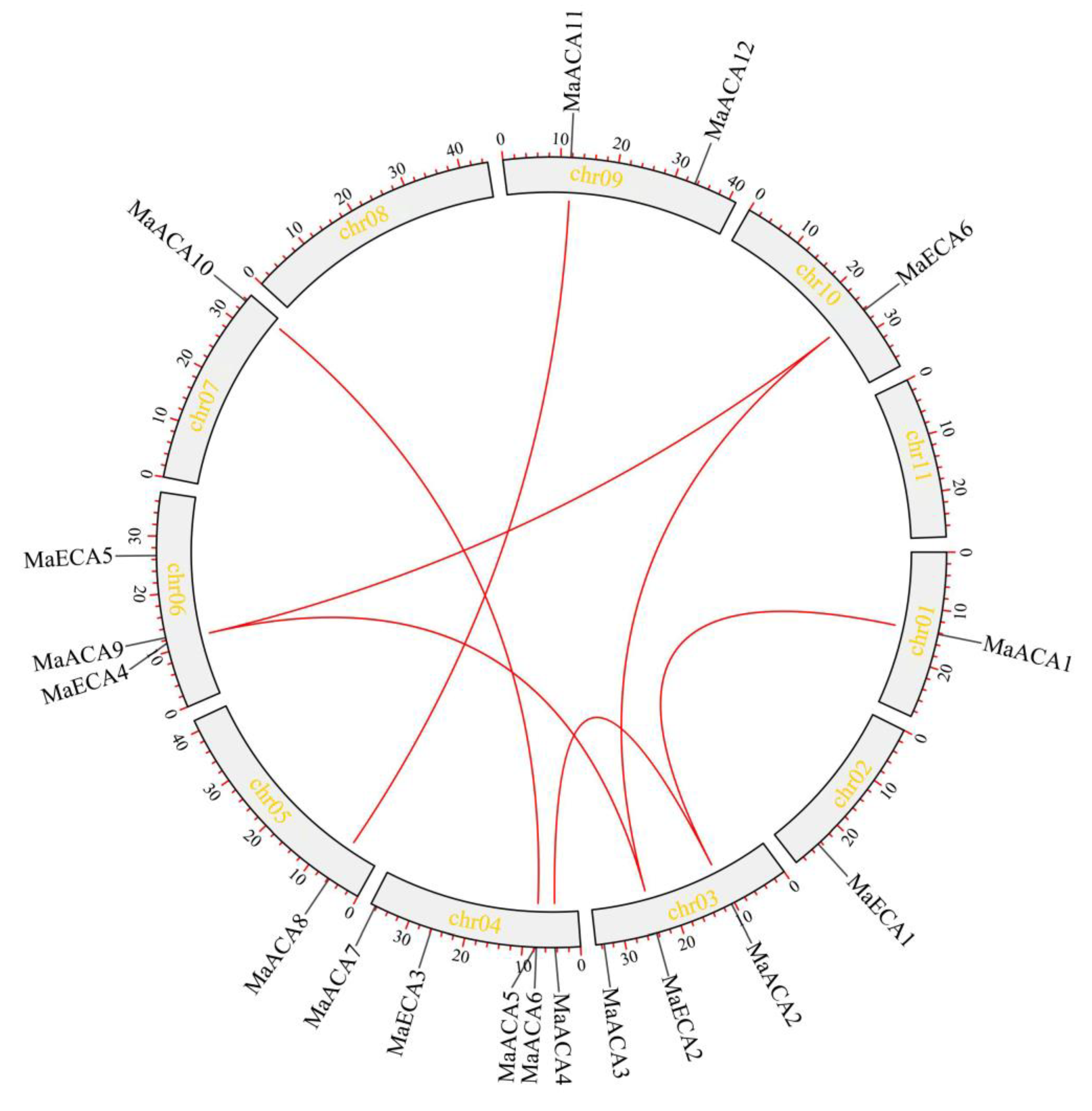
2.1. Identification and Characterization of Banana Ca2+-ATPase Gene Family Members
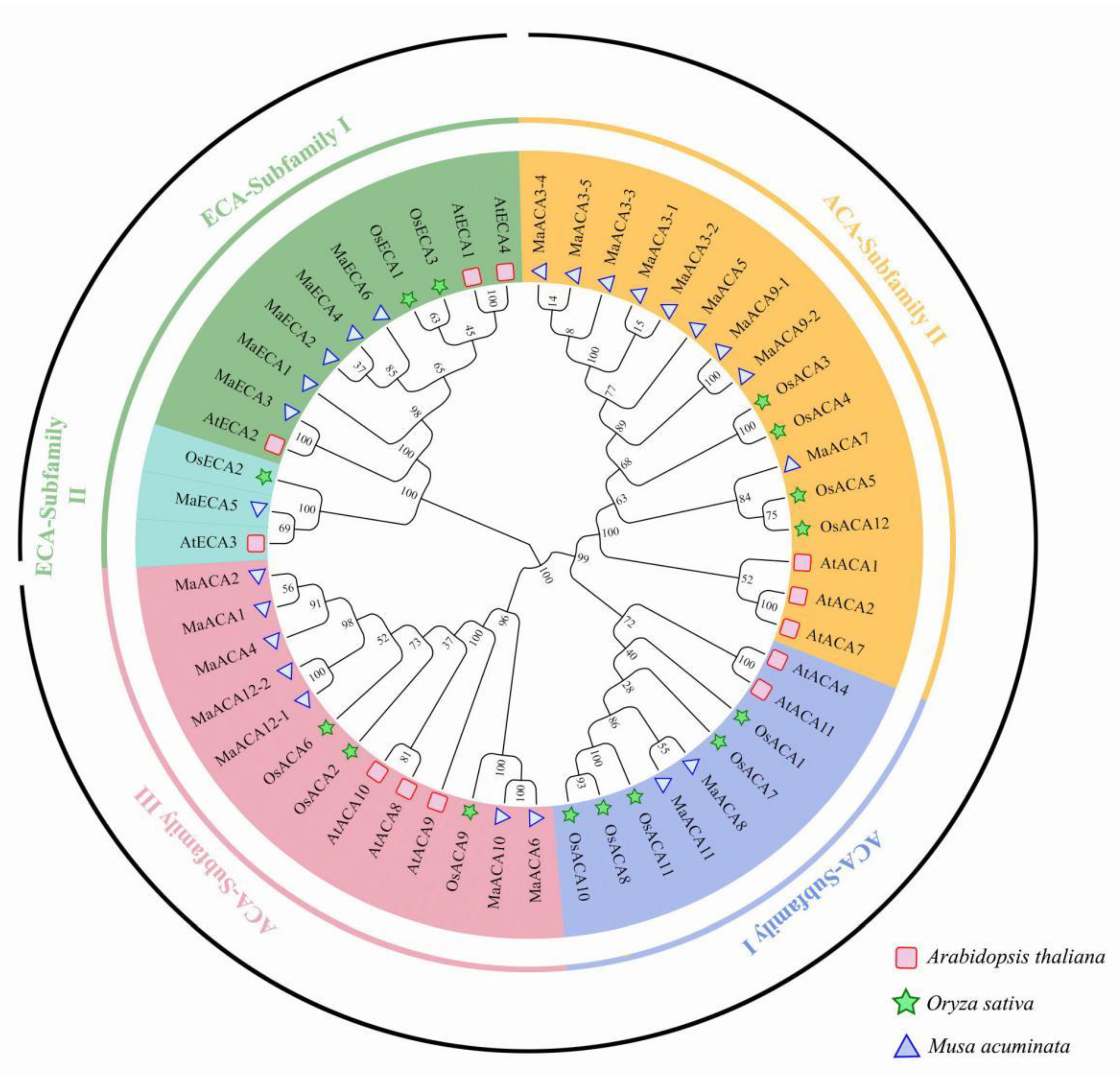
2.2. Phylogenetic Analysis of Ca2+-ATPase Genes
2.3. Conserved Motif and Gene Structure Analysis Results
2.4. Transcription Factor Binding Site (TFBS) and Cis-Acting Elements in Banana Ca2+-ATPase Promoters
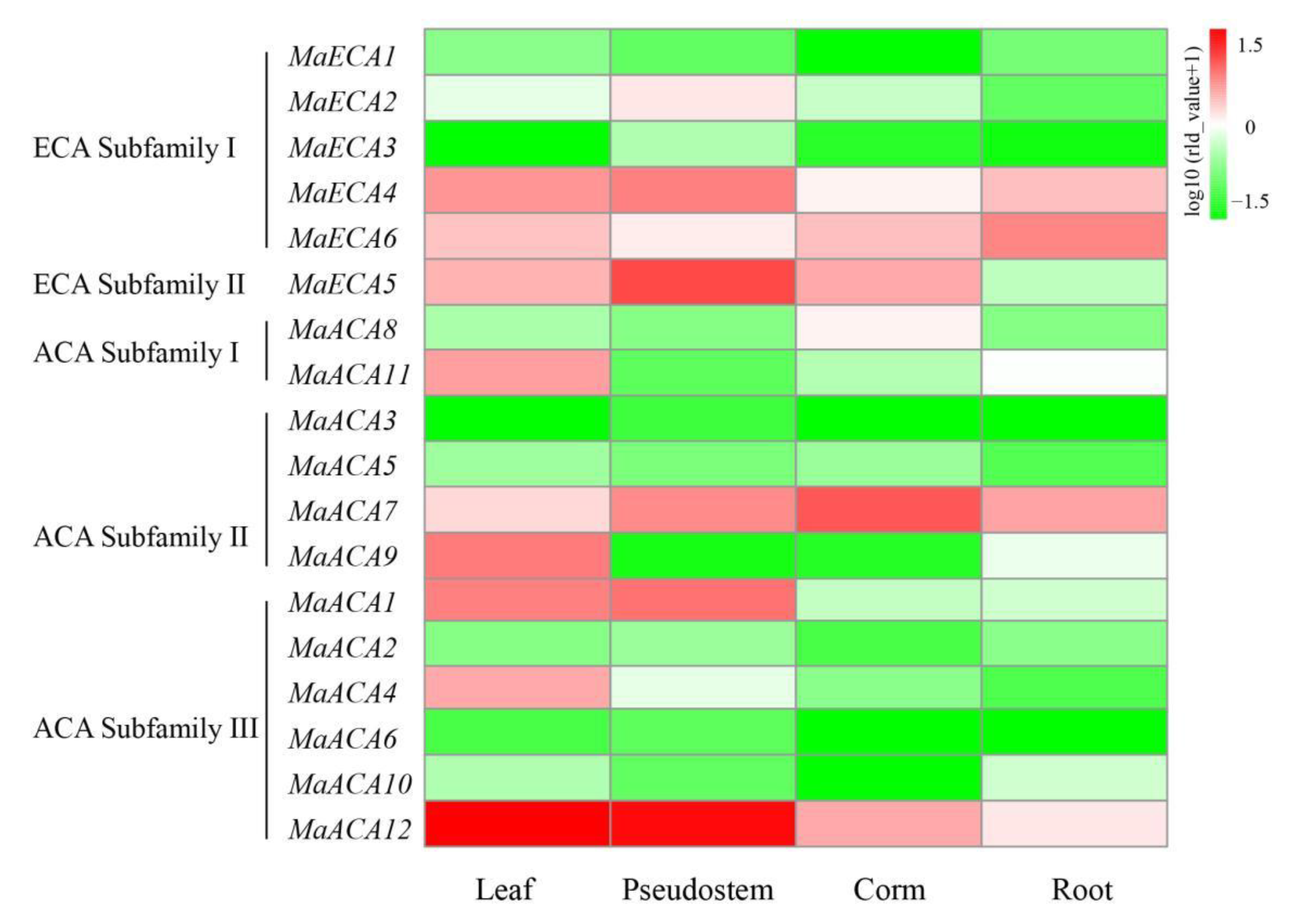
2.5. Expression Profiles of Ca2+-ATPase Genes in Different Banana Parts
2.6. Influences of Different Concentrations of Ca2+ Treatments on the Growth of Banana Seedling
2.7. Influences of Different Concentrations of Ca2+ Treatments on the P, K, Ca, Mg and S Contents in Banana Root, Leaf and Pseudostem
2.8. The Expression Patterns of Banana Ca2+-ATPase Genes under Different Concentrations of Ca2+ Treatments
3. Discussion
3.1. Comprehensive Genome-Wide Identification and Characterization of Banana Ca2+-ATPase Gene Family
3.2. Ca2+ Deficiency Impairs Banana Growth and Nutrient Homeostasis
3.3. Low Ca2+ and Ca2+ Deficiency Treatments Significantly Inhibited the Expression of Many Ca2+-ATPase Genes
4. Materials and Methods
4.1. Identification of Banana Ca2+-ATPase Genes
4.2. Phylogenetic Analysis
4.3. Gene Structure and Conserved Motifs Analysis
4.4. Promoter Analysis of the Banana Ca2+-ATPase Gene Promoters
4.5. Plant Materials and Treatment
4.6. Measurements of Growth-Related Parameters and Microelement Contents
4.7. Gene Expression Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tang, R.; Luan, S. Regulation of calcium and magnesium homeostasis in plants: From transporters to signaling network. Curr. Opin. Plant Biol. 2017, 39, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Case, R.; Eisner, D.; Gurney, A.; Jones, O.; Muallem, S.; Verkhratsky, A. Evolution of calcium homeostasis: From birth of the first cell to an omnipresent signalling system. Cell Calcium 2007, 42, 345–350. [Google Scholar] [CrossRef] [PubMed]
