Roux-En-Y Gastric Bypass (RYGB) Surgery during High Liquid Sucrose Diet Leads to Gut Microbiota-Related Systematic Alterations
Abstract
:1. Introduction
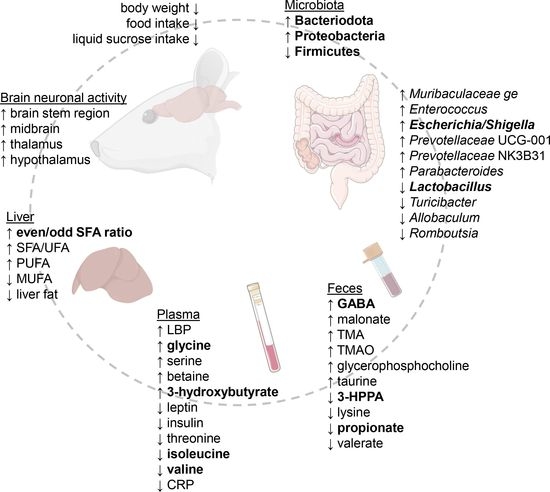
2. Results
2.1. RYGB Surgery Leads to Reduced Body Weight, Food, and Liquid Sucrose Intake
2.2. RYGB Surgery Leads to Reduced Relative Abundance of Firmicutes and Increased Bacteroidota and Proteobacteria Species
2.3. Fecal Metabolomics Show Altered Metabolite Production Post-Surgery
2.4. RYGB Surgery-Altered Microbiota Composition Correlates with the Feces Metabolite Changes
2.5. RYGB Surgery Changes the Branched-Chain Amino Acid, Serine, and Glycine Metabolism in Plasma
2.6. RYGB Animals Have Reduced C-Reactive and Increased Lipopolysaccharide-Binding Proteins in Plasma
2.7. The Hepatic Fatty Acid Profile Changes with Liquid Sucrose Diet after RYGB Surgery
2.8. The Correlation of Metabolomics Data with Plasma Metabolomics, Immune, Hormonal, and Liver Parameters Indicates Surgery-Specific and Food-Related Patterns
2.9. Brain Imaging Reveals Increased Neuronal Activity after RYGB Surgery
3. Discussion
3.1. Liquid Sucrose-Induced Obesity Is Reverted by RYGB Surgery
3.2. Microbiota Perturbations Lead to Upregulated Fecal GABA and Lower Fiber Fermentation
3.3. RYGB Surgery Leads to Lowered Plasma BCAA and Increased Glycine
3.4. RYGB Surgery Results in Elevated LBP Not as a Result of Inflammation but Possibly from Increased Lipolysis as Seen by Plasma Ketone Body Levels
3.5. The Even/Odd Saturated FA Ratio Is Increased after RYGB Surgery
3.6. RYGB Surgery Leads to Altered Neuronal Activity in the Rat Brain
3.7. Key Findings and Future Implications
4. Materials and Methods
4.1. Animals
4.2. Experimental Design and Surgical Technique
4.3. [18F]FDG-PET with Glucose Stimulation
4.4. [18F]FDG-PET Data Analysis
4.5. Tissue and Feces Collection
4.6. Immunohistochemistry
4.7. Confocal Imaging
4.8. 16S rRNA Gene Sequencing-Based Microbiome Analysis
4.9. 1H-NMR Spectroscopy-Based Metabolomics Analysis of Plasma and Feces
4.10. Immune and Hormonal Parameter Profiling
4.11. Liver Gene Expression
4.12. Liver Fatty Acid Composition
4.13. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, Y.-J.; Ni, Y.-H. Gut microbiota and pediatric obesity/non-alcoholic fatty liver disease. J. Formos. Med. Assoc. 2019, 118, S55–S61. [Google Scholar] [CrossRef]
- Singer-Englar, T.; Barlow, G.; Mathur, R. Obesity, diabetes, and the gut microbiome: An updated review. Expert Rev. Gastroenterol. Hepatol. 2019, 13, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, A.D.; Manson, J.E.; Rifai, N.; Buring, J.E.; Ridker, P.M. C-reactive protein, interleukin 6, and risk of developing type 2 diabetes mellitus. JAMA-J. Am. Med. Assoc. 2001, 286, 327–334. [Google Scholar] [CrossRef] [PubMed]
- Jacka, F.N.; Cherbuin, N.; Anstey, K.J.; Sachdev, P.; Butterworth, P. Western diet is associated with a smaller hippocampus: A longitudinal investigation. BMC Med. 2015, 13, 215. [Google Scholar] [CrossRef] [Green Version]
