Role of EmaSR in Ethanol Metabolism by Acinetobacter baumannii
Abstract
:1. Introduction
2. Results
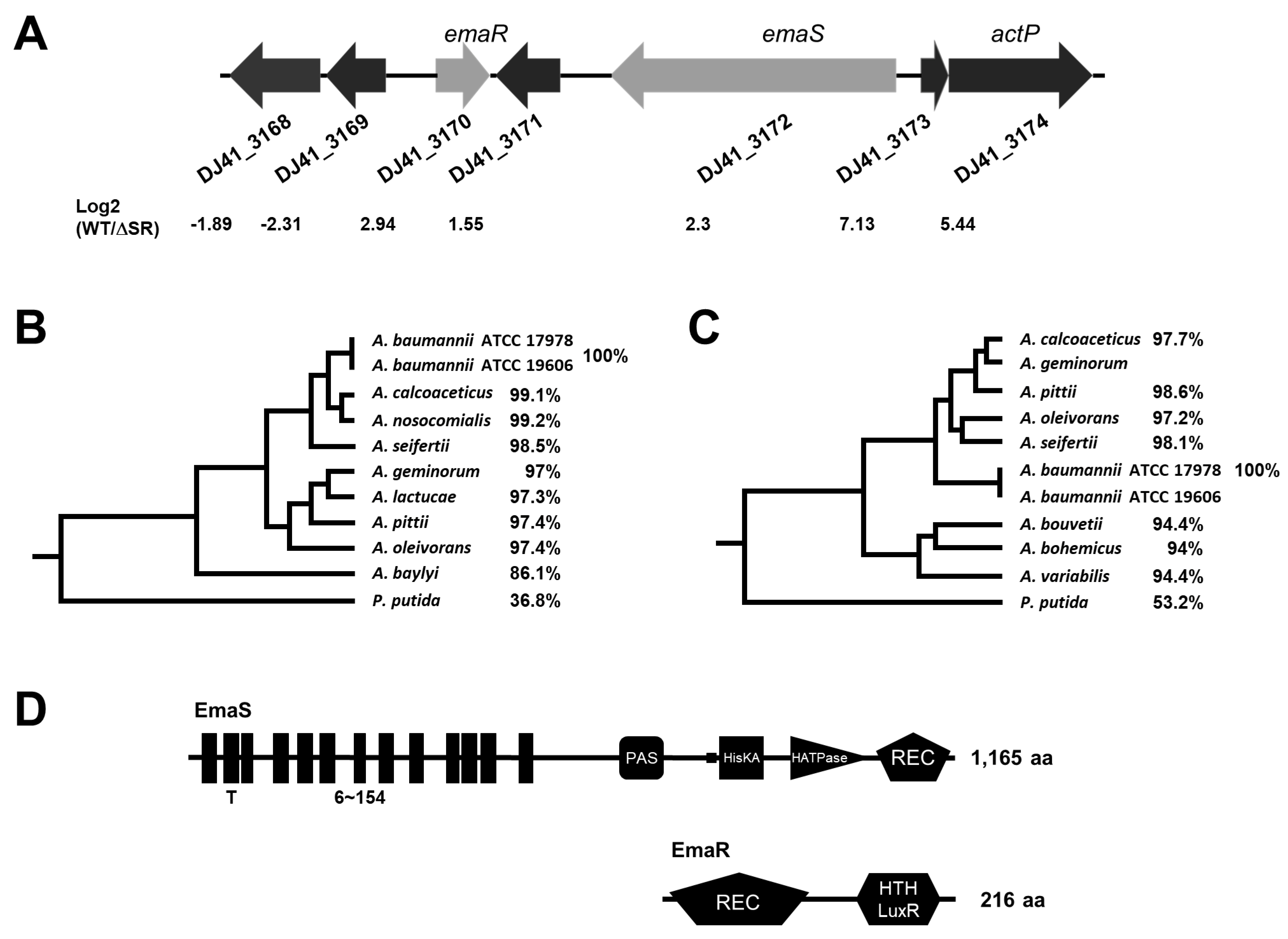
2.1. The EmaSR TCS in A. baumannii
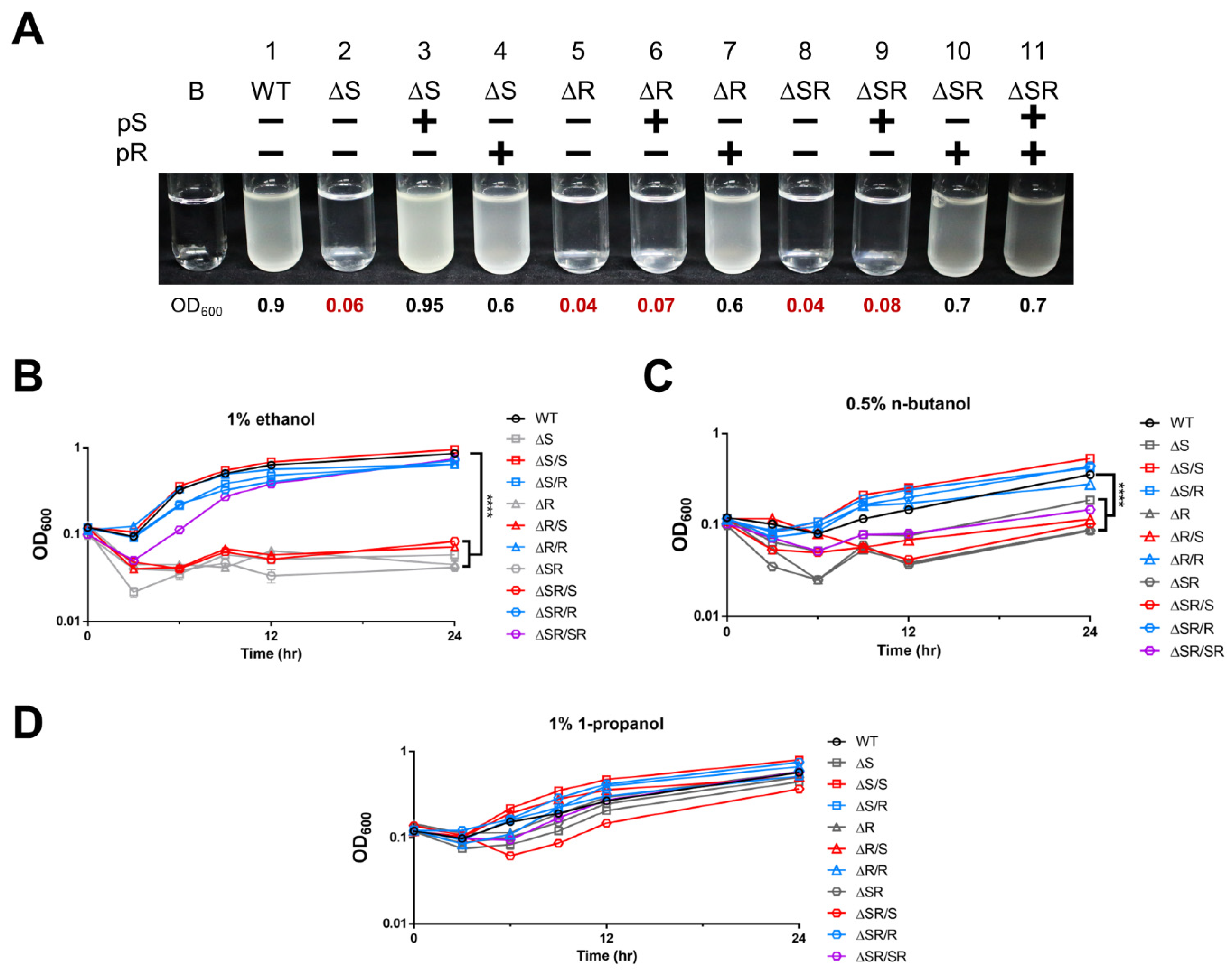
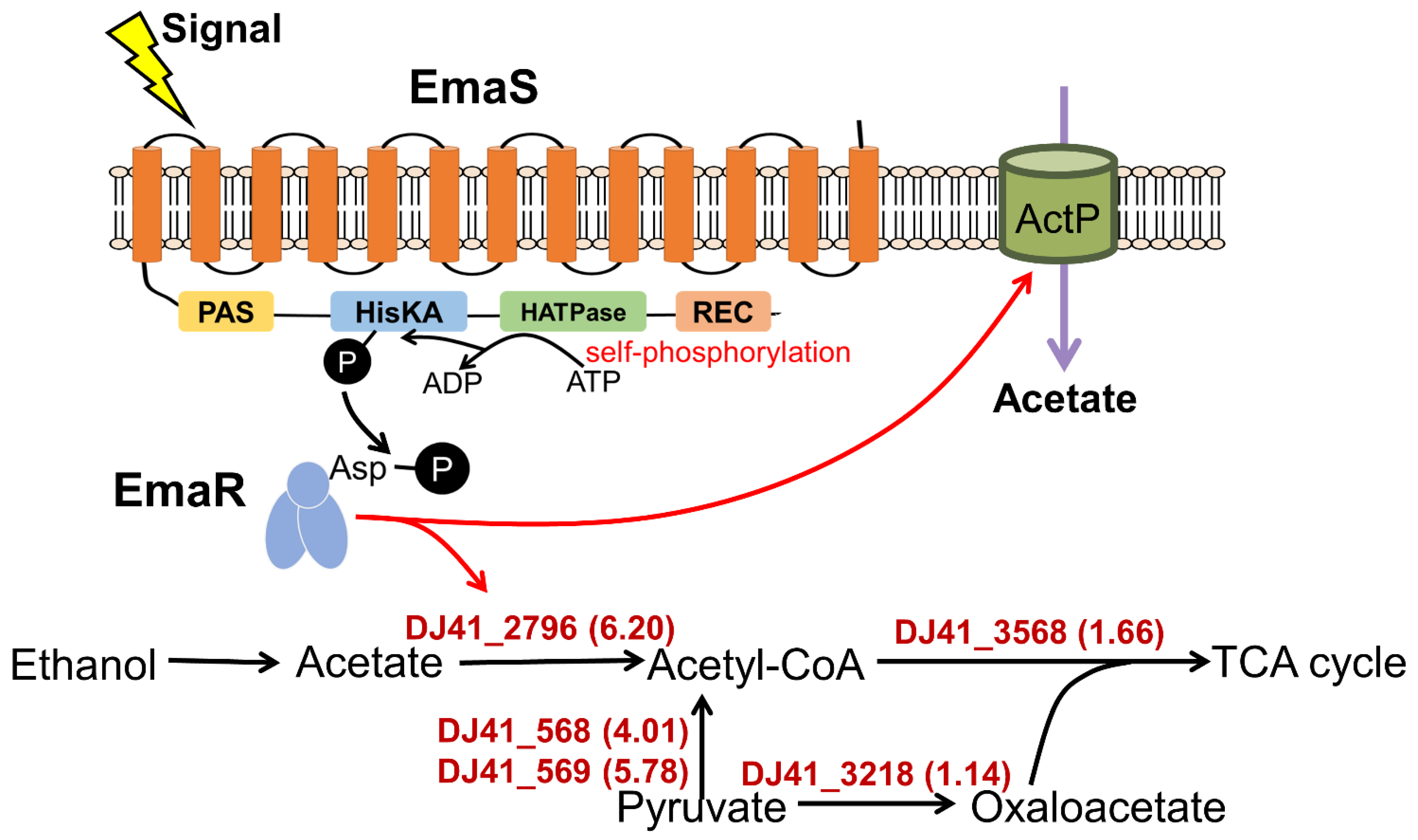
2.2. EmaSR Functions in Ethanol Metabolism
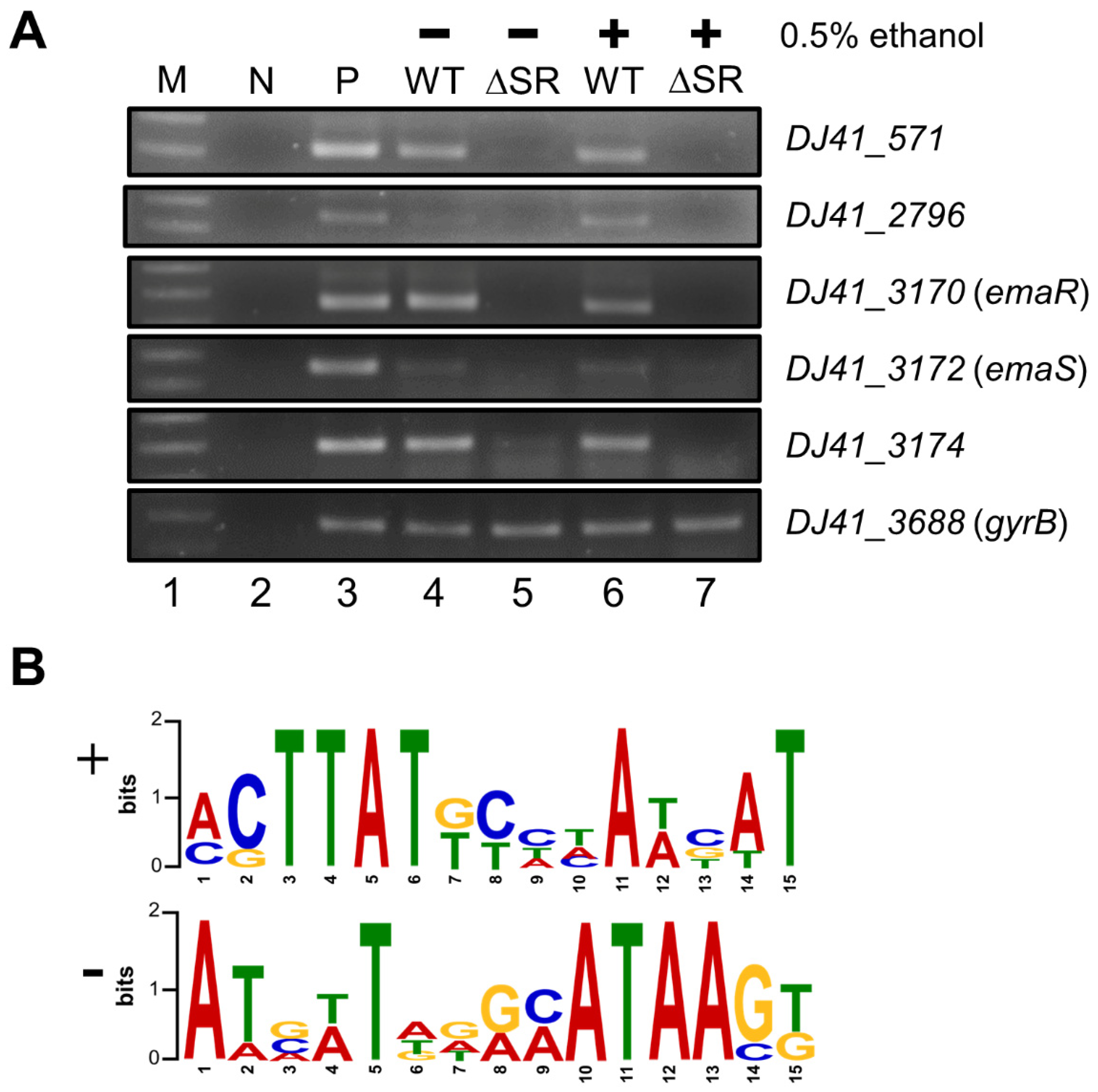
2.3. EmaR Serves as the Cognate Regulator of EmaS
2.4. Loss of EmaSR Increased Oxidative Resistance but Reduced Motility and Biofilm Formation Ability
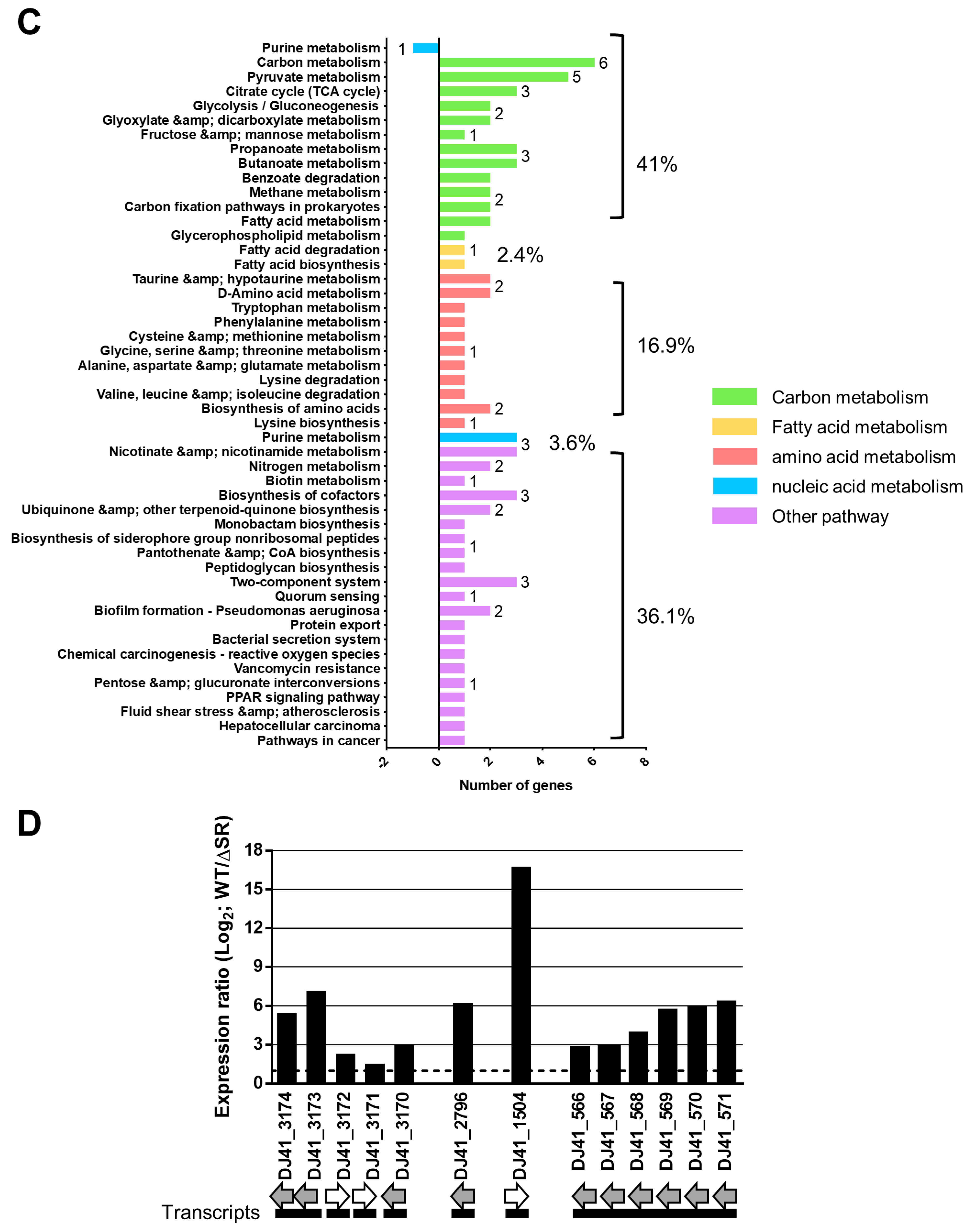
2.5. Gene Expression Changes Respectively Caused by Knockout of EmaSR, EmaS, and EmaR
2.6. EmaS and EmaR Are Associated with Virulence against G. mellonella
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Plasmids, and Primers
4.2. Marker-Less Mutation
4.3. Construction of Complement Plasmid and Strains
4.4. Recombinant EmaR and Antibody Production
4.5. Phos-tagTM Analysis of EmaR
4.6. Motility
4.7. Inorganic Stress Resistance
4.8. Biofilm Formation
4.9. RNA Sequencing
4.10. RNA Extraction and RT-PCR
4.11. Virulence Assay with G. mellonella
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Almasaudi, S.B. Acinetobacter spp. as nosocomial pathogens: Epidemiology and resistance features. Saudi J. Biol. Sci. 2018, 25, 586–596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez-Villoria, A.M.; Valverde-Garduno, V. Antibiotic-resistant Acinetobacter baumannii increasing success remains a challenge as a nosocomial pathogen. J. Pathog. 2016, 2016, 7318075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, H.R.; Shu, H.Y.; Lin, G.H. Biological roles of indole-3-acetic acid in Acinetobacter baumannii. Microbiol. Res. 2018, 216, 30–39. [Google Scholar] [CrossRef]