- Aslam, R.; Williams, L.; Bhatti, M.; Virk, N. Genome-wide analysis of wheat calcium ATPases and potential role of selected ACAs and ECAs in calcium stress. BMC Plant Biol. 2017, 17, 174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonza, M.; De Michelis, M. The plant Ca2+-ATPase repertoire: Biochemical features and physiological functions. Plant Biol. 2011, 13, 421–430. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, C.; Axelsen, K.; Harper, J.; Palmgren, M. Evolution of plant P-type ATPases. Front. Plant Sci. 2012, 3, 31. [Google Scholar] [CrossRef] [Green Version]
- Geisler, M.; Axelsen, K.; Harper, J.; Palmgren, M. Molecular aspects of higher plant P-Type Ca2+-ATPases. Biochim. Biophys. Acta 2000, 1465, 52–78. [Google Scholar] [CrossRef] [Green Version]
- Tuteja, N.; Mahajan, S. Calcium Signaling Network in Plants: An Overview. Plant Signal. Behav. 2007, 2, 79–85. [Google Scholar] [CrossRef] [Green Version]
- Axelsen, K.; Palmgren, M. Inventory of the superfamily of P-type ion pumps in Arabidopsis. Plant Physiol. 2001, 126, 696–706. [Google Scholar] [CrossRef] [Green Version]
- Wimmers, L.; Ewing, N.; Bennett, A. Higher plant Ca2+-ATPase: Primary structure and regulation of mRNA abundance by salt. Proc. Natl. Acad. Sci. USA 1992, 89, 9205–9209. [Google Scholar] [CrossRef] [Green Version]
- Schiøtt, M.; Romanowsky, S.; Bækgaard, L.; Jakobsen, M.; Palmgren, M.; Harper, J. A plant plasma membrane Ca2+ pump is required for normal pollen tube growth and fertilization. PNAS 2004, 101, 9502–9507. [Google Scholar] [CrossRef]
- George, L.; Romanowsky, S.; Harper, J.; Sharrock, R. The ACA10 Ca2+-ATPase Regulates Adult Vegetative Development and Inflorescence Architecture in Arabidopsis. Plant Physiol. 2008, 146, 716–728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Z.; Liang, F.; Hong, B.; Young, J.; Sussman, M.; Harper, J.; Sze, H. An endoplasmic reticulum-bound Ca2+/Mn2+ pump, ECA1, supports plant growth and confers tolerance to Mn2+ stress. Plant Physiol. 