- Andersen, C.J.; Murphy, K.E.; Fernandez, M.L. Impact of Obesity and Metabolic Syndrome on Immunity. Adv. Nutr. 2016, 7, 66–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaspar, J.M.; Baptista, F.I.; Paula Macedo, M.; Ambrosio, A.F. Inside the Diabetic Brain: Role of Different Players Involved in Cognitive Decline. ACS Chem. Neurosci. 2016, 7, 131–142. [Google Scholar] [CrossRef]
- Hotamisligil, G.S. Inflammation, metaflammation and immunometabolic disorders. Nature 2017, 542, 177–185. [Google Scholar] [CrossRef]
- Pinhel, M.A.S.; Noronha, N.Y.; Nicoletti, C.F.; Pereira, V.A.B.; de Oliveira, B.A.P.; Cortes-Oliveira, C.; Salgado, W., Jr.; Barbosa, F.; Marchini, J.S.; Souza, D.R.S.; et al. Changes in DNA Methylation and Gene Expression of Insulin and Obesity-Related GenePIK3R1after Roux-en-Y Gastric Bypass. Int. J. Mol. Sci. 2020, 21, 4476. [Google Scholar] [CrossRef]
- Hu, F.B.; van Dam, R.M.; Liu, S. Diet and risk of Type II diabetes: The role of types of fat and carbohydrate. Diabetologia 2001, 44, 805–817. [Google Scholar] [CrossRef]
- Di Rienzi, S.C.; Britton, R.A. Adaptation of the Gut Microbiota to Modern Dietary Sugars and Sweeteners. Adv. Nutr. 2020, 11, 616–629. [Google Scholar] [CrossRef] [Green Version]
- Shalev, D.; Arbuckle, M.R. Metabolism and Memory: Obesity, Diabetes, and Dementia. Biol. Psychiatry 2017, 82, e81–e83. [Google Scholar] [CrossRef] [Green Version]
- Graham, L.C.; Harder, J.M.; Soto, I.; de Vries, W.N.; John, S.W.M.; Howell, G.R. Chronic consumption of a western diet induces robust glial activation in aging mice and in a mouse model of Alzheimer’s disease. Sci. Rep. 2016, 6, 21568. [Google Scholar] [CrossRef]
- Almby, K.E.; Lundqvist, M.H.; Abrahamsson, N.; Kvernby, S.; Fahlstrom, M.; Pereira, M.J.; Gingnell, M.; Karlsson, F.A.; Fanni, G.; Sundbom, M.; et al. Effects of Gastric Bypass Surgery on the Brain: Simultaneous Assessment of Glucose Uptake, Blood Flow, Neural Activity, and Cognitive Function During Normo- and Hypoglycemia. Diabetes 2021, 70, 1265–1277. [Google Scholar] [CrossRef]
- Khayyatzadeh, S.S.; Bagherniya, M.; Fazeli, M.; Khorasanchi, Z.; Bidokhti, M.S.; Ahmadinejad, M.; Khoshmohabbat, S.; Arabpour, M.; Afkhamizadeh, M.; Ferns, G.A.; et al. A Western dietary pattern is associated with elevated level of high sensitive C-reactive protein among adolescent girls. Eur. J. Clin. Investig. 2018, 48, e12897. [Google Scholar] [CrossRef] [PubMed]
- Swinburn, B.A.; Sacks, G.; Hall, K.D.; McPherson, K.; Finegood, D.T.; Moodie, M.L.; Gortmaker, S.L. Obesity 1 The global obesity pandemic: Shaped by global drivers and local environments. Lancet 2011, 378, 804–814. [Google Scholar] [CrossRef]
- World Health Organization. Obesity and Overweight. Available online: https://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight/ (accessed on 20 October 2021).
