Complementary Regulation of BfmRS Two-Component and AbaIR Quorum Sensing Systems to Express Virulence-Associated Genes in Acinetobacter baumannii
Abstract
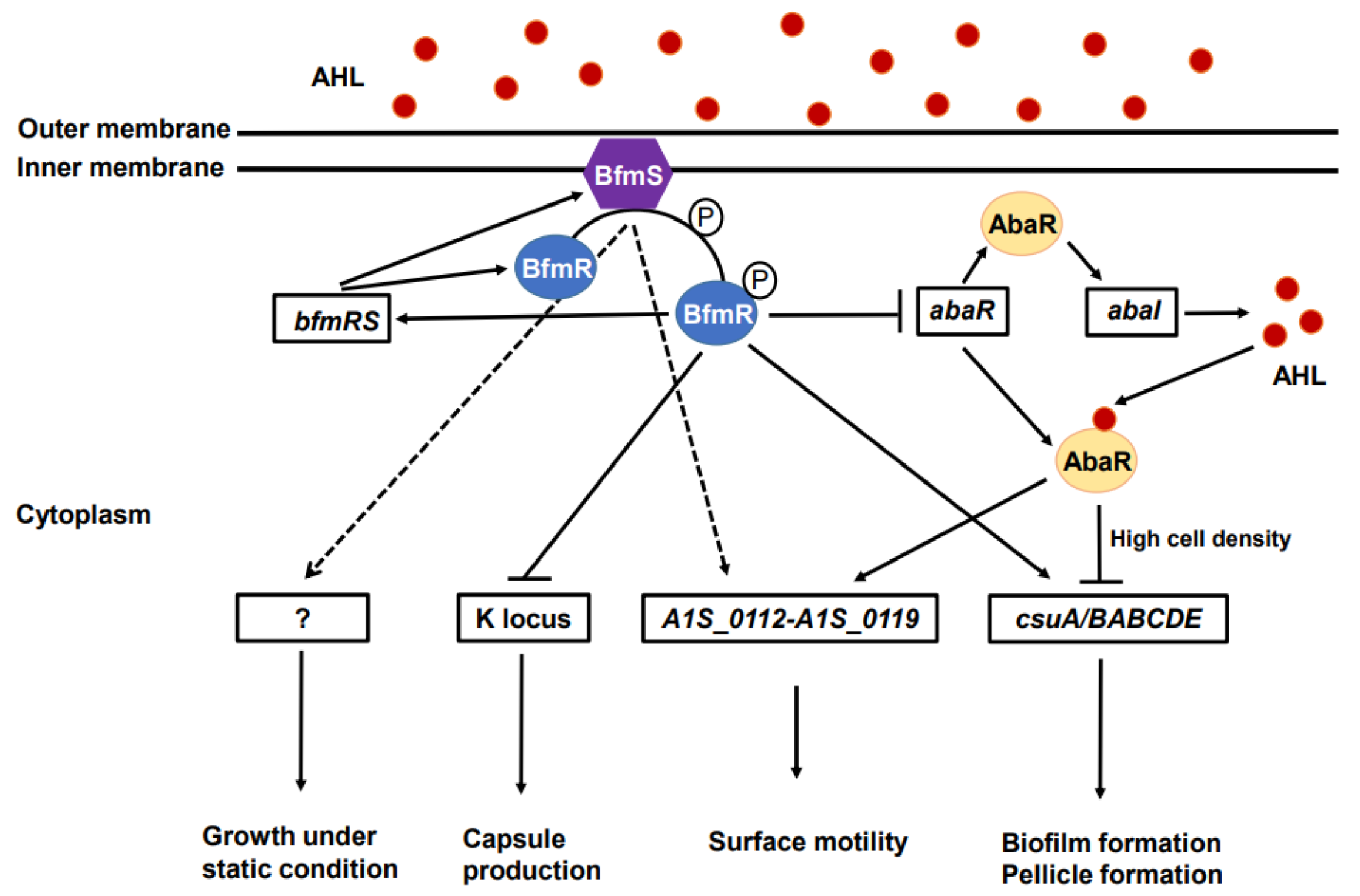
:1. Introduction
2. Results
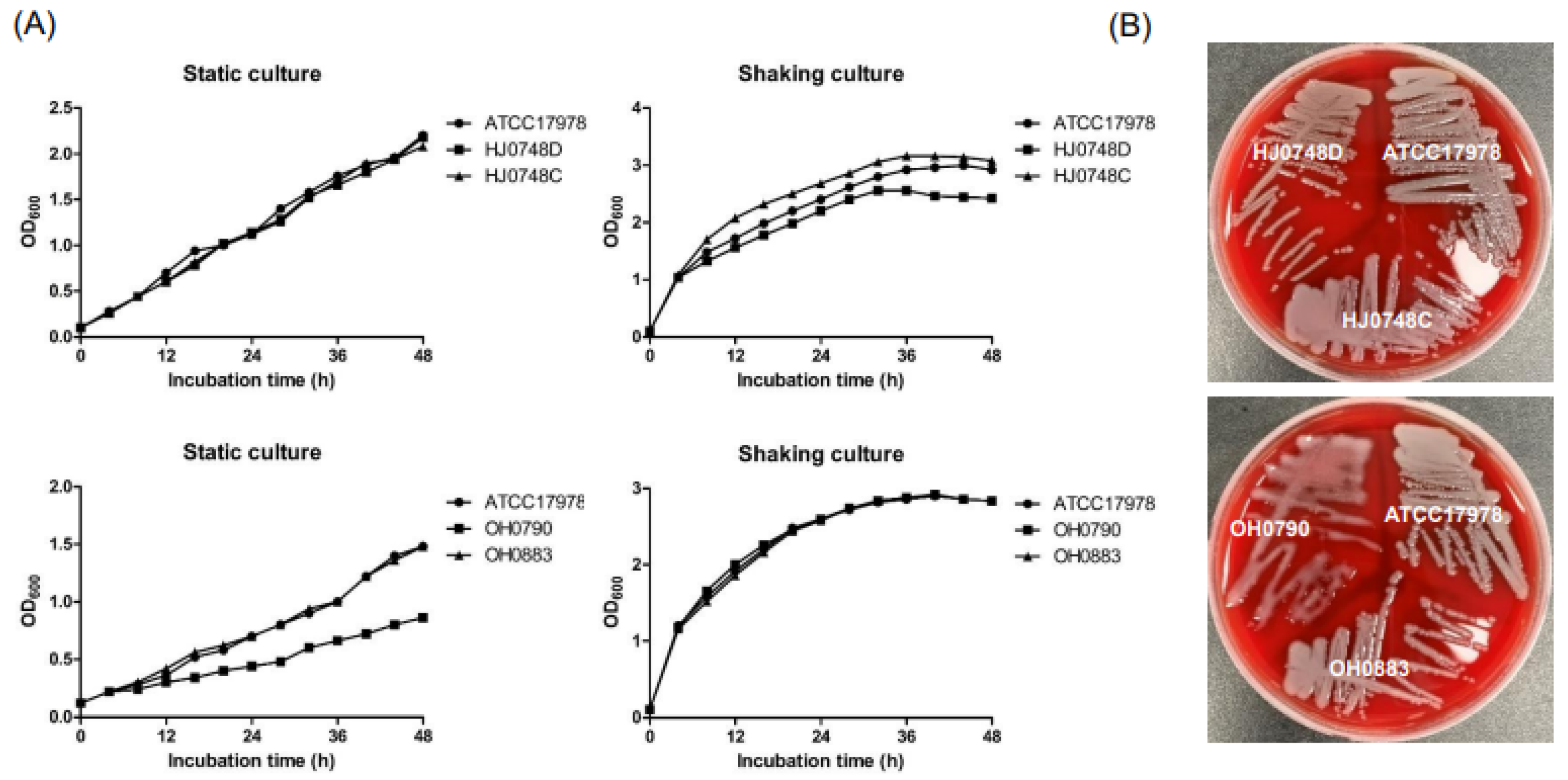
2.1. Effect of BfmRS on Bacterial Growth and Capsule Production
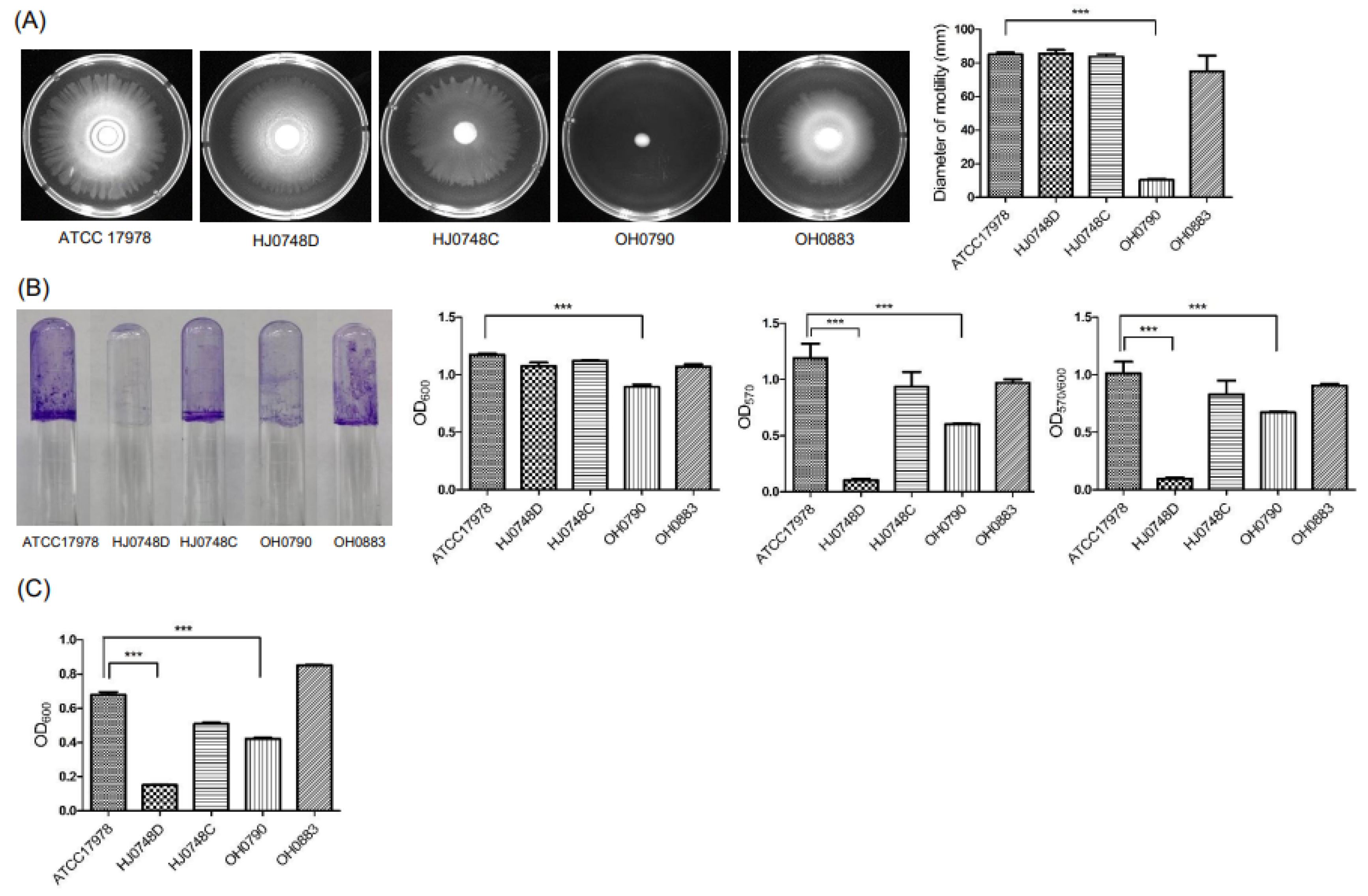
2.2. Effect of BfmRS on the Surface Motility, Twitching Motility, Biofilm Formation, and Pellicle Formation
2.3. Regulation of Surface-Motility-, Twitching-Motility-, and Biofilm-Associated Genes by BfmRS
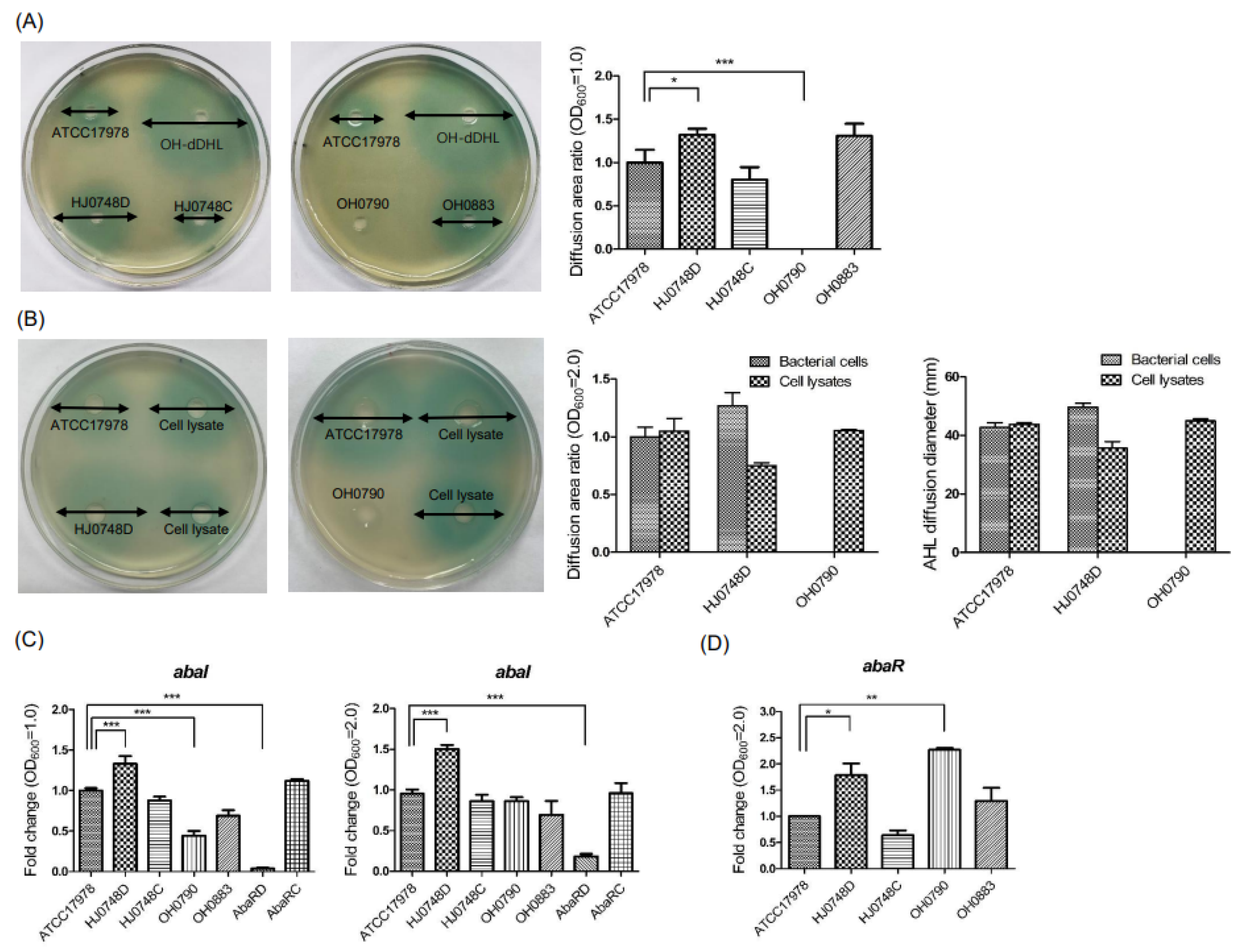
2.4. Effect of BfmRS on the Production of Autoinducers
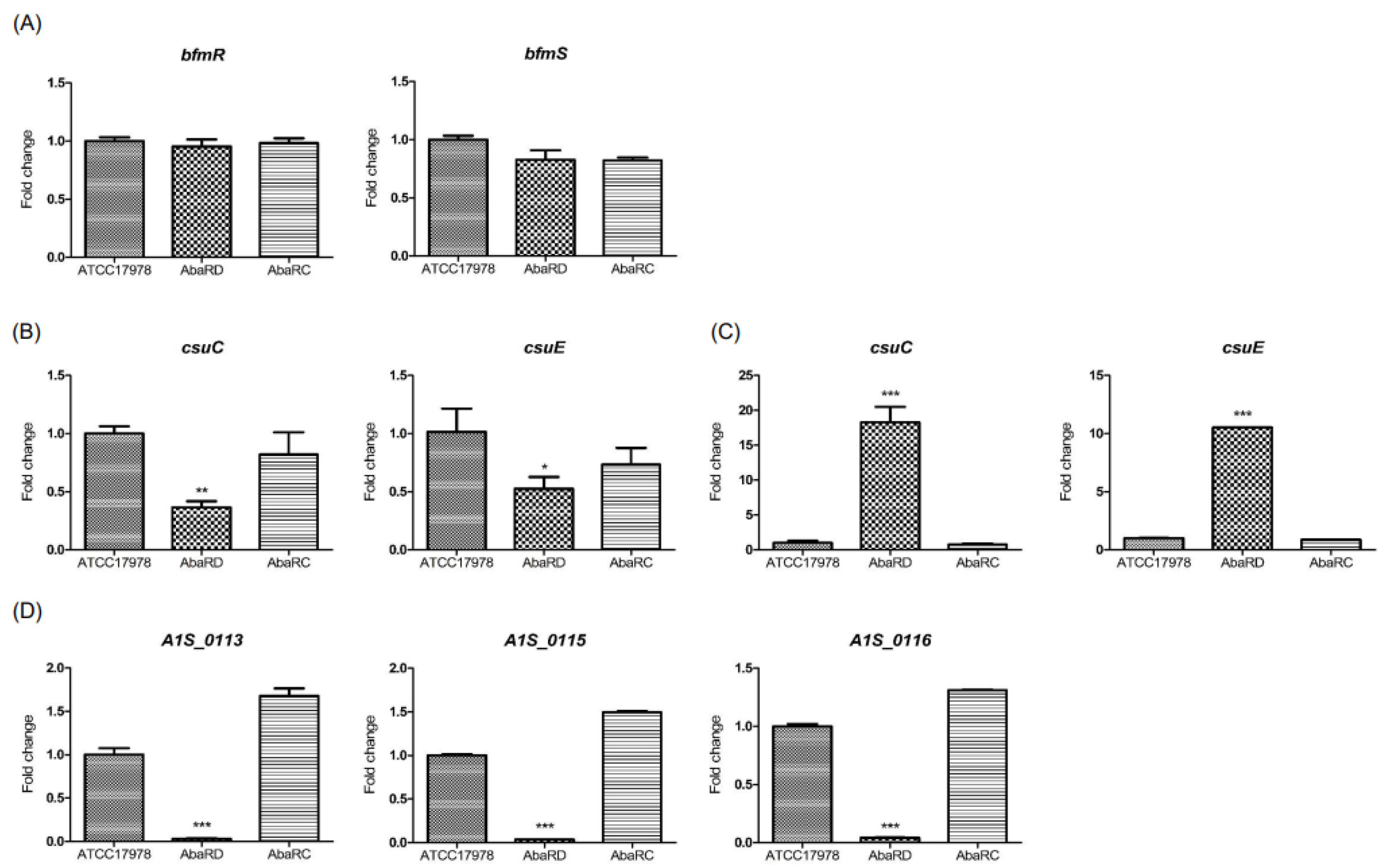
2.5. Effect of AbaR on the Expression of Biofilm- and Surface-Motility-Associated Genes
3. Discussion
4. Materials and Methods
4.1. Bacteria, Plasmids, Culture Media, and Growth Conditions
4.2. Construction of the ΔbfmR Mutant and bfmR-Complemented Strains
4.3. Bacterial Growth Studies
4.4. RNA Isolation and Gene Expression Assay Using qPCR
4.5. Surface Motility Assay
4.6. Biofilm and Pellicle Formation Assays
4.7. Bioassay for the Detection of Autoinducers
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mancuso, G.; Midiri, A.; Gerace, E.; Biondo, C. Bacterial antibiotic resistance: The most critical pathogens. Pathogens 2021, 10, 1310. [Google Scholar] [CrossRef] [PubMed]