2002, 130, 128–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mills, R.; Doherty, M.; Lopez-Marques, R.; Weimar, T.; Dupree, P.; Palmgren, M.; Pittman, J.; Williams, L. ECA3, a Golgi-localized P2A-Type ATPase, plays a crucial role in manganese nutrition in Arabidopsis. Plant Physiol. 2008, 146, 116–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Li, W. Molecular mechanism of Ca2+-ATPase ACA8 gene in response to low temperature stress in Arabidopsis Thaliana. In Proceedings of the 15th General Assembly and the 80th Annual Conference of the Chinese Botanical Society, Nanchang, China, 13 October 2013. [Google Scholar]
- Huda, K.; Banu, M.; Garg, B.; Tula, S.; Tuteja, R.; Tuteja, N. OsACA6, a P-Type IIB Ca2+ATPase promotes salinity and drought stress tolerance in tobacco by ROS scavenging and enhancing the expression of stress-responsive genes. Plant J. 2013, 76, 997–1015. [Google Scholar] [CrossRef]
- Sun, M.; Jia, B.; Cui, N.; Wen, Y.; Duanmu, H.; Yu, Q.; Xiao, J.; Sun, X.; Zhu, Y. Functional characterization of a Glycine Soja Ca2+ATPase in salt-alkaline stress responses. Plant Mol. Biol. 2016, 90, 419–434. [Google Scholar] [CrossRef]
- Jordine, C. Metal deficiency in banana. Nature 1962, 194, 1161–1163. [Google Scholar] [CrossRef]
- Freitas, A.; Pozza, E.; Pozza, A.; Oliveira, M.; Silva, H.; Rocha, H.; Galvão, L. Impact of nutritional deficiency on Yellow Sigatoka of banana. Australas. Plant Pathol. 2015, 44, 583–590. [Google Scholar] [CrossRef]
- Chen, H. Study on Banana Nutrition Characteristics under N, P, K, Ca, Mg, S Deficiency and Banana Nutrition Diagnosis. Ph.D. Dissertation, South China Agricultural University, Guangdong, China, 2017. [Google Scholar]
- Yang, B.; Li, J.; Yao, L.; Li, G.; He, Z.; Tu, S. Effects of potassium, calcium and magnesium nutrition on growth and leaf physiological characteristics of banana. Soils Fertil. Sci. China 2010, 1, 29–36. [Google Scholar]