- Lin, G.H.; Hsieh, M.C.; Shu, H.Y. Role of iron-containing alcohol dehydrogenases in Acinetobacter baumannii ATCC 19606 stress resistance and virulence. Int. J. Mol. Sci. 2021, 22, 9921. [Google Scholar] [CrossRef]
- Roy, S.; Junghare, V.; Dutta, S.; Hazra, S.; Basu, S. Differential binding of carbapenems with the AdeABC efflux pump and modulation of the expression of AdeB linked to novel mutations within two-component system AdeRS in carbapenem-resistant Acinetobacter baumannii. mSystems 2022, 7, e0021722. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.F.; Lin, Y.Y.; Yeh, H.W.; Lan, C.Y. Role of the BaeSR two-component system in the regulation of Acinetobacter baumannii adeAB genes and its correlation with tigecycline susceptibility. BMC Microbiol. 2014, 14, 119. [Google Scholar] [CrossRef] [Green Version]
- Palethorpe, S.; Farrow, J.M., 3rd; Wells, G.; Milton, M.E.; Actis, L.A.; Cavanagh, J.; Pesci, E.C. Acinetobacter baumannii regulates its stress responses via the BfmRS two-component regulatory system. J. Bacteriol. 2022, 204, e0049421. [Google Scholar] [CrossRef]
- Cerqueira, G.M.; Kostoulias, X.; Khoo, C.; Aibinu, I.; Qu, Y.; Traven, A.; Peleg, A.Y. A global virulence regulator in Acinetobacter baumannii and its control of the phenylacetic acid catabolic pathway. J. Infect. Dis. 2014, 210, 46–55. [Google Scholar] [CrossRef] [Green Version]