- Apovian, C.M.; Aronne, L.J.; Bessesen, D.H.; McDonnell, M.E.; Murad, M.H.; Pagotto, U.; Ryan, D.H.; Still, C.D. Pharmacological Management of Obesity: An Endocrine Society Clinical Practice Guideline. J. Clin. Endocrinol. Metab. 2015, 100, 342–362. [Google Scholar] [CrossRef] [PubMed]
- Bauer, K.; Lau, T.; Schwille-Kiuntke, J.; Schild, S.; Hauner, H.; Stengel, A.; Zipfel, S.; Mack, I. Conventional weight loss interventions across the different BMI obesity classes: A systematic review and quantitative comparative analysis. Eur. Eat. Disord. Rev. 2020, 28, 492–512. [Google Scholar] [CrossRef]
- Abdeen, G.; le Roux, C.W. Mechanism Underlying the Weight Loss and Complications of Roux-en-Y Gastric Bypass. Review. Obes. Surg. 2016, 26, 410–421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lutz, T.A.; Bueter, M. The Use of Rat and Mouse Models in Bariatric Surgery experiments. Front. Nutr. 2016, 3, 25. [Google Scholar] [CrossRef] [Green Version]
- Olivo, G.; Zhou, W.; Sundbom, M.; Zhukovsky, C.; Hogenkamp, P.; Nikontovic, L.; Stark, J.; Wiemerslage, L.; Larsson, E.M.; Benedict, C.; et al. Resting-state brain connectivity changes in obese women after Roux-en-Y gastric bypass surgery: A longitudinal study. Sci. Rep. 2017, 7, 6616. [Google Scholar] [CrossRef] [Green Version]
- Näslund, E.; Melin, I.; Grybäck, P.; Hägg, A.; Hellström, P.M.; Jacobsson, H.; Theodorsson, E.; Rössner, S.; Backman, L. Reduced food intake after jejunoileal bypass: A possible association with prolonged gastric emptying and altered gut hormone patterns. Am. J. Clin. Nutr. 1997, 66, 26–32. [Google Scholar] [CrossRef] [Green Version]
- Olbers, T.; Björkman, S.; Lindroos, A.; Maleckas, A.; Lönn, L.; Sjöström, L.; Lönroth, H. Body composition, dietary intake, and energy expenditure after laparoscopic Roux-en-Y gastric bypass and laparoscopic vertical banded gastroplasty: A randomized clinical trial. Ann. Surg. 2006, 244, 715–722. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Shin, A.C.; Lenard, N.R.; Townsend, R.L.; Patterson, L.M.; Sigalet, D.L.; Berthoud, H.R. Meal patterns, satiety, and food choice in a rat model of Roux-en-Y gastric bypass surgery. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2009, 297, R1273–R1282. [Google Scholar] [CrossRef] [PubMed]
- Schauer, P.R.; Mingrone, G.; Ikramuddin, S.; Wolfe, B. Clinical Outcomes of Metabolic Surgery: Efficacy of Glycemic Control, Weight Loss, and Remission of Diabetes. Diabetes Care 2016, 39, 902–911. [Google Scholar] [CrossRef] [Green Version]
- Schauer, P.R.; Bhatt, D.L.; Kirwan, J.P.; Wolski, K.; Aminian, A.; Brethauer, S.A.; Navaneethan, S.D.; Singh, R.P.; Pothier, C.E.; Nissen, S.E.; et al. Bariatric Surgery versus Intensive Medical Therapy for Diabetes—5-Year Outcomes. N. Engl. J. Med. 2017, 376, 641–651. [Google Scholar] [CrossRef] [Green Version]
- Kwon, I.G.; Kang, C.W.; Park, J.P.; Oh, J.H.; Wang, E.K.; Kim, T.Y.; Sung, J.S.; Park, N.; Lee, Y.J.; Sung, H.J.; et al. Serum glucose excretion after Roux-en-Y gastric bypass: A potential target for diabetes treatment. Gut 2021, 70, 1847–1856. [Google Scholar] [CrossRef]
- Saeidi, N.; Meoli, L.; Nestoridi, E.; Gupta, N.K.; Kvas, S.; Kucharczyk, J.; Bonab, A.A.; Fischman, A.J.; Yarmush, M.L.; Stylopoulos, N. Reprogramming of Intestinal Glucose Metabolism and Glycemic Control in Rats After Gastric Bypass. Science 2013, 341, 406–410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cavin, J.B.; Couvelard, A.; Lebtahi, R.; Ducroc, R.; Arapis, K.; Voitellier, E.; Cluzeaud, F.; Gillard, L.; Hourseau, M.; Mikail, N.; et al. Differences in Alimentary Glucose Absorption and Intestinal Disposal of Blood Glucose After Roux-en-Y Gastric Bypass vs. Sleeve Gastrectomy. Gastroenterology 2016, 150, 454–464. [Google Scholar] [CrossRef] [Green Version]