- Kamrul Huda, K.; Yadav, S.; Akhter Banu, M.; Trivedi, D.; Tuteja, N. Genome-wide analysis of plant-type II Ca2+ATPases gene family from rice and Arabidopsis: Potential role in abiotic stresses. Plant Physiol. Biochem. 2013, 65, 32–47. [Google Scholar] [CrossRef]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S. Evolution of gene duplication in plants. Plant Physiol. 2016, 4, 2294–2316. [Google Scholar] [CrossRef] [Green Version]
- Goel, A.; Taj, G.; Pandey, D.; Gupta, S.; Kumar, A. Genome-wide comparative in silico analysis of calcium transporters of rice and sorghum. Genom. Proteom. Bioinf. 2011, 9, 138–150. [Google Scholar] [CrossRef]
- Møller, J.; Juul, B.; Le Maire, M. Structural organization, ion transport, and energy transduction of P-type ATPases. Biochim. Biophys. Acta 1996, 1286, 1–51. [Google Scholar] [CrossRef]
- Liu, Y.; Shu, Y.; Zhang, N.; Chen, X.; Wang, A. Identification and analysis of Ca2+-ATPase gene family in Solanaceae. Mol. Plant Breed. 2021, 19, 4268–4277. [Google Scholar] [CrossRef]
- Zhang, M.; Yang, J.; Sun, M.; Jia, B.; Sun, X. Screening and identification of environmental stress responsive Medicago sativa Ca2+ATPases based on gene family analyses. Plant Physiol. J. 2017, 53, 198–208. [Google Scholar] [CrossRef]
- Wang, J.; Wu, X.; Yang, L.; Ying, Q.; Huang, J. Genome-wide identification and expression analysis of ACA gene family in Brassica rapa. Scientia Agric. Sinica 2021, 54, 4851–4868. [Google Scholar] [CrossRef]
- Baillo, E.; Kimotho, R.; Zhang, Z.; Xu, P. Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement. Genes 2019, 10, 771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shivani; Awasthi, P.; Sharma, V.; Kaur, N.; Kaur, N.; Pandey, P.; Tiwari, S. Genome-wide analysis of transcription factors during somatic embryogenesis in banana (Musa spp.) cv. Grand Naine. PLoS ONE 2017, 12, e0182242. [Google Scholar] [CrossRef] [Green Version]
- Kuang, J.; Wu, C.; Guo, Y.; Walther, D.; Shan, W.; Chen, J.; Chen, L.; Lu, W. Deciphering transcriptional regulators of banana fruit ripening by regulatory network analysis. Plant Biotechnol. J. 2021, 19, 477–489. [Google Scholar] [CrossRef]