- Charretier, Y.; Diene, S.M.; Baud, D.; Chatellier, S.; Santiago-Allexant, E.; van Belkum, A.; Guigon, G.; Schrenzel, J. Colistin Heteroresistance and involvement of the PmrAB regulatory system in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2018, 62, e00788-18. [Google Scholar] [CrossRef] [Green Version]
- Giles, S.K.; Stroeher, U.H.; Papudeshi, B.; Edwards, R.A.; Carlson-Jones, J.A.; Roach, M.; Brown, M.H. The StkSR two-component system influences colistin resistance in Acinetobacter baumannii. Microorganisms 2022, 10, 985. [Google Scholar] [CrossRef]
- Chen, R.; Lv, R.; Xiao, L.; Wang, M.; Du, Z.; Tan, Y.; Cui, Y.; Yan, Y.; Luo, Y.; Yang, R.; et al. A1S_2811, a CheA/Y-like hybrid two-component regulator from Acinetobacter baumannii ATCC 17978, is involved in surface motility and biofilm formation in this bacterium. Microbiologyopen 2017, 6, e00510. [Google Scholar] [CrossRef] [PubMed]
- De Silva, P.M.; Kumar, A. Signal transduction proteins in Acinetobacter baumannii: Role in antibiotic resistance, virulence, and potential as drug targets. Front. Microbiol. 2019, 10, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henriquez, T.; Jung, H. Involvement of the MxtR/ErdR (CrbS/CrbR) two-component system in acetate metabolism in Pseudomonas putida KT2440. Microorganisms 2021, 9, 1558. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.J.; Shu, H.Y.; Lin, G.H. Regulation of tert-butyl hydroperoxide resistance by chromosomal OhrR in A. baumannii ATCC 19606. Microorganisms 2021, 9, 629. [Google Scholar] [CrossRef]