- Grayson, B.E.; Schneider, K.M.; Woods, S.C.; Seeley, R.J. Improved Rodent Maternal Metabolism but Reduced Intrauterine Growth after Vertical Sleeve Gastrectomy. Sci. Transl. Med. 2013, 5, 199ra112. [Google Scholar] [CrossRef] [Green Version]
- Fernandes-Lima, F.; Monte, T.; Nascimento, F.A.D.; Gregorio, B.M. Short Exposure to a High-Sucrose Diet and the First ‘Hit’ of Nonalcoholic Fatty Liver Disease in Mice. Cells Tissues Organs 2015, 201, 464–472. [Google Scholar] [CrossRef]
- Palleja, A.; Kashani, A.; Allin, K.H.; Nielsen, T.; Zhang, C.; Li, Y.; Brach, T.; Liang, S.; Feng, Q.; Jorgensen, N.B.; et al. Roux-en-Y gastric bypass surgery of morbidly obese patients induces swift and persistent changes of the individual gut microbiota. Genome Med. 2016, 8, 67. [Google Scholar] [CrossRef] [Green Version]
- Liou, A.P.; Paziuk, M.; Luevano, J.-M., Jr.; Machineni, S.; Turnbaugh, P.J.; Kaplan, L.M. Conserved Shifts in the Gut Microbiota Due to Gastric Bypass Reduce Host Weight and Adiposity. Sci. Transl. Med. 2013, 5, 178ra41. [Google Scholar] [CrossRef] [Green Version]
- Kirchner, H.; Nylen, C.; Laber, S.; Barres, R.; Yan, J.; Krook, A.; Zierath, J.R.; Naslund, E. Altered promoter methylation of PDK4, IL1 B, IL6, and TNF after Roux-en Y gastric bypass. Surg. Obes. Relat. Dis. 2014, 10, 671–678. [Google Scholar] [CrossRef]
- Gupta, A.; Osadchiy, V.; Mayer, E.A. Brain-gut-microbiome interactions in obesity and food addiction. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 655–672. [Google Scholar] [CrossRef] [PubMed]
- Luo, P.; Yu, H.; Zhao, X.; Bao, Y.; Hong, C.S.; Zhang, P.; Tu, Y.; Yin, P.; Gao, P.; Wei, L.; et al. Metabolomics Study of Roux-en-Y Gastric Bypass Surgery (RYGB) to Treat Type 2 Diabetes Patients Based on Ultraperformance Liquid Chromatography-Mass Spectrometry. J. Proteome Res. 2016, 15, 1288–1299. [Google Scholar] [CrossRef] [PubMed]
- Cerreto, M.; Santopaolo, F.; Gasbarrini, A.; Pompili, M.; Ponziani, F.R. Bariatric Surgery and Liver Disease: General Considerations and Role of the Gut-Liver Axis. Nutrients 2021, 13, 2649. [Google Scholar] [CrossRef] [PubMed]
- Seyfried, F.; Phetcharaburanin, J.; Glymenaki, M.; Nordbeck, A.; Hankir, M.; Nicholson, J.K.; Holmes, E.; Marchesi, J.R.; Li, J.V. Roux-en-Y gastric bypass surgery in Zucker rats induces bacterial and systemic metabolic changes independent of caloric restriction-induced weight loss. Gut Microbes 2021, 13, 1875108. [Google Scholar] [CrossRef]
- Ha, J.; Kwon, Y.; Park, S. Metabolomics in Bariatric Surgery: Towards Identification of Mechanisms and Biomarkers of Metabolic Outcomes. Obes. Surg. 2021, 31, 4564–4574. [Google Scholar] [CrossRef] [PubMed]
- Keller, A.; Della Torre, S.B. Sugar-Sweetened Beverages and Obesity among Children and Adolescents: A Review of Systematic Literature Reviews. Child. Obes. 2015, 11, 338–346. [Google Scholar] [CrossRef]
- Ritze, Y.; Bárdos, G.; D’Haese, J.G.; Ernst, B.; Thurnheer, M.; Schultes, B.; Bischoff, S.C. Effect of High Sugar Intake on Glucose Transporter and Weight Regulating Hormones in Mice and Humans. PLoS ONE 2014, 9, e101702. [Google Scholar] [CrossRef] [PubMed]
- Hao, Z.; Townsend, R.L.; Mumphrey, M.B.; Morrison, C.D.; Munzberg, H.; Berthoud, H.-R. RYGB Produces more Sustained Body Weight Loss and Improvement of Glycemic Control Compared with VSG in the Diet-Induced Obese Mouse Model. Obes. Surg. 2017, 27, 2424–2433. [Google Scholar] [CrossRef] [Green Version]
