Systematic Identification of Methyl Jasmonate-Responsive Long Noncoding RNAs and Their Nearby Coding Genes Unveils Their Potential Defence Roles in Tobacco BY-2 Cells
Abstract
:1. Introduction
2. Results
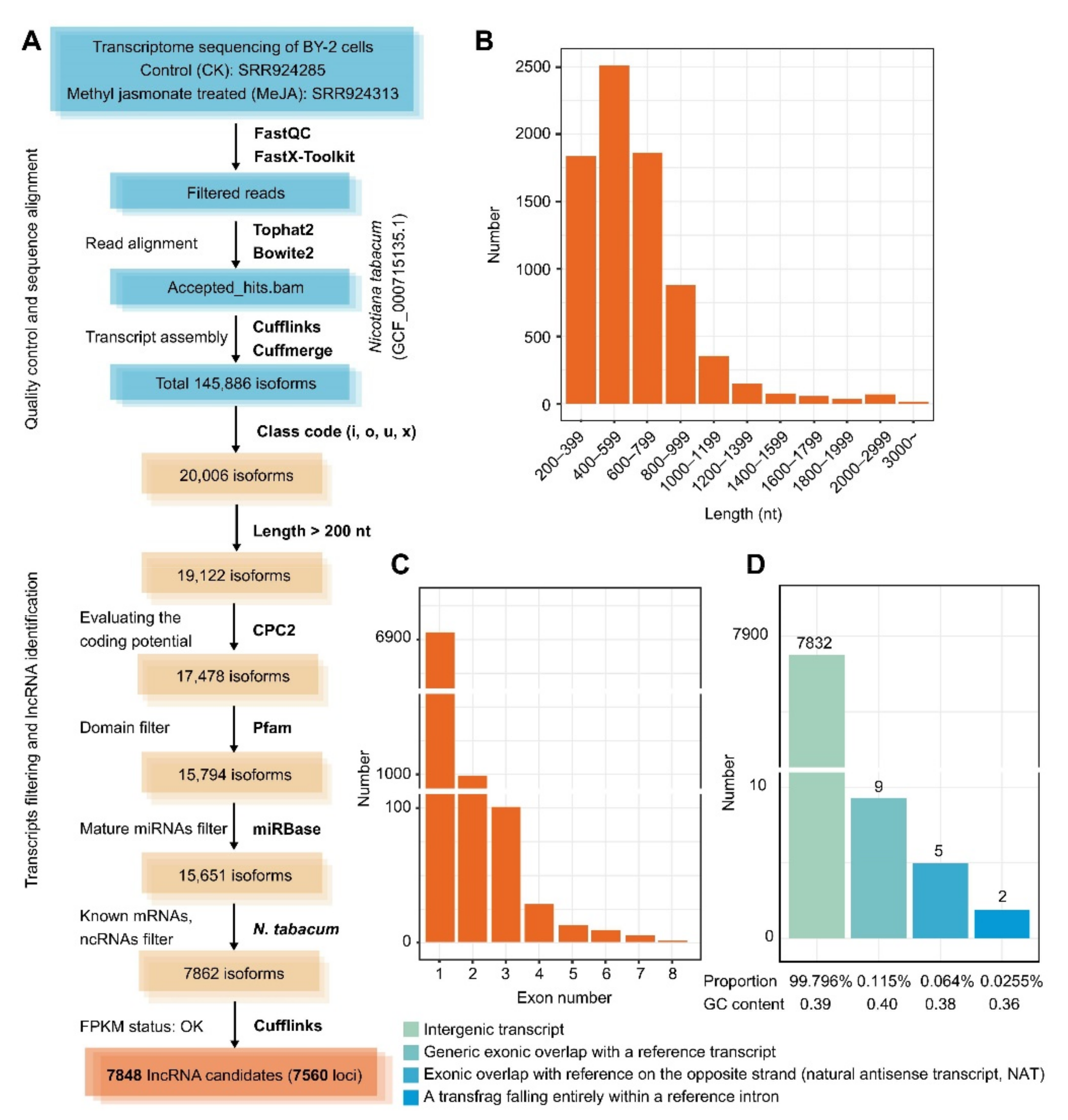
2.1. Reliable Mining for lncRNAs Indicates That Intergenic Transcripts Are the Most Prominent Group
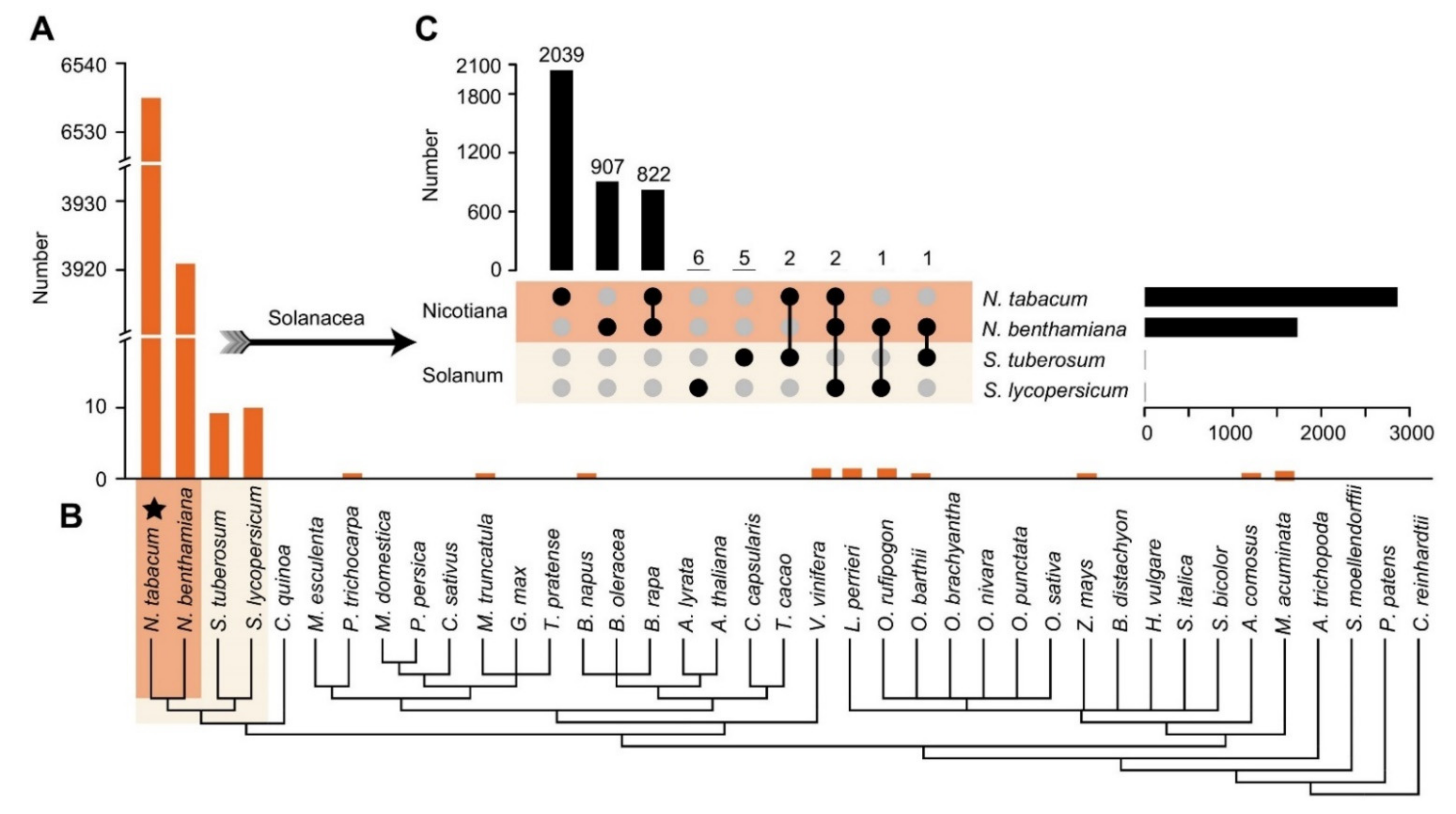
2.2. The lncRNAs in BY-2 Cells Have Extreme Genus Specificity for Nicotiana
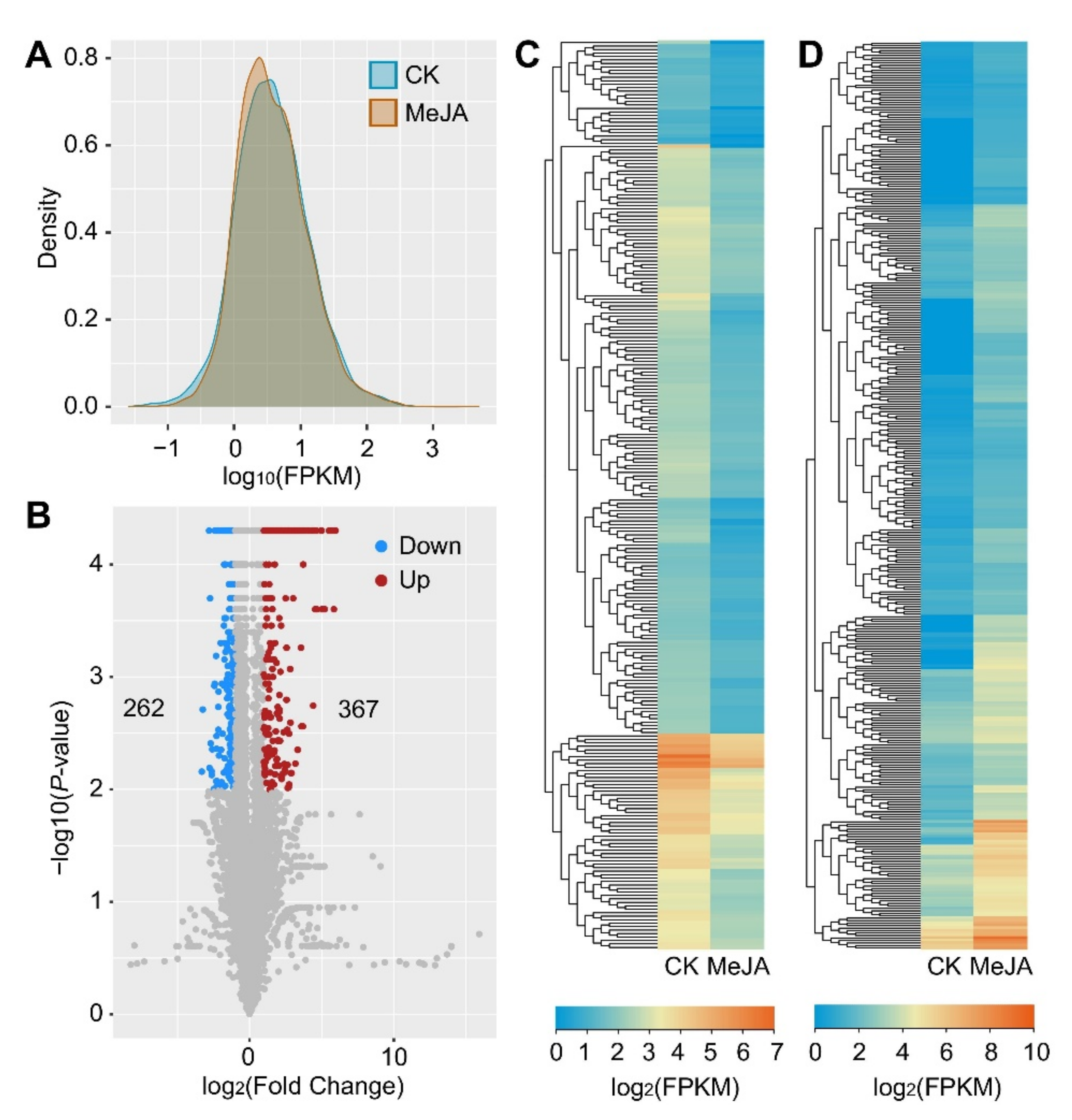
2.3. The Upregulated lncRNAs Constitute a Large Majority of the MeJA-Responsive lncRNAs
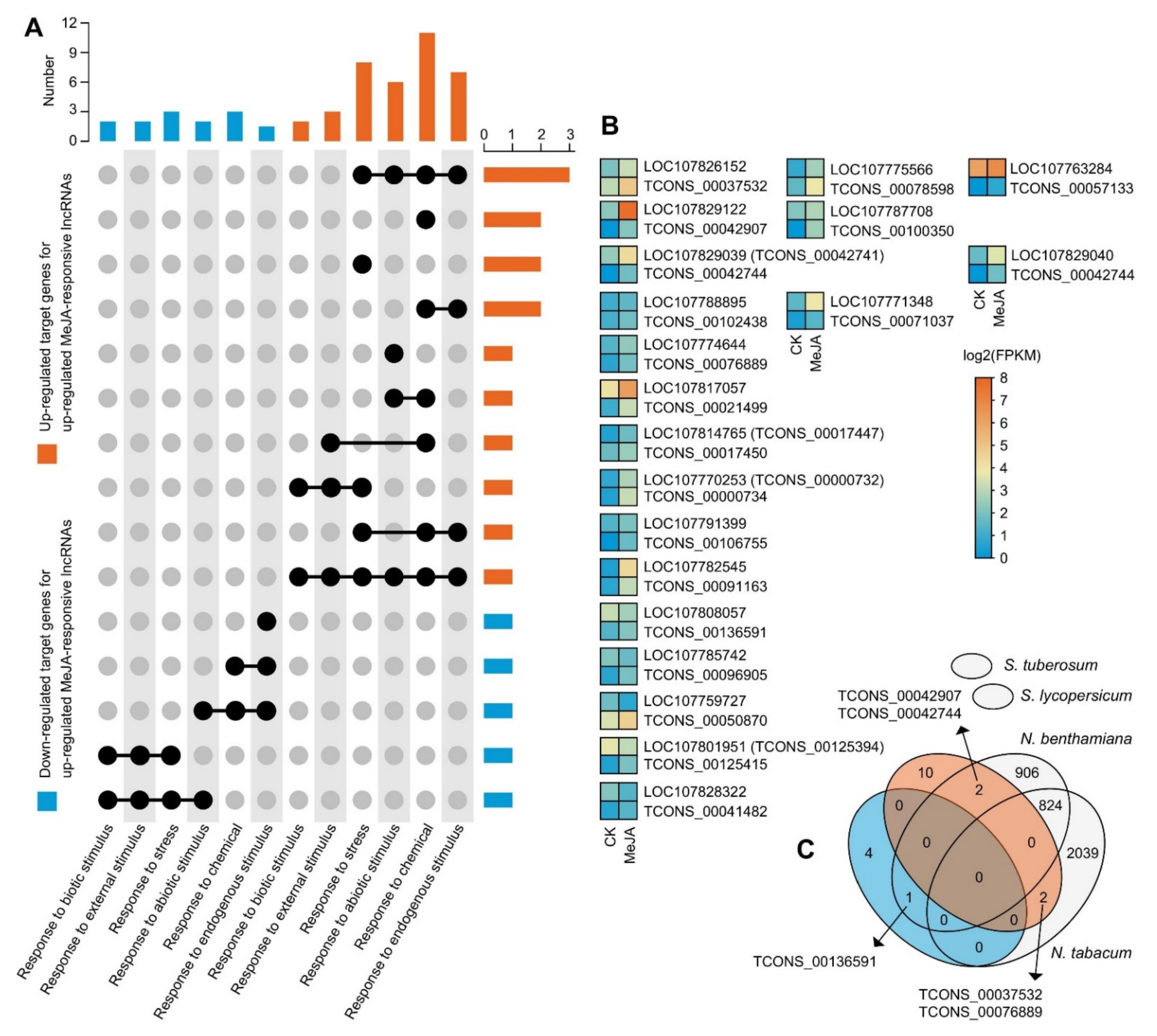
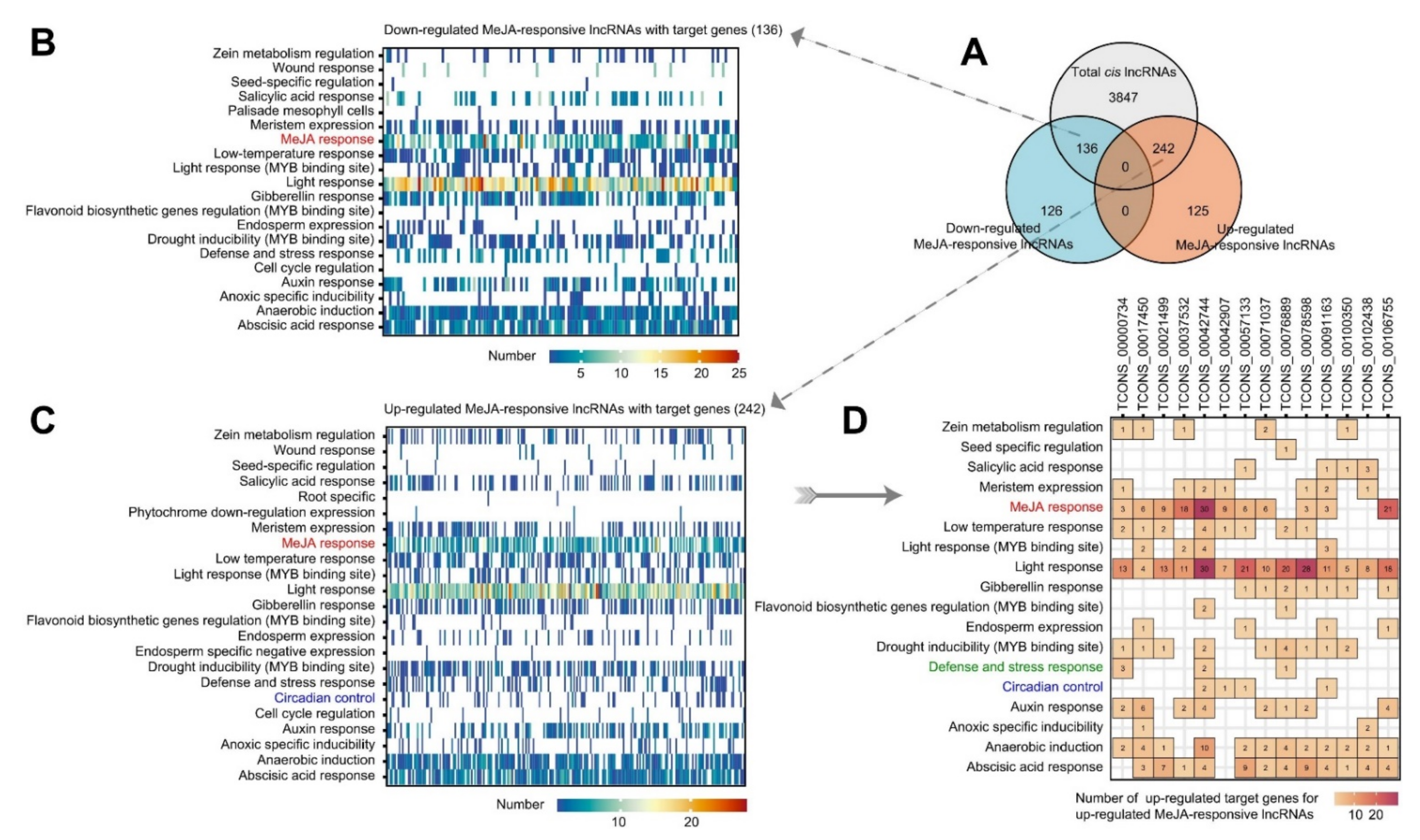
2.4. Potential Up- and Downregulated Target Genes of MeJA-Upregulated lncRNAs Are Responsible for Different Biological Functions and Metabolic Patterns
2.5. Response-Associated Target Genes of Some Conserved MeJA-Responsive lncRNAs Are Involved in Plant Defence and Stress Resistance
2.6. MeJA-Responsive Stress- or Defence-Related Cis-Regulatory Elements May Be Responsible for the Upregulation of lncRNAs and Be Involved in Plant Defence through Their Target Genes
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. General RNA-Seq Read Mapping and Assembly
4.3. lncRNA Identification and Prediction
4.4. Differential Expression Analysis of MeJA-Responsive lncRNAs
4.5. Prediction of Cis-Regulating Target Genes of MeJA-Responsive lncRNAs
4.6. Expression Pattern Comparison of Cis-Regulated Target Genes with Their MeJA-Responsive lncRNAs
4.7. Gene Ontology and Pathway Enrichment Analysis
4.8. Cis-Regulatory Element Prediction in the Promoter Sequences
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pauli, A.; Rinn, J.L.; Schier, A.F. Non-Coding RNAs as Regulators of Embryogenesis. Nat. Rev. Genet. 2011, 12, 136–149. [Google Scholar] [CrossRef] [PubMed]
- Rai, M.I.; Alam, M.; Lightfoot, D.A.; Gurha, P.; Afzal, A.J. Classification and Experimental Identification of Plant Long Non-Coding RNAs. Genomics 2019, 111, 997–1005. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Zhang, Y.; Chen, X.; Chen, Y. Plant Noncoding RNAs: Hidden Players in Development and Stress Responses. Annu. Rev. Cell Dev. Biol. 2019, 35, 407–431. [Google Scholar] [CrossRef]