- Schäfer, A.; Tauch, A.; Jäger, W.; Kalinowski, J.; Thierbach, G.; Pühler, A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: Selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 1994, 145, 69–73. [Google Scholar] [CrossRef]
- Hunger, M.; Schmucker, R.; Kishan, V.; Hillen, W. Analysis and nucleotide sequence of an origin of DNA replication in Acinetobacter calcoaceticus and its use for Escherichia coli shuttle plasmids. Gene 1990, 87, 45–51. [Google Scholar] [CrossRef]
- Bouvet, P.J.; Grimont, P.A. Identification and biotyping of clinical isolates of Acinetobacter. Ann. Inst. Pasteur Microbiol. 1987, 138, 569–578. [Google Scholar] [CrossRef]
- Nwugo, C.C.; Arivett, B.A.; Zimbler, D.L.; Gaddy, J.A.; Richards, A.M.; Actis, L.A. Effect of ethanol on differential protein production and expression of potential virulence functions in the opportunistic pathogen Acinetobacter baumannii. PLoS ONE 2012, 7, e51936. [Google Scholar] [CrossRef] [Green Version]
- Lucaßen, K.; Xanthopoulou, K.; Wille, J.; Wille, T.; Wen, Y.; Hua, X.; Seifert, H.; Higgins, P.G. Characterization of amino acid substitutions in the two-component regulatory system AdeRS identified in multidrug-resistant Acinetobacter baumannii. mSphere 2021, 6, e0070921. [Google Scholar] [CrossRef]
- Honma, S.; Ito, S.; Yajima, S.; Sasaki, Y. Nitric oxide signaling for actinorhodin production in Streptomyces coelicolor A3(2) via the DevS/R Two-Component System. Appl. Environ. Microbiol. 2021, 87, e0048021. [Google Scholar] [CrossRef]