- Baheeg, M.; El-Din, M.T.; Labib, M.F.; Elgohary, S.A.; Hasan, A. Long-term durability of weight loss after bariatric surgery; a retrospective study. Int. J. Surg. Open 2021, 28, 37–40. [Google Scholar] [CrossRef]
- Guo, Y.; Huang, Z.-P.; Liu, C.-Q.; Qi, L.; Sheng, Y.; Zou, D.-J. Modulation of the gut microbiome: A systematic review of the effect of bariatric surgery. Eur. J. Endocrinol. 2018, 178, 43–56. [Google Scholar] [CrossRef]
- Cook, J.; Lehne, C.; Weiland, A.; Archid, R.; Ritze, Y.; Bauer, K.; Zipfel, S.; Penders, J.; Enck, P.; Mack, I. Gut Microbiota, Probiotics and Psychological States and Behaviors after Bariatric Surgery-A Systematic Review of Their Interrelation. Nutrients 2020, 12, 2396. [Google Scholar] [CrossRef] [PubMed]
- Strandwitz, P.; Kim, K.H.; Terekhova, D.; Liu, J.K.; Sharma, A.; Levering, J.; McDonald, D.; Dietrich, D.; Ramadhar, T.R.; Lekbua, A.; et al. GABA-modulating bacteria of the human gut microbiota. Nat. Microbiol. 2019, 4, 396–403. [Google Scholar] [CrossRef]
- Strandwitz, P. Neurotransmitter modulation by the gut microbiota. Brain Res. 2018, 1693, 128–133. [Google Scholar] [CrossRef]
- Chen, O.; Mah, E.; Dioum, E.; Marwaha, A.; Shanmugam, S.; Malleshi, N.; Sudha, V.; Gayathri, R.; Unnikrishnan, R.; Anjana, R.M.; et al. The Role of Oat Nutrients in the Immune System: A Narrative Review. Nutrients 2021, 13, 1048. [Google Scholar] [CrossRef]
- Krantis, A. GABA in the mammalian enteric nervous system. News Physiol. Sci. 2000, 15, 284–290. [Google Scholar] [CrossRef] [PubMed]
- Inotsuka, R.; Uchimura, K.; Yamatsu, A.; Kim, M.; Katakura, Y. Gamma-Aminobutyric acid (GABA) activates neuronal cells by inducing the secretion of exosomes from intestinal cells. Food Funct. 2020, 11, 9285–9290. [Google Scholar] [CrossRef]
- Auteri, M.; Zizzo, M.G.; Serio, R. The GABAergic System and the Gastrointestinal Physiopathology. Curr. Pharm. Des. 2015, 21, 4996–5016. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.K.; Holmes, E.; Kinross, J.; Burcelin, R.; Gibson, G.; Jia, W.; Pettersson, S. Host-Gut Microbiota Metabolic Interactions. Science 2012, 336, 1262–1267. [Google Scholar] [CrossRef] [Green Version]
- Lagkouvardos, I.; Lesker, T.R.; Hitch, T.C.A.; Galvez, E.J.C.; Smit, N.; Neuhaus, K.; Wang, J.; Baines, J.F.; Abt, B.; Stecher, B.; et al. Sequence and cultivation study of Muribaculaceae reveals novel species, host preference, and functional potential of this yet undescribed family. Microbiome 2019, 7, 28. [Google Scholar] [CrossRef] [Green Version]
- Rowland, I.; Gibson, G.; Heinken, A.; Scott, K.; Swann, J.; Thiele, I.; Tuohy, K. Gut microbiota functions: Metabolism of nutrients and other food components. Eur. J. Nutr. 2018, 57, 1–24. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Wang, Q.; Bai, J.; Zhao, N.; Wang, Y.; Zhou, R.; Kong, W.; Zeng, T.; Tao, K.; Wang, G.; et al. Upregulation of Intestinal NLRP6 Inflammasomes After Roux-en-Y Gastric Bypass Promotes Gut Immune Homeostasis. Obes. Surg. 2020, 30, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Jameson, E.; Crosatti, M.; Schaefer, H.; Rajakumar, K.; Bugg, T.D.H.; Chen, Y. Carnitine metabolism to trimethylamine by an unusual Rieske-type oxygenase from human microbiota. Proc. Natl. Acad. Sci. USA 2014, 111, 4268–4273. [Google Scholar] [CrossRef] [Green Version]
- Craciun, S.; Balskus, E.P. Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc. Natl. Acad. Sci. USA 2012, 109, 21307–21312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.-R.; Fang, S.-T.; Liu, H.-Y.; Zheng, H.-M.; He, Y.; Chen, Z.-W.; Chen, M.-X.; Zhang, G.-X.; Zhou, H.-W. Degradation of trimethylamine in vitro and in vivo by Enterococcus faecalis isolated from healthy human gut. Int. Biodeterior. Biodegrad. 2018, 135, 24–32. [Google Scholar] [CrossRef]