- Pendleton, J.N.; Gorman, S.P.; Gilmore, B.F. Clinical relevance of the ESKAPE pathogens. Expert Rev. Anti. Infect. Ther. 2013, 11, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Antunes, L.C.S.; Visca, P.; Towner, K.J. Acinetobacter baumannii: Evolution of a global pathogen. Pathog. Dis. 2014, 71, 292–301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajamohan, G.; Srinivasan, V.B.; Gebreyes, W.A. Molecular and functional characterization of a novel efflux pump, AmvA, mediating antimicrobial and disinfectant resistance in Acinetobacter baumannii. J. Antimicrob. Chemother. 2010, 65, 1919–1925. [Google Scholar] [CrossRef] [Green Version]
- Wendt, C.; Dietze, B.; Dietz, E.; Rüden, H. Survival of Acinetobacter baumannii on dry surfaces. J. Clin. Microbiol. 1997, 35, 1394–1397. [Google Scholar] [CrossRef] [Green Version]
- Harding, C.M.; Hennon, S.W.; Feldman, M.F. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat. Rev. Microbiol. 2018, 16, 91–102. [Google Scholar] [CrossRef]
- Morris, F.C.; Dexter, C.; Kostoulias, X.; Uddin, M.I.; Peleg, A.Y. The mechanisms of disease caused by Acinetobacter baumannii. Front. Microbiol. 2019, 10, 1601. [Google Scholar] [CrossRef] [Green Version]
- De Silva, P.M.; Kumar, A. Signal transduction proteins in Acinetobacter baumannii: Role in antibiotic resistance, virulence, and potential as drug targets. Front. Microbiol. 2019, 10, 49. [Google Scholar] [CrossRef] [Green Version]