- Shan, W.; Kan, J.; Cai, X.; Yin, M. Insights into mucoid Acinetobacter baumannii: A review of microbiological characteristics, virulence, and pathogenic mechanisms in a threatening nosocomial pathogen. Microbiol. Res. 2022, 261, 127057. [Google Scholar] [CrossRef] [PubMed]
- Hirschmann, S.; Gómez-Mejia, A.; Mäder, U.; Karsunke, J.; Driesch, D.; Rohde, M.; Häussler, S.; Burchhardt, G.; Hammerschmidt, S. The two-component system 09 regulates Pneumococcal carbohydrate metabolism and capsule expression. Microorganisms 2021, 9, 468. [Google Scholar] [CrossRef] [PubMed]
- Andreassen, P.R.; Trappetti, C.; Minhas, V.; Nielsen, F.D.; Pakula, K.; Paton, J.C.; Jørgensen, M.G. Host-glycan metabolism is regulated by a species-conserved two-component system in Streptococcus pneumoniae. PLoS Pathog. 2020, 16, e1008332. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Mohsin, A.; Yu, J.; Hu, Y.; Ali, M.F.; Chen, Z.; Zhuang, Y.; Chu, J.; Guo, M. Two-component-system RspA1/A2-dependent regulation on primary metabolism in Streptomyces albus A30 cultivated with glutamate as the sole nitrogen source. Front. Microbiol. 2020, 11, 1658. [Google Scholar] [CrossRef]
- Zhong, C.; Zhang, P.; Liu, C.; Liu, M.; Chen, W.; Fu, J.; Qi, X.; Cao, G. The PolS-PolR two-component system regulates genes involved in Poly-P metabolism and phosphate transport in Microlunatus phosphovorus. Front. Microbiol. 2019, 10, 2127. [Google Scholar] [CrossRef]
- Krause, A.; Julich, H.; Mankar, M.; Reinhold-Hurek, B. The regulatory network controlling ethanol-induced expression of alcohol dehydrogenase in the endophyte Azoarcus sp. Strain BH72. Mol. Plant Microbe Interact. 2017, 30, 778–785. [Google Scholar] [CrossRef] [Green Version]