- Sarropoulos, I.; Marin, R.; Cardoso-Moreira, M.; Kaessmann, H. Developmental Dynamics of LncRNAs across Mammalian Organs and Species. Nature 2019, 571, 510–514. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Wang, C.; Bao, H.; Chen, H.; Wang, Y. Genome-Wide Identification and Characterization of Novel LncRNAs in Populus under Nitrogen Deficiency. Mol. Genet. Genom. 2016, 291, 1663–1680. [Google Scholar] [CrossRef] [PubMed]
- Böhmdorfer, G.; Wierzbicki, A.T. Control of Chromatin Structure by Long Noncoding RNA. Trends Cell Biol. 2015, 25, 623–632. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Yang, L.; Chen, L.L. The Diversity of Long Noncoding RNAs and Their Generation. Trends Genet. 2017, 33, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Penfield, S. Feedback Regulation of COOLAIR Expression Controls Seed Dormancy and Flowering Time. Science 2018, 360, 1014–1017. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.H.; Sung, S. Coordination of the Vernalization Response through a VIN3 and FLC Gene Family Regulatory Network in Arabidopsis. Plant Cell 2013, 25, 454–469. [Google Scholar] [CrossRef] [Green Version]
- Bardou, F.; Ariel, F.; Simpson, C.G.; Romero-Barrios, N.; Laporte, P.; Balzergue, S.; Brown, J.W.S.; Crespi, M. Long Noncoding RNA Modulates Alternative Splicing Regulators in Arabidopsis. Dev. Cell 2014, 30, 166–176. [Google Scholar] [CrossRef]
- Röhrig, H.; Schmidt, J.; Miklashevichs, E.; Schell, J.; John, M. Soybean ENOD40 Encodes Two Peptides That Bind to Sucrose Synthase. Proc. Natl. Acad. Sci. USA 2002, 99, 1915–1920. [Google Scholar] [CrossRef] [Green Version]
- Sousa, C.; Johansson, C.; Charon, C.; Manyani, H.; Sautter, C.; Kondorosi, A.; Crespi, M. Translational and Structural Requirements of the Early Nodulin Gene Enod40, a Short-Open Reading Frame-Containing RNA, for Elicitation of a Cell-Specific Growth Response in the Alfalfa Root Cortex. Mol. Cell Biol. 2001, 21, 354–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Begcy, K.; Dresselhaus, T. Epigenetic Responses to Abiotic Stresses during Reproductive Development in Cereals. Plant Reprod. 2018, 31, 343–355. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, C.; Brennan, R.M.; Graham, J.; Karley, A.J. Plant Defense against Herbivorous Pests: Exploiting Resistance and Tolerance Traits for Sustainable Crop Protection. Front. Plant Sci. 2016, 7, 1132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stenberg, J.A.; Heil, M.; Åhman, I.; Björkman, C. Optimizing Crops for Biocontrol of Pests and Disease. Trends Plant Sci. 2015, 20, 698–712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, C.; Eigenbrode, S.D. Effects of Surface Wax Variation in Pisum sativum on Herbivorous and Entomophagous Insects in the Field. Environ. Entomol. 2000, 29, 773–780. [Google Scholar] [CrossRef]