- Saipriya, K.; Swathi, C.H.; Ratnakar, K.S.; Sritharan, V. Quorum-sensing system in Acinetobacter baumannii: A potential target for new drug development. J. Appl. Microbiol. 2020, 128, 15–27. [Google Scholar] [CrossRef] [Green Version]
- Eijkelkamp, B.A.; Stroeher, U.H.; Hassan, K.A.; Elbourne, L.D.H.; Paulsen, I.T.; Brown, M.H. H-NS plays a role in expression of Acinetobacter baumannii virulence features. Infect. Immun. 2013, 81, 2574–2583. [Google Scholar] [CrossRef]
- Kim, N.; Son, J.H.; Kim, K.; Kim, H.J.; Kim, Y.J.; Shin, M.; Lee, J.C. Global regulator DksA modulates virulence of Acinetobacter baumannii. Virulence 2021, 12, 2750–2763. [Google Scholar] [CrossRef]
- Laub, M.T.; Goulian, M. Specificity in two-component signal transduction pathways. Annu. Rev. Genet. 2007, 41, 121–145. [Google Scholar] [CrossRef] [Green Version]
- Geisinger, E.; Isberg, R.R. Antibiotic modulation of capsular exopolysaccharide and virulence in Acinetobacter baumannii. PLoS Pathog. 2015, 11, e1004691. [Google Scholar] [CrossRef] [Green Version]
- Kaushik, V.; Tiwari, M.; Joshi, R.; Tiwari, V. Therapeutic strategies against potential antibiofilm targets of multidrug-resistant Acinetobacter baumannii. J. Cell. Physiol. 2022, 237, 2045–2063. [Google Scholar] [CrossRef]
- Russo, T.A.; Manohar, A.; Beanan, J.M.; Olson, R.; MacDonald, U.; Graham, J.; Umland, T.C. The response regulator BfmR is a potential drug target for Acinetobacter baumannii. mSphere 2016, 1, e00082-16. [Google Scholar] [CrossRef] [Green Version]