- Mern, D.S.; Ha, S.W.; Khodaverdi, V.; Gliese, N.; Görisch, H. A complex regulatory network controls aerobic ethanol oxidation in Pseudomonas aeruginosa: Indication of four levels of sensor kinases and response regulators. Microbiology 2010, 156, 1505–1516. [Google Scholar] [CrossRef] [Green Version]
- Xie, Y.; Shao, X.; Zhang, Y.; Liu, J.; Wang, T.; Zhang, W.; Hua, C.; Deng, X. Pseudomonas savastanoi two-component system RhpRS switches between virulence and metabolism by tuning phosphorylation state and sensing nutritional conditions. mBio 2019, 10, e02838-18. [Google Scholar] [CrossRef] [Green Version]
- Kinoshita, E.; Takahashi, M.; Takeda, H.; Shiro, M.; Koike, T. Recognition of phosphate monoester dianion by an alkoxide-bridged dinuclear zinc(II) complex. Dalton Trans. 2004, 8, 1189–1193. [Google Scholar] [CrossRef]
- Li, R.; Yu, C.; Li, Y.; Lam, T.W.; Yiu, S.M.; Kristiansen, K.; Wang, J. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics 2009, 25, 1966–1967. [Google Scholar] [CrossRef]
- Marioni, J.C.; Mason, C.E.; Mane, S.M.; Stephens, M.; Gilad, Y. RNA-seq: An assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008, 18, 1509–1517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Drai, D.; Elmer, G.; Kafkafi, N.; Golani, I. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 2001, 125, 279–284. [Google Scholar] [CrossRef] [Green Version]
- Tsai, C.J.; Loh, J.M.; Proft, T. Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence 2016, 7, 214–229. [Google Scholar] [CrossRef] [PubMed]