- Jia, C.; Wang, Z.; Wang, J.; Miao, H.; Zhang, J.; Xu, B.; Liu, J.; Jin, Z.; Liu, J. Genome-wide analysis of the banana WRKY transcription factor gene family closely related to fruit ripening and stress. Plants 2022, 11, 662. [Google Scholar] [CrossRef]
- El Habbasha, S.; Faten, M. Calcium: Physiological function, deficiency and absorption. Int. J. Chemtech Res. 2015, 8, 196–202. [Google Scholar]
- Wang, Y.; Martins, L.; Sermons, S.; Balint-Kurti, P. Genetic and physiological characterization of a calcium deficiency phenotype in maize. G3 Bethesda 2020, 10, 1963–1970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Da Silva, D.; De Mello Prado, R.; Tenesaca, L.; Da Silva, J.; Mattiuz, B. Silicon attenuates calcium deficiency by increasing ascorbic acid content, growth and quality of cabbage leaves. Sci. Rep. 2021, 11, 1770. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Riaz, M.; Yan, L.; Zeng, Y.; Cuncang, J. Boron and calcium deficiency disturbing the growth of trifoliate rootstock seedlings (Poncirus trifoliate L.) by changing root architecture and cell wall. Plant Physiol. Biochem. 2019, 144, 345–354. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Yin, Y.; Feng, L.; Wu, C.; Tang, H.; Jiao, Q. Effects of exogenous calcium on mineral contents in pomegranate leaves and pericarp during fruit development. J. Fruit Sci. 2017, 34, 111–118. [Google Scholar] [CrossRef]
- White, P.; Broadley, M. Calcium in plants. Ann. Bot. 2003, 92, 487–511. [Google Scholar] [CrossRef]
- Schiott, M.; Palmgren, M. Two plant Ca2+ pumps expressed in stomatal guard cells show opposite expression patterns during cold stress. Physiol. Plant. 2005, 124, 278–283. [Google Scholar] [CrossRef]
- Wheeler, T.; Eddy, S. nhmmer: DNA homology search with profile HMMs. Bioinformatics 2013, 29, 2487–2489. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Tang, H.; Debarry, J.; Tan, X.; Li, J.; Wang, X.; Lee, T.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [Green Version]