- Tomaras, A.P.; Flagler, M.J.; Dorsey, C.W.; Gaddy, J.A.; Actis, L.A. Characterization of a two-component regulatory system from Acinetobacter baumannii that controls biofilm formation and cellular morphology. Microbiology 2008, 154, 3398–3409. [Google Scholar] [CrossRef] [Green Version]
- Geisinger, E.; Mortman, N.J.; Vargas-Cuebas, G.; Tai, A.K.; Isberg, R.R. A global regulatory system links virulence and antibiotic resistance to envelope homeostasis in Acinetobacter baumannii. PLoS Pathog. 2018, 14, e1007030. [Google Scholar] [CrossRef] [Green Version]
- Krasauskas, R.; Skerniškytė, J.; Armalytė, J.; Sužiedėlienė, E. The role of Acinetobacter baumannii response regulator BfmR in pellicle formation and competitiveness via contact-dependent inhibition system. BMC Microbiol. 2019, 19, 241. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.Y.; Kim, M.H.; Kim, S.I.; Son, J.H.; Kim, S.; Lee, Y.C.; Shin, M.; Oh, M.H.; Lee, J.C. The sensor kinase BfmS controls production of outer membrane vesicles in Acinetobacter baumannii. BMC Microbiol. 2019, 19, 301. [Google Scholar] [CrossRef]
- Liou, M.L.; Soo, P.C.; Ling, S.R.; Kuo, H.Y.; Yang, C.Y.; Chang, K.C. The sensor kinase BfmS mediates virulence in Acinetobacter baumannii. J. Microbiol. Immunol. Infect. 2014, 47, 275–281. [Google Scholar] [CrossRef]
- Mayer, C.; Muras, A.; Parga, A.; Romero, M.; Feal, S.R.; Poza, M.; Ramos-Vivas, J.; Otero, A. Quorum sensing as a target for controlling surface associated motility and biofilm formation in Acinetobacter baumannii ATCC® 17978TM. Front. Microbiol. 2020, 11, 565548. [Google Scholar] [CrossRef] [PubMed]