- Liu, Y.; Dai, M. Trimethylamine N-Oxide Generated by the Gut Microbiota Is Associated with Vascular Inflammation: New Insights into Atherosclerosis. Mediat. Inflamm. 2020, 2020, 15. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.; Li, X.; Yang, F.; Zhao, R.; Pan, X.; Liang, J.; Tian, L.; Li, X.; Liu, L.; Xing, Y.; et al. Gut Microbiota-Dependent Marker TMAO in Promoting Cardiovascular Disease: Inflammation Mechanism, Clinical Prognostic, and Potential as a Therapeutic Target. Front. Pharmacol. 2019, 10, 1360. [Google Scholar] [CrossRef]
- Rath, S.; Heidrich, B.; Pieper, D.H.; Vital, M. Uncovering the trimethylamine-producing bacteria of the human gut microbiota. Microbiome 2017, 5, 54. [Google Scholar] [CrossRef] [Green Version]
- Wu, Q.; Li, J.V.; Seyfried, F.; le Roux, C.W.; Ashrafian, H.; Athanasiou, T.; Fenske, W.; Darzi, A.; Nicholson, J.K.; Holmes, E.; et al. Metabolic phenotype-microRNA data fusion analysis of the systemic consequences of Roux-en-Y gastric bypass surgery. Int. J. Obes. 2015, 39, 1126–1134. [Google Scholar] [CrossRef] [Green Version]
- Siddik, M.A.B.; Shin, A.C. Recent Progress on Branched-Chain Amino Acids in Obesity, Diabetes, and Beyond. Endocrinol. Metab. 2019, 34, 234–246. [Google Scholar] [CrossRef]
- White, P.J.; Lapworth, A.L.; An, J.; Wang, L.; McGarrah, R.W.; Stevens, R.D.; Ilkayeva, O.; George, T.; Muehlbauer, M.J.; Bain, J.R.; et al. Branched-chain amino acid restriction in Zucker-fatty rats improves muscle insulin sensitivity by enhancing efficiency of fatty acid oxidation and acyl-glycine export. Mol. Metab. 2016, 5, 538–551. [Google Scholar] [CrossRef]
- Rosenzweig, A.; Blenis, J.; Gomes, A.P. Beyond the Warburg Effect: How Do Cancer Cells Regulate One-Carbon Metabolism? Front. Cell Dev. Biol. 2018, 6, 90. [Google Scholar] [CrossRef]
- Alves, A.; Bassot, A.; Bulteau, A.-L.; Pirola, L.; Morio, B. Glycine Metabolism and Its Alterations in Obesity and Metabolic Diseases. Nutrients 2019, 11, 1356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, P.S.; Chang, Y.-K.; Wu, T.-K. Serum Lipopolysaccharide-Binding Protein is Associated with Chronic Inflammation and Metabolic Syndrome in Hemodialysis Patients. Blood Purif. 2019, 47, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Parlesak, A.; Schaeckeler, S.; Moser, L.; Bode, C. Conjugated primary bile salts reduce permeability of endotoxin through intestinal epithelial cells and synergize with phosphatidylcholine in suppression of inflammatory cytokine production. Crit. Care Med. 2007, 35, 2367–2374. [Google Scholar] [CrossRef] [Green Version]
- Lin, T.-L.; Shu, C.-C.; Chen, Y.-M.; Lu, J.-J.; Wu, T.-S.; Lai, W.-F.; Tzeng, C.-M.; Lai, H.-C.; Lu, C.-C. Like Cures Like: Pharmacological Activity of Anti-Inflammatory Lipopolysaccharides From Gut Microbiome. Front. Pharmacol. 2020, 11, 554. [Google Scholar] [CrossRef] [PubMed]
- Whipp, A.M.; Vuoksimaa, E.; Korhonen, T.; Pool, R.; But, A.; Ligthart, L.; Hagenbeek, F.A.; Bartels, M.; Bogl, L.H.; Pulkkinen, L.; et al. Ketone body 3-hydroxybutyrate as a biomarker of aggression. Sci. Rep. 2021, 11, 5813. [Google Scholar] [CrossRef]