- Zúñiga, G.; Corcuera, L.J. Effect of Gramine in the Resistance of Barley Seedlings to the Aphid Rhopalosiphum Padi. Entomol. Exp. Appl. 2011, 40, 259–262. [Google Scholar] [CrossRef]
- Steppuhn, A.; Gase, K.; Krock, B.; Halitschke, R.; Baldwin, I.T. Nicotine’s Defensive Function in Nature. PLoS Biol. 2004, 2, e217. [Google Scholar] [CrossRef]
- Memelink, J.; Verpoorte, R.; Kijne, J.W. ORCAnization of Jasmonate-Responsive Gene Expression in Alkaloid Metabolism. Trends Plant Sci. 2001, 6, 212–219. [Google Scholar] [CrossRef]
- Kutchan, T.M. Alkaloid Biosynthesis [Mdash] The Basis for Metabolic Engineering of Medicinal Plants. Plant Cell 1995, 7, 1059. [Google Scholar] [CrossRef] [PubMed]
- Oksman-Caldentey, K.M. Tropane and Nicotine Alkaloid Biosynthesis-Novel Approaches towards Biotechnological Production of Plant-Derived Pharmaceuticals. Curr. Pharm. Biotechnol. 2007, 8, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Shoji, T.; Ogawa, T.; Hashimoto, T. Jasmonate-Induced Nicotine Formation in Tobacco Is Mediated by Tobacco COI1 and JAZ Genes. Plant Cell Physiol. 2008, 49, 1003–1012. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.; Li, C.; Zhang, F. Nicotine Synthesis in Nicotiana Tabacum L. Induced by Mechanical Wounding Is Regulated by Auxin. J. Exp. Bot. 2006, 57, 2899–2907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Geyter, N.; Gholami, A.; Goormachtig, S.; Goossens, A. Transcriptional Machineries in Jasmonate-Elicited Plant Secondary Metabolism. Trends Plant Sci. 2012, 17, 349–359. [Google Scholar] [CrossRef]
- Ryan, C.A.; Lamb, C.J.; Jagendorf, A.T.; Kolattukudy, P.E.; Doares, S.H.; Syrovetst, T.; Weilert, E.W. Oligogalacturonides and Chitosan Activate Plant Defensive Genes through the Octadecanoid Pathway. Proc. Natl. Acad. Sci. USA 1995, 92, 4095–4098. [Google Scholar] [CrossRef] [Green Version]
- Baldwin, I.T.; Schmelz, E.A.; Ohnmeiss, T.E. Wound-Induced Changes in Root and Shoot Jasmonic Acid Pools Correlate with Induced Nicotine Synthesis in Nicotiana Sylvestris Spegazzini and Comes. J. Chem. Ecol. 1994, 20, 2139–2157. [Google Scholar] [CrossRef]