- Oh, M.H.; Han, K. AbaR is a LuxR type regulator essential for motility and the formation of biofilm and pellicle in Acinetobacter baumannii. Genes Genom. 2020, 42, 1339–1346. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Chen, Y.; Wang, X.; Ding, Y.; Sun, X.; Ni, Z. Contribution of the AbaI/AbaR quorum sensing system to resistance and virulence of Acinetobacter baumannii; clinical strains. Infect. Drug Resist. 2020, 13, 4273–4281. [Google Scholar] [CrossRef]
- Niu, C.; Clemmer, K.M.; Bonomo, R.A.; Rather, P.N. Isolation and characterization of an autoinducer synthase from Acinetobacter baumannii. J. Bacteriol. 2008, 190, 3386–3392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stacy, D.M.; Welsh, M.A.; Rather, P.N.; Blackwell, H.E. Attenuation of quorum sensing in the pathogen Acinetobacter baumannii using non-native N-acyl homoserine lactones. ACS Chem. Biol. 2012, 7, 1719–1728. [Google Scholar] [CrossRef] [Green Version]
- Rumbo-Feal, S.; Pérez, A.; Ramelot, T.A.; Álvarez-Fraga, L.; Vallejo, J.A.; Beceiro, A.; Ohneck, E.J.; Arivett, B.A.; Merino, M.; Fiester, S.E.; et al. Contribution of the A. baumannii A1S_0114 gene to the interaction with eukaryotic cells and virulence. Front. Cell. Infect. Microbiol. 2017, 7, 108. [Google Scholar] [CrossRef] [Green Version]