- He, Z.; Zhang, H.; Gao, S.; Lercher, M.; Chen, W.; Hu, S. Evolview v2: An online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 2016, 44, W236–W241. [Google Scholar] [CrossRef]
- Bailey, T.; Boden, M.; Buske, F.; Frith, M.; Grant, C.; Clementi, L.; Ren, J.; Li, W.; Noble, W. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thormas, H.; Frank, M.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Tian, N.; Liu, F.; Sun, X.; Che, J.; Xiang, L.; Lai, Z.; Cheng, C. Effects of hydroponic culture hardening on the growth and photosynthetic characteristics of banana seedlings. Chin. J. Appl. Environ. Biol. 2020, 26, 582–589. [Google Scholar] [CrossRef]
- Cheng, C.; Li, D.; Qi, Q.; Sun, X.; Anue, M.; David, B.; Zhang, Y.; Hao, X.; Zhang, Z.; Lai, Z. The root endophytic fungus Serendipita indica improves resistance of banana to Fusarium oxysporum f. sp. cubense tropical race 4. Eur. J. Plant Pathol. 2020, 156, 87–100. [Google Scholar] [CrossRef]
- Sitko, K.; Gieron, Z.; Szopinski, M.; Zieleznik-Rusinowska, P.; Rusinowski, S.; Pogrzeba, M.; Daszkowska-Golec, A.; Kalaji, H.; Malkowski, E. Influence of short-term macronutrient deprivation in maize on photosynthetic characteristics, transpiration and pigment content. Sci. Rep. 2019, 9, 14181. [Google Scholar] [CrossRef] [Green Version]
- He, H.; Jin, X.; Ma, H.; Deng, Y.; Huang, J.; Yin, L. Changes of plant biomass partitioning, tissue nutrients and carbohydrates status in magnesium-deficient banana seedlings and remedy potential by foliar application of magnesium. Sci. Hortic. 2020, 268, 109377. [Google Scholar] [CrossRef]
- Chen, L.; Zhong, H.; Kuang, J.; Li, J.; Lu, W.; Chen, J. Validation of reference genes for RT-qPCR studies of gene expression in banana fruit under different experimental conditions. Planta 2011, 234, 377–390. [Google Scholar] [CrossRef]





| Gene ID | Gene Name | Chromosome Location | Length (aa) | Molecular Weight/Da | pI | Instability Index | GRAVY | Signal Peptide | Transmembrane Structure Number | Protein Subcellular Localization Prediction |