- Creelman, R.A.; Tierney, M.L.; Mullet, J.E. Jasmonic Acid/Methyl Jasmonate Accumulate in Wounded Soybean Hypocotyls and Modulate Wound Gene Expression. Proc. Natl. Acad. Sci. USA 1992, 89, 4938–4941. [Google Scholar] [CrossRef] [Green Version]
- Papazian, S.; Girdwood, T.; Wessels, B.A.; Poelman, E.H.; Dicke, M.; Moritz, T.; Albrectsen, B.R. Leaf Metabolic Signatures Induced by Real and Simulated Herbivory in Black Mustard (Brassica nigra). Metabolomics 2019, 15, 130. [Google Scholar] [CrossRef] [Green Version]
- Boddu, J.; Jiang, C.; Sangar, V.; Olson, T.; Peterson, T.; Chopra, S. Comparative Structural and Functional Characterization of Sorghum and Maize Duplications Containing Orthologous Myb Transcription Regulators of 3-Deoxyflavonoid Biosynthesis. Plant Mol. Biol. 2006, 60, 185–199. [Google Scholar] [CrossRef]
- Gao, Y.; Liu, J.; Chen, Y.; Tang, H.; Wang, Y.; He, Y.; Ou, Y.; Sun, X.; Wang, S.; Yao, Y. Tomato SlAN11 Regulates Flavonoid Biosynthesis and Seed Dormancy by Interaction with BHLH Proteins but Not with MYB Proteins. Hort. Res. 2018, 5, 27. [Google Scholar] [CrossRef]
- vom Endt, D.; Kijne, J.W.; Memelink, J. Transcription Factors Controlling Plant Secondary Metabolism: What Regulates the Regulators? Phytochemistry 2002, 61, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Sears, M.T.; Zhang, H.; Rushton, P.J.; Wu, M.; Han, S.; Spano, A.J.; Timko, M.P. NtERF32: A Non-NIC2 Locus AP2/ERF Transcription Factor Required in Jasmonate-Inducible Nicotine Biosynthesis in Tobacco. Plant Mol. Biol. 2013, 84, 49–66. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Yang, Y.; Li, R.; Fu, D.; Wen, L.; Luo, Y.; Zhu, H. RNA Sequencing and Functional Analysis Implicate the Regulatory Role of Long Non-Coding RNAs in Tomato Fruit Ripening. J. Exp. Bot. 2015, 66, 4483–4495. [Google Scholar] [CrossRef] [Green Version]
- de Boer, K.; Tilleman, S.; Pauwels, L.; vanden Bossche, R.; de Sutter, V.; Vanderhaeghen, R.; Hilson, P.; Hamill, J.D.; Goossens, A. Apetala2/Ethylene Response Factor and Basic Helix-Loop-Helix Tobacco Transcription Factors Cooperatively Mediate Jasmonate-Elicited Nicotine Biosynthesis. Plant J. 2011, 66, 1053–1065. [Google Scholar] [CrossRef]