- Harding, C.M.; Tracy, E.N.; Carruthers, M.D.; Rather, P.N.; Actis, L.A.; Munson, R.S., Jr. Acinetobacter baumannii strain M2 produces type IV pili which play a role in natural transformation and twitching motility but not surface-associated motility. mBio 2013, 4, e00360-13. [Google Scholar] [CrossRef] [Green Version]
- Giles, S.K.; Stroeher, U.H.; Eijkelkamp, B.A.; Brown, M.H. Identification of genes essential for pellicle formation in Acinetobacter baumannii. BMC Microbiol. 2015, 15, 116. [Google Scholar] [CrossRef] [Green Version]
- Gaddy, J.A.; Actis, L.A. Regulation of Acinetobacter baumannii biofilm formation. Future Microbiol. 2009, 4, 273–278. [Google Scholar] [CrossRef] [Green Version]
- Clemmer, K.M.; Bonomo, R.A.; Rather, P.N. Genetic analysis of surface motility in Acinetobacter baumannii. Microbiology 2011, 157, 2534–2544. [Google Scholar] [CrossRef]
- Draughn, G.L.; Milton, M.E.; Feldmann, E.A.; Bobay, B.G.; Roth, B.M.; Olson, A.L.; Thompson, R.J.; Actis, L.A.; Davies, C.; Cavanagh, J. The structure of the biofilm-controlling response regulator BfmR from Acinetobacter baumannii reveals details of its DNA-binding mechanism. J. Mol. Biol. 2018, 430, 806–821. [Google Scholar] [CrossRef]
- Upmayu, K.; Haq, Q.M.R.; Singh, R. Factors mediating Acinetobacter baumannii biofilm formation: Opportunities for developing therapeutics. Curr. Res. Microb. Sci. 2002, 3, 100131. [Google Scholar]