- Ghoshal, S.; Witta, J.; Zhong, J.; de Villiers, W.; Eckhardt, E. Chylomicrons promote intestinal absorption of lipopolysaccharides. J. Lipid Res. 2009, 50, 90–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halinski, L.P.; Pakiet, A.; Jablonska, P.; Kaska, L.; Proczko-Stepaniak, M.; Slominska, E.; Sledzinski, T.; Mika, A. One Anastomosis Gastric Bypass Reconstitutes the Appropriate Profile of Serum Amino Acids in Patients with Morbid Obesity. J. Clin. Med. 2020, 9, 100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kindt, A.; Liebisch, G.; Clavel, T.; Haller, D.; Hoermannsperger, G.; Yoon, H.; Kolmeder, D.; Sigruener, A.; Krautbauer, S.; Seeliger, C.; et al. The gut microbiota promotes hepatic fatty acid desaturation and elongation in mice. Nat. Commun. 2018, 9, 3760. [Google Scholar] [CrossRef] [Green Version]
- Weitkunat, K.; Schumann, S.; Nickel, D.; Hornemann, S.; Petzke, K.J.; Schulze, M.B.; Pfeiffer, A.F.H.; Klaus, S. Odd-chain fatty acids as a biomarker for dietary fiber intake: A novel pathway for endogenous production from propionate. Am. J. Clin. Nutr. 2017, 105, 1544–1551. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Sun, G.Y.; Eckert, G.P.; Lee, J.C.M. Cellular Membrane Fluidity in Amyloid Precursor Protein Processing. Mol. Neurobiol. 2014, 50, 119–129. [Google Scholar] [CrossRef]
- Vlaeminck, B.; Fievez, V.; Cabrita, A.R.J.; Fonseca, A.J.M.; Dewhurst, R.J. Factors affecting odd- and branched-chain fatty acids in milk: A review. Anim. Feed. Sci. Technol. 2006, 131, 389–417. [Google Scholar] [CrossRef]
- Fonteh, A.N.; Cipolla, M.; Chiang, J.; Arakaki, X.; Harrington, M.G. Human Cerebrospinal Fluid Fatty Acid Levels Differ between Supernatant Fluid and Brain-Derived Nanoparticle Fractions, and Are Altered in Alzheimer’s Disease. PLoS ONE 2014, 9, e100519. [Google Scholar] [CrossRef] [Green Version]
- Lam, T.K.T.; Pocai, A.; Gutierrez-Juarez, R.; Obici, S.; Bryan, J.; Aguilar-Bryan, L.; Schwartz, G.J.; Rossetti, L. Hypothalamic sensing of circulating fatty acids is required for glucose homeostasis. Nat. Med. 2005, 11, 320–327. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Hernandez, T.; Barroso-Chinea, P.; de la Cruz, M.A.P.; Valera, P.; Dopico, J.G.; Rodriguez, M. Response of gabaergic cells in the deep mesencephalic nucleus to dopaminergic cell degeneration: An electrophysiological and in situ hybridization study. Neuroscience 2002, 113, 311–321. [Google Scholar] [CrossRef]
- Hankir, M.K.; Seyfried, F.; Miras, A.D.; Cowley, M.A. Brain Feeding Circuits after Roux-en-Y Gastric Bypass. Trends Endocrinol. Metab. 2018, 29, 218–237. [Google Scholar] [CrossRef] [Green Version]
- Singh, Y.; Trautwein, C.; Dhariwal, A.; Salker, M.S.; Alauddin, M.; Zizmare, L.; Pelzl, L.; Feger, M.; Admard, J.; Casadei, N.; et al. DJ-1 (Park7) affects the gut microbiome, metabolites and the development of innate lymphoid cells (ILCs). Sci. Rep. 2020, 10, 16131. [Google Scholar] [CrossRef]
- Bueter, M.; Abegg, K.; Seyfried, F.; Lutz, T.A.; le Roux, C.W. Gastric Bypass Operation in Rats. J. Vis. Exp. 2012, 64, e3940. [Google Scholar] [CrossRef] [Green Version]
- Wehrl, H.F.; Hossain, M.; Lankes, K.; Liu, C.-C.; Bezrukov, I.; Martirosian, P.; Schickt, F.; Reischl, G.; Pichler, B.J. Simultaneous PET-MRI reveals brain function in activated and resting state on metabolic, hemodynamic and multiple temporal scales. Nat. Med. 2013, 19, 1184–1189. [Google Scholar] [CrossRef]
- Schiffer, W.K.; Mirrione, M.M.; Biegon, A.; Alexoff, D.L.; Patel, V.; Dewey, S.L. Serial microPET measures of the metabolic reaction to a microdialysis probe implant. J. Neurosci. Methods 2006, 155, 272–284. [Google Scholar] [CrossRef]
- Ionescu, T.M.; Amend, M.; Hafiz, R.; Biswal, B.B.; Wehrl, H.F.; Herfert, K.; Pichler, B.J. Elucidating the complementarity of resting-state networks derived from dynamic F-18 FDG and hemodynamic fluctuations using simultaneous small-animal PET/MRI. Neuroimage 2021, 236, 118045. [Google Scholar] [CrossRef]