- Chen, D.; Mubeen, B.; Hasnain, A.; Rizwan, M.; Adrees, M.; Naqvi, S.A.H.; Iqbal, S.; Kamran, M.; El-Sabrout, A.M.; Elansary, H.O.; et al. Role of Promising Secondary Metabolites to Confer Resistance Against Environmental Stresses in Crop Plants: Current Scenario and Future Perspectives. Front. Plant Sci. 2022, 13, 881032. [Google Scholar] [CrossRef]
- Proost, S.; Pattyn, P.; Gerats, T.; van de Peer, Y. Journey through the Past: 150 Million Years of Plant Genome Evolution. Plant J. 2011, 66, 58–65. [Google Scholar] [CrossRef]
- Leitch, I.J.; Hanson, L.; Lim, K.Y.; Kovarik, A.; Chase, M.W.; Clarkson, J.J.; Leitch, A.R. The Ups and Downs of Genome Size Evolution in Polyploid Species of Nicotiana (Solanaceae). Ann. Bot. 2008, 101, 805–814. [Google Scholar] [CrossRef] [Green Version]
- Hochholdinger, F.; Baldauf, J.A. Current Biology Heterosis in Plants. Curr. Biol. 2018, 28, 1075–1095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mo, Z.; Luo, W.; Pi, K.; Duan, L.; Wang, P.; Ke, Y.; Zeng, S.; Jia, R.; Liang, T.; Huang, Y.; et al. Comparative Transcriptome Analysis between Inbred Lines and Hybrids Provides Molecular Insights into K+ Content Heterosis of Tobacco (Nicotiana Tabacum L.). Front Plant Sci. 2022, 13, 2598. [Google Scholar] [CrossRef]
- Shi, C.; Zhao, L.; Zhang, X.; Lv, G.; Pan, Y.; Chen, F. Gene Regulatory Network and Abundant Genetic Variation Play Critical Roles in Heading Stage of Polyploidy Wheat. BMC Plant Biol. 2019, 19, 6. [Google Scholar] [CrossRef]
- Howe, G.A.; Schilmiller, A.L. Oxylipin Metabolism in Response to Stress. Curr. Opin. Plant Biol. 2002, 5, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Green, T.R.; Ryan, C.A. Wound-Induced Proteinase Inhibitor in Plant Leaves: A Possible Defense Mechanism against Insects. Science 1972, 175, 776–777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryan, S.M.; Cane, K.A.; DeBoer, K.D.; Sinclair, S.J.; Brimblecombe, R.; Hamill, J.D. Structure and Expression of the Quinolinate Phosphoribosyltransferase (QPT) Gene Family in Nicotiana. Plant Sci. 2012, 188–189, 102–110. [Google Scholar] [CrossRef]
- Lee, H.K.; Cho, S.K.; Son, O.; Xu, Z.; Hwang, I.; Kim, W.T. Drought Stress-Induced Rma1H1, a RING Membrane-Anchor E3 Ubiquitin Ligase Homolog, Regulates Aquaporin Levels via Ubiquitination in Transgenic Arabidopsis Plants. Plant Cell 2009, 21, 622–641. [Google Scholar] [CrossRef] [Green Version]
- Nagata, T.; Nemoto, Y.; Hasezawa, S. Tobacco BY-2 Cell Line as the “HeLa” Cell in the Cell Biology of Higher Plants. Int. Rev. Cytol. 1992, 132, 1–30. [Google Scholar] [CrossRef]