- Cao, Q.; Wang, Y.; Chen, F.; Xia, Y.; Lou, J.; Zhang, X.; Yang, N.; Sun, X.; Zhang, Q.; Zhuo, C.; et al. A novel signal transduction pathway that modulates quorum sensing and bacterial virulence in Pseudomonas aeruginosa. PLoS Pathog. 2014, 10, e1004340. [Google Scholar] [CrossRef] [Green Version]
- Cui, C.; Yang, C.; Song, S.; Fu, S.; Sun, X.; Yang, L.; He, F.; Zhang, L.H.; Zhang, Y.; Deng, Y. A novel two-component system modulates quorum sensing and pathogenicity in Burkholderia cenocepacia. Mol. Microbiol. 2018, 108, 32–44. [Google Scholar] [CrossRef] [Green Version]
- Law, S.K.K.; Tan, H.S. The role of quorum sensing, biofilm formation, and iron acquisition as key virulence mechanisms in Acinetobacter baumannii and the corresponding anti-virulence strategies. Microbiol. Res. 2022, 260, 127032. [Google Scholar] [CrossRef]
- Sun, X.; Ni, Z.; Tang, J.; Ding, Y.; Wnag, X.; Li, F. The abaI/abaR quorum sensing system effects on pathogenicity in Acinetobacter baumannii. Front. Microbiol. 2021, 12, 679241. [Google Scholar] [CrossRef]
- Luo, L.M.; Wu, L.J.; Xiao, Y.L.; Zhao, D.; Chen, Z.X.; Kang, M.; Zhang, Q.; Xie, Y. Enhancing pili assembly and biofilm formation in Acinetobacter baumannii ATCC19606 using non-native acyl-homoserine lactones. BMC Microbiol. 2015, 15, 62. [Google Scholar] [CrossRef] [Green Version]
- Oh, M.H.; Choi, C.H. Role of LuxIR homologue AnoIR in Acinetobacter nosocomialis and the effect of virstatin on the expression of anoR gene. J. Microbiol. Biotechnol. 2015, 25, 1390–1400. [Google Scholar] [CrossRef]