| P-type Ca2+-ATPaseIIA (ECA) | ||||||||||
| Ma02_g17850 | MaECA1 | chr02:24603794…24609861(+) | 1064 | 116,317.60 | 5.28 | 35.82 | 0.089 | NO | 7 | Plasma membrane |
| Ma03_g18840 | MaECA2 | chr03:24338682…24347596(−) | 1059 | 115,754.37 | 5.36 | 37.30 | 0.095 | NO | 9 | Plasma membrane |
| Ma04_g23800 | MaECA3 | chr04:25894068…25901262(+) | 1051 | 116,158.69 | 5.40 | 33.61 | 0.131 | NO | 8 | Plasma membrane |
| Ma06_g16960 | MaECA4 | chr06:11494289…11502009(−) | 1058 | 115,980.52 | 5.19 | 36.63 | 0.078 | NO | 7 | Plasma membrane |
| Ma06_g26060 | MaECA5 | chr06:26671312…26740153(+) | 1000 | 109,753.56 | 5.87 | 36.37 | 0.222 | NO | 8 | Plasma membrane |
| Ma10_g13570 | MaECA6 | chr10:26320460…26327228(+) | 1059 | 115,636.05 | 5.26 | 36.29 | 0.098 | NO | 9 | Plasma membrane |
| P-type Ca2+-ATPaseIIB (ACA) | ||||||||||
| Ma01_g18810 | MaACA1 | chr01:14159363…14175495(+) | 1095 | 119,281.68 | 8.36 | 38.10 | 0.050 | NO | 8 | Plasma membrane |
| Ma03_g13430 | MaACA2 | chr03:10547060…10561232(−) | 1090 | 119,195.56 | 8.27 | 36.89 | 0.040 | NO | 11 | Plasma membrane |
| Ma03_g31290 | MaACA3-1 | chr03:33555522…33560125(+) | 942 | 102,943.61 | 5.98 | 35.91 | 0.221 | NO | 6 | Plasma membrane |
| MaACA3-2 | 1016 | 110,612.29 | 5.84 | 34.99 | 0.207 | NO | 6 | Plasma membrane | ||
| MaACA3-3 | 1012 | 110,180.84 | 5.84 | 34.60 | 0.216 | NO | 6 | Plasma membrane | ||
| MaACA3-4 | 1017 | 110,723.47 | 5.84 | 35.53 | 0.213 | NO | 6 | Plasma membrane | ||
| MaACA3-5 | 1034 | 112,541.57 | 5.87 | 35.13 | 0.220 | NO | 6 | Plasma membrane | ||
| Ma04_g05840 | MaACA4 | chr04:4360063…4378280(+) | 1082 | 118,244.80 | 8.47 | 40.33 | 0.066 | NO | 7 | Plasma membrane |
| Ma04_g10940 | MaACA5 | chr04:7723372…7730223(+) | 1020 | 111,212.01 | 5.88 | 35.51 | 0.200 | NO | 6 | Plasma membrane |
| Ma04_g10640 | MaACA6 | chr04:7560089…7563361(+) | 1017 | 111,767.31 | 5.70 | 31.68 | 0.121 | NO | 7 | Plasma membrane |