| Plasmid | Description | Antibiotic Resistance (µg/mL) | Reference/Source |
|---|---|---|---|
| pK18mobsacB | Suicide vector for homologous recombination | Kan50 | [15] |
| pK18DemaS | pK18mobsacB contains the upstream and downstream region of emaS | Kan50 | This study |
| pK18DemaR | pK18mobsacB contains the upstream and downstream region of emaR | Kan50 | This study |
| pWH1266 | Apr; Tcr; shuttle vector for E. coli and A. baumannii Apr; fluorescent cytosolic biosensor | Amp50, Tc12.5 | [16] |
| pS | pWH1266 contains PemaS- emaS | Amp50 | This study |
| pR | pWH1266 contains PemaR- emaR | Amp50, Tc12.5 | This study |
| pSR | pWH1266 contains PemaS- emaS and PemaR- emaR | Amp50 | This study |
| pQE80L | Expression vector with colE1 origin for His-tag fusion protein purification | Amp50 | Qiagen |
| pQE80L-emaR | Apr; emaR cloned into the BamHI-SmaI site of pQE80L | Amp50 | This study |
| Strain | Description | Reference/Source | |
| E. coli DH5α | F-, supE44, hsdR17, recA1, gyrA96, endA1, thi-1, relA1, deoR, λ- | ATCC 53868 | |
| Acinetobacter baumannii ATCC 19606 | Primary strain used in this study | [17] | |
| ΔemaS (ΔS) | Marker-less emaS deletion mutant | This study | |
| ΔemaS/pS (ΔS/S) | ΔemaS containing pS; Apr | This study | |
| ΔemaS/pR (ΔS/R) | ΔemaS containing pR; Apr, Tcr | This study | |
| ΔemaR (ΔR) | Marker-less emaR deletion mutant | This study | |
| ΔemaR/pS (ΔR/S) | ΔemaR containing pS; Apr | This study | |
| ΔemaR/pR (ΔR/S) | ΔemaR containing pR; Apr, Tcr | This study | |