- Kwon, H.I.; Kim, S.; Oh, M.H.; Na, S.H.; Kim, Y.J.; Jeon, Y.H.; Lee, J.C. Outer membrane protein A contributes to antimicrobial resistance of Acinetobacter baumannii through the OmpA-like domain. J. Antimicrob. Chemother. 2017, 72, 3013015. [Google Scholar] [CrossRef]
- Oh, M.H.; Lee, J.C.; Kim, J.; Choi, C.H.; Han, K. Simple method for markerless gene deletion in multidrug-resistant Acinetobacter baumannii. Appl. Environ. Microbiol. 2015, 81, 3357–3368. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.-J.; Kim, N.-Y.; Ko, S.-Y.; Park, S.-Y.; Oh, M.-H.; Shin, M.-S.; Lee, Y.-C.; Lee, J.-C. Complementary Regulation of BfmRS Two-Component and AbaIR Quorum Sensing Systems to Express Virulence-Associated Genes in Acinetobacter baumannii. Int. J. Mol. Sci. 2022, 23, 13136. https://doi.org/10.3390/ijms232113136
Kim H-J, Kim N-Y, Ko S-Y, Park S-Y, Oh M-H, Shin M-S, Lee Y-C, Lee J-C. Complementary Regulation of BfmRS Two-Component and AbaIR Quorum Sensing Systems to Express Virulence-Associated Genes in Acinetobacter baumannii. International Journal of Molecular Sciences. 2022; 23(21):13136. https://doi.org/10.3390/ijms232113136
Chicago/Turabian StyleKim, Hyo-Jeong, Na-Yeong Kim, Seo-Yeon Ko, Seong-Yong Park, Man-Hwan Oh, Min-Sang Shin, Yoo-Chul Lee, and Je-Chul Lee. 2022. "Complementary Regulation of BfmRS Two-Component and AbaIR Quorum Sensing Systems to Express Virulence-Associated Genes in Acinetobacter baumannii" International Journal of Molecular Sciences 23, no. 21: 13136. https://doi.org/10.3390/ijms232113136