- Chen, X.; Sun, S.; Liu, F.; Shen, E.; Liu, L.; Ye, C.; Xiao, B.; Timko, M.P.; Zhu, Q.H.; Fan, L.; et al. A Transcriptomic Profile of Topping Responsive Non-Coding RNAs in Tobacco Roots (Nicotiana tabacum). BMC Genom. 2019, 20, 856. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Gao, J.; Wang, C.; Xu, Y.; Li, X.; Yang, J.; Chen, K.; Kang, Y.; Wang, Y.; Cao, P.; et al. Comprehensive Analysis of Long Non-Coding RNA Modulates Axillary Bud Development in Tobacco (Nicotiana tabacum L.). Front. Plant Sci. 2022, 13, 59. [Google Scholar] [CrossRef]
- Gil, N.; Ulitsky, I. Regulation of Gene Expression by Cis-Acting Long Non-Coding RNAs. Nat. Rev. Genet. 2020, 21, 102–117. [Google Scholar] [CrossRef] [PubMed]
- Johnsson, P.; Lipovich, L.; Grandér, D.; Morris, K.V. Evolutionary Conservation of Long Non-Coding RNAs; Sequence, Structure, Function. Biochim. Biophys. Acta 2014, 1840, 1063–1071. [Google Scholar] [CrossRef] [Green Version]
- Song, X.; Hu, J.; Wu, T.; Yang, Q.; Feng, X.; Lin, H.; Feng, S.; Cui, C.; Yu, Y.; Zhou, R.; et al. Comparative Analysis of Long Noncoding RNAs in Angiosperms and Characterization of Long Noncoding RNAs in Response to Heat Stress in Chinese Cabbage. Hort. Res. 2021, 8, 48. [Google Scholar] [CrossRef]
- Benfey, P.N.; Chua, N.H. The Cauliflower Mosaic Virus 35S Promoter: Combinatorial Regulation of Transcription in Plants. Science 1990, 250, 959–966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Salasini, B.C.; Khan, M.; Devi, B.; Bush, M.; Subramaniam, R.; Hepworth, S.R. Clade I TGACG-Motif Binding Basic Leucine Zipper Transcription Factors Mediate Blade-on-Petiole-Dependent Regulation of Development. Plant Physiol. 2019, 180, 937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Idrovo Espín, F.M.; Peraza-Echeverria, S.; Fuentes, G.; Santamaría, J.M. In Silico Cloning and Characterization of the TGA (Tgacg Motif-Binding Factor) Transcription Factors Subfamily in Carica papaya. Plant Physiol. Biochem. 2012, 54, 113–122. [Google Scholar] [CrossRef]
- Zhang, J.-Y.; Qu, S.-C.; Du, X.-L.; Qiao, Y.-S.; Cai, B.-H.; Guo, Z.-R.; Zhang, Z. Overexpression of the Malus Hupehensis MhTGA2 Gene, a Novel BZIP Transcription Factor for Increased Tolerance to Salt and Osmotic Stress in Transgenic Tobacco. Int. J. Plant Sci. 2012, 173, 441–453. [Google Scholar] [CrossRef]