| Ma04_g39050 | MaACA7 | chr04:36293787…36300960(+) | 1020 | 111,518.14 | 5.91 | 32.30 | 0.158 | NO | 6 | Plasma membrane |
| Ma05_g07830 | MaACA8 | chr05:5750563…5759010(+) | 1034 | 114,138.10 | 6.74 | 31.15 | 0.169 | NO | 10 | Plasma membrane |
| Ma06_g18390 | MaACA9-1 | chr06:12494351…12501092(−) | 1019 | 111,002.40 | 5.26 | 32.57 | 0.201 | NO | 6 | Plasma membrane |
| MaACA9-2 | 1012 | 109,834.23 | 5.22 | 34.65 | 0.242 | NO | 6 | Plasma membrane | ||
| Ma07_g27150 | MaACA10 | chr07:33645382…33649713(+) | 1024 | 112,321.63 | 5.85 | 34.05 | 0.091 | NO | 8 | Plasma membrane |
| Ma09_g16370 | MaACA11 | chr09:11747981…11758304(−) | 1035 | 114,334.58 | 8.30 | 30.90 | 0.144 | NO | 10 | Plasma membrane |
| Ma09_g21740 | MaACA12-1 | chr09:33595832…33627266(+) | 1078 | 117,956.48 | 7.28 | 38.83 | 0.070 | NO | 9 | Plasma membrane |
| MaACA12-2 | 1103 | 120,673.88 | 8.87 | 35.90 | 0.072 | NO | 9 | Plasma membrane | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, R.; Tian, N.; Wang, J.; Fan, M.; Wang, B.; Qu, P.; Xu, S.; Xu, Y.; Cheng, C.; Lü, P. Genome-Wide Identification and Characterization of Banana Ca2+-ATPase Genes and Expression Analysis under Different Concentrations of Ca2+ Treatments. Int. J. Mol. Sci. 2022, 23, 11914. https://doi.org/10.3390/ijms231911914
Ma R, Tian N, Wang J, Fan M, Wang B, Qu P, Xu S, Xu Y, Cheng C, Lü P. Genome-Wide Identification and Characterization of Banana Ca2+-ATPase Genes and Expression Analysis under Different Concentrations of Ca2+ Treatments. International Journal of Molecular Sciences. 2022; 23(19):11914. https://doi.org/10.3390/ijms231911914
Chicago/Turabian StyleMa, Ronghui, Na Tian, Jiashui Wang, Minlei Fan, Bin Wang, Pengyan Qu, Shiyao Xu, Yanbing Xu, Chunzhen Cheng, and Peitao Lü. 2022. "Genome-Wide Identification and Characterization of Banana Ca2+-ATPase Genes and Expression Analysis under Different Concentrations of Ca2+ Treatments" International Journal of Molecular Sciences 23, no. 19: 11914. https://doi.org/10.3390/ijms231911914






