Mitochondrial Sirtuins in Chronic Degenerative Diseases: New Metabolic Targets in Colorectal Cancer
Abstract
:1. Introduction
2. Sirtuins as Modulators of Chronic Degenerative Diseases
3. Sirtuins and Cancer