- Paxinos, G.; Watson, C. The Rat Brain in Stereotaxic Coordinates, 4th ed.; Academic Press: Cambridge, MA, USA, 1998. [Google Scholar]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walters, W.; Hyde, E.R.; Berg-Lyons, D.; Ackermann, G.; Humphrey, G.; Parada, A.; Gilbert, J.A.; Jansson, J.K.; Caporaso, J.G.; Fuhrman, J.A.; et al. Improved Bacterial 16S rRNA Gene (V4 and V4-5) and Fungal Internal Transcribed Spacer Marker Gene Primers for Microbial Community Surveys. Msystems 2016, 1, e00009-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a Dual-Index Sequencing Strategy and Curation Pipeline for Analyzing Amplicon Sequence Data on the MiSeq Illumina Sequencing Platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Callahan, B.J.; McMurdie, P.J.; Holmes, S.P. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017, 11, 2639–2643. [Google Scholar] [CrossRef] [Green Version]
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [Green Version]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [Green Version]
- Ecker, J.; Scherer, M.; Schmitz, G.; Liebisch, G. A rapid GC-MS method for quantification of positional and geometric isomers of fatty acid methyl esters. Journal of chromatography. B. Anal. Technol. Biomed. Life Sci. 2012, 897, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Gille, A.; Hollenbach, R.; Trautmann, A.; Gomez, M.R.; Kruger, R.; Bischoff, S.C.; Posten, C.; Briviba, K. Lipophilic compounds, but not fucoxanthin, mediate the genotoxic effect of photoautotrophic grown Phaeodactylum tricomutum in Caco-2 and HT-29 cells. J. Funct. Foods 2020, 64, 103671. [Google Scholar] [CrossRef]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef] [PubMed]









| Primer Name | Target | Sequence |
|---|---|---|
| b-Act-F | beta-Actin | 5′-ccc gcg agt aca acc ttc t-3′ |
| b-Act-R | beta-Actin | 5′-cgt cat cca tgg cga act-3′ |
| HPRT-F | HPRT | 5′-tag cac ctc ctc cgc cag-3′ |
| HPRT-R | HPRT | 5′-cac taa tca cga cgc tgg ga-3′ |
| FAS-F | FAS | 5′-ggc cac ctc agt cct gtt at-3′ |
| FAS-R | FAS | 5′-agg gtc cag cta gag ggt aca-3′ |
| rFABP1-F | FABP1 | 5′-ctt ctc cgg caa gta cca ag-3′ |
| rFABP1-R | FABP1 | 5′-ttc cct ttc tgg atg agg tc-3′ |
| rLPL-1294-F | LPL | 5′-aca gtg gct gag aac a-3′ |
| rLPL-1445-R | LPL | 5′-tct gac cag cta gga g-3′ |
| PPARa-180-F | PPAR alpha | 5′-CACAGCGTGGTGCATTTGG-3′ |
| PPARa-412-R | PPAR alpha | 5′-GAGAGAGGACAGATGGGGCT-3′ |
| SREBP1-F | SREBP1 | 5′-gta cag cgt ggc tgg gaa c-3′ |
| SREBP1-R | SREBP1 | 5′-ggc tga gcg ata cag ttc aa-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zizmare, L.; Boyle, C.N.; Buss, S.; Louis, S.; Kuebler, L.; Mulay, K.; Krüger, R.; Steinhauer, L.; Mack, I.; Rodriguez Gomez, M.; et al. Roux-En-Y Gastric Bypass (RYGB) Surgery during High Liquid Sucrose Diet Leads to Gut Microbiota-Related Systematic Alterations. Int. J. Mol. Sci. 2022, 23, 1126. https://doi.org/10.3390/ijms23031126
Zizmare L, Boyle CN, Buss S, Louis S, Kuebler L, Mulay K, Krüger R, Steinhauer L, Mack I, Rodriguez Gomez M, et al. Roux-En-Y Gastric Bypass (RYGB) Surgery during High Liquid Sucrose Diet Leads to Gut Microbiota-Related Systematic Alterations. International Journal of Molecular Sciences. 2022; 23(3):1126. https://doi.org/10.3390/ijms23031126
Chicago/Turabian StyleZizmare, Laimdota, Christina N. Boyle, Sabrina Buss, Sandrine Louis, Laura Kuebler, Ketki Mulay, Ralf Krüger, Lara Steinhauer, Isabelle Mack, Manuel Rodriguez Gomez, and et al. 2022. "Roux-En-Y Gastric Bypass (RYGB) Surgery during High Liquid Sucrose Diet Leads to Gut Microbiota-Related Systematic Alterations" International Journal of Molecular Sciences 23, no. 3: 1126. https://doi.org/10.3390/ijms23031126