- Zander, M.; la Camera, S.; Lamotte, O.; Métraux, J.P.; Gatz, C. Arabidopsis Thaliana Class-II TGA Transcription Factors Are Essential Activators of Jasmonic Acid/Ethylene-Induced Defense Responses. Plant J. 2010, 61, 200–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Yan, P.; Yi, C.; Li, W.; Chai, Y.; Fei, L.; Gao, P.; Zhao, H.; Wang, Y.; Timko, M.P.; et al. Transcriptome-Wide Analysis of Jasmonate-Treated BY-2 Cells Reveals New Transcriptional Regulators Associated with Alkaloid Formation in Tobacco. J. Plant Physiol. 2017, 215, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Guo, J.; Yan, P.; Li, Y.; Liu, K.; Gao, P.; Zhao, H.; Chen, Y.; Wang, Y.; Timko, M.P.; et al. Transcriptome Profiling Identified Multiple Jasmonate ZIM-Domain Proteins Involved in the Regulation of Alkaloid Biosynthesis in Tobacco BY-2 Cells. Plant Mol. Biol. Rep. 2014, 33, 153–166. [Google Scholar] [CrossRef]
- Andersen, R.E.; Hong, S.J.; Lim, J.J.; Cui, M.; Harpur, B.A.; Hwang, E.; Delgado, R.N.; Ramos, A.D.; Liu, S.J.; Blencowe, B.J.; et al. The Long Noncoding RNA Pnky Is a Trans-Acting Regulator of Cortical Development In Vivo. Dev. Cell 2019, 49, 632–642.e7. [Google Scholar] [CrossRef]
- Huarte, M.; Guttman, M.; Feldser, D.; Garber, M.; Koziol, M.J.; Kenzelmann-Broz, D.; Khalil, A.M.; Zuk, O.; Amit, I.; Rabani, M.; et al. A Large Intergenic Noncoding RNA Induced by P53 Mediates Global Gene Repression in the P53 Response. Cell 2010, 142, 409–419. [Google Scholar] [CrossRef] [Green Version]
- Ding, M.; Liu, Y.; Liao, X.; Zhan, H.; Liu, Y.; Huang, W. Enhancer RNAs (ERNAs): New Insights into Gene Transcription and Disease Treatment. J. Cancer 2018, 9, 2334. [Google Scholar] [CrossRef]
- Li, F.; Wang, W.; Zhao, N.; Xiao, B.; Cao, P.; Wu, X.; Ye, C.; Shen, E.; Qiu, J.; Zhu, Q.H.; et al. Regulation of Nicotine Biosynthesis by an Endogenous Target Mimicry of MicroRNA in Tobacco. Plant Physiol. 2015, 169, 1062–1071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zenkner, F.F.; Margis-Pinheiro, M.; Cagliari, A. Nicotine Biosynthesis in Nicotiana: A Metabolic Overview. Tobacco Sci. 2019, 56, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vellosillo, T.; Martínez, M.; López, M.A.; Vicente, J.; Cascón, T.; Dolan, L.; Hamberg, M.; Castresana, C. Oxylipins Produced by the 9-Lipoxygenase Pathway in Arabidopsis Regulate Lateral Root Development and Defense Responses