| ΔemaSR (ΔSR) | Marker-less emaSR double-deletion mutant | This study | |
| ΔemaSR/pS (ΔSR/S) | ΔemaSR containing pS; Apr | This study | |
| ΔemaSR/pR (ΔSR/R) | ΔemaSR containing pR; Apr, Tcr | This study | |
| ΔemaSR/pSR (ΔSR/SR) | ΔemaSR containing pSR; Apr | This study | |
| Name | Strand | Position * | p-Value | Sequence (5′-3′) |
|---|---|---|---|---|
| DJ41_568 | + | −114 to −128 | 2.4 × 10−6 | AAAAA ACTTATTTAAAACTT TTTAG |
| DJ41_1503 | - | −107 to −121 | 1.5 × 10−6 | TGTAA ATATTTGGAATAACT CAAAA |
| DJ41_2796 | - | −107 to −121 | 1.1 × 10−6 | TAAAA ATCATAAAAATAAGT TATAC |
| DJ41_3173 | + | −150 to −164 | 2.3 × 10−9 | TGAAA CCTTATGCCTATCAT AACCC |
| emaR | + | −15 to −29 | 1.6 × 10−7 | GCCAT ACTTATGCTCAAGAT TACTT |
| emaS | - | −20 to −34 | 2.3 × 10−9 | GGGTT ATGATAGGCATAAGG TTTCA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shu, H.-Y.; Huang, Y.-W.; Tsai, P.-Y.; Hsieh, K.-S.; Lin, G.-H. Role of EmaSR in Ethanol Metabolism by Acinetobacter baumannii. Int. J. Mol. Sci. 2022, 23, 12606. https://doi.org/10.3390/ijms232012606
Shu H-Y, Huang Y-W, Tsai P-Y, Hsieh K-S, Lin G-H. Role of EmaSR in Ethanol Metabolism by Acinetobacter baumannii. International Journal of Molecular Sciences. 2022; 23(20):12606. https://doi.org/10.3390/ijms232012606
Chicago/Turabian StyleShu, Hung-Yu, Yu-Wen Huang, Ping-Yi Tsai, Kun-Sheng Hsieh, and Guang-Huey Lin. 2022. "Role of EmaSR in Ethanol Metabolism by Acinetobacter baumannii" International Journal of Molecular Sciences 23, no. 20: 12606. https://doi.org/10.3390/ijms232012606