through a Specific Signaling Cascade. Plant Cell 2007, 19, 831–846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Göbel, C.; Feussner, I.; Schmidt, A.; Scheel, D.; Sanchez-Serrano, J.; Hamberg, M.; Rosahl, S. Oxylipin Profiling Reveals the Preferential Stimulation of the 9-Lipoxygenase Pathway in Elicitor-Treated Potato Cells. J. Biol. Chem. 2001, 276, 6267–6273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikeda, T.; Matsumoto, T.; Noguchi, M. Effects of Nutritional Factors on the Formation of Ubiquinone by Tobacco Plant Cells in Suspension Culture. Agric. Biol. Chem. 1976, 40, 1765–1770. [Google Scholar] [CrossRef] [Green Version]
- Sierro, N.; Battey, J.N.D.; Ouadi, S.; Bakaher, N.; Bovet, L.; Willig, A.; Goepfert, S.; Peitsch, M.C.; Ivanov, N.V. The Tobacco Genome Sequence and Its Comparison with Those of Tomato and Potato. Nat. Commun. 2014, 5, 3833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential Gene and Transcript Expression Analysis of RNA-Seq Experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [Green Version]
- Jia, H.; Osak, M.; Bogu, G.K.; Stanton, L.W.; Johnson, R.; Lipovich, L. Genome-Wide Computational Identification and Manual Annotation of Human Long Noncoding RNA Genes. RNA 2010, 16, 1478–1487. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, K.; Wang, Z.; Pang, L.; Song, Z.; Zhao, H.; Wang, Y.; Wang, B.; Han, S. Systematic Identification of Methyl Jasmonate-Responsive Long Noncoding RNAs and Their Nearby Coding Genes Unveils Their Potential Defence Roles in Tobacco BY-2 Cells. Int. J. Mol. Sci. 2022, 23, 15568. https://doi.org/10.3390/ijms232415568
Zheng K, Wang Z, Pang L, Song Z, Zhao H, Wang Y, Wang B, Han S. Systematic Identification of Methyl Jasmonate-Responsive Long Noncoding RNAs and Their Nearby Coding Genes Unveils Their Potential Defence Roles in Tobacco BY-2 Cells. International Journal of Molecular Sciences. 2022; 23(24):15568. https://doi.org/10.3390/ijms232415568
Chicago/Turabian StyleZheng, Kaifeng, Zitao Wang, Lu Pang, Zhongbang Song, Heping Zhao, Yingdian Wang, Bingwu Wang, and Shengcheng Han. 2022. "Systematic Identification of Methyl Jasmonate-Responsive Long Noncoding RNAs and Their Nearby Coding Genes Unveils Their Potential Defence Roles in Tobacco BY-2 Cells" International Journal of Molecular Sciences 23, no. 24: 15568. https://doi.org/10.3390/ijms232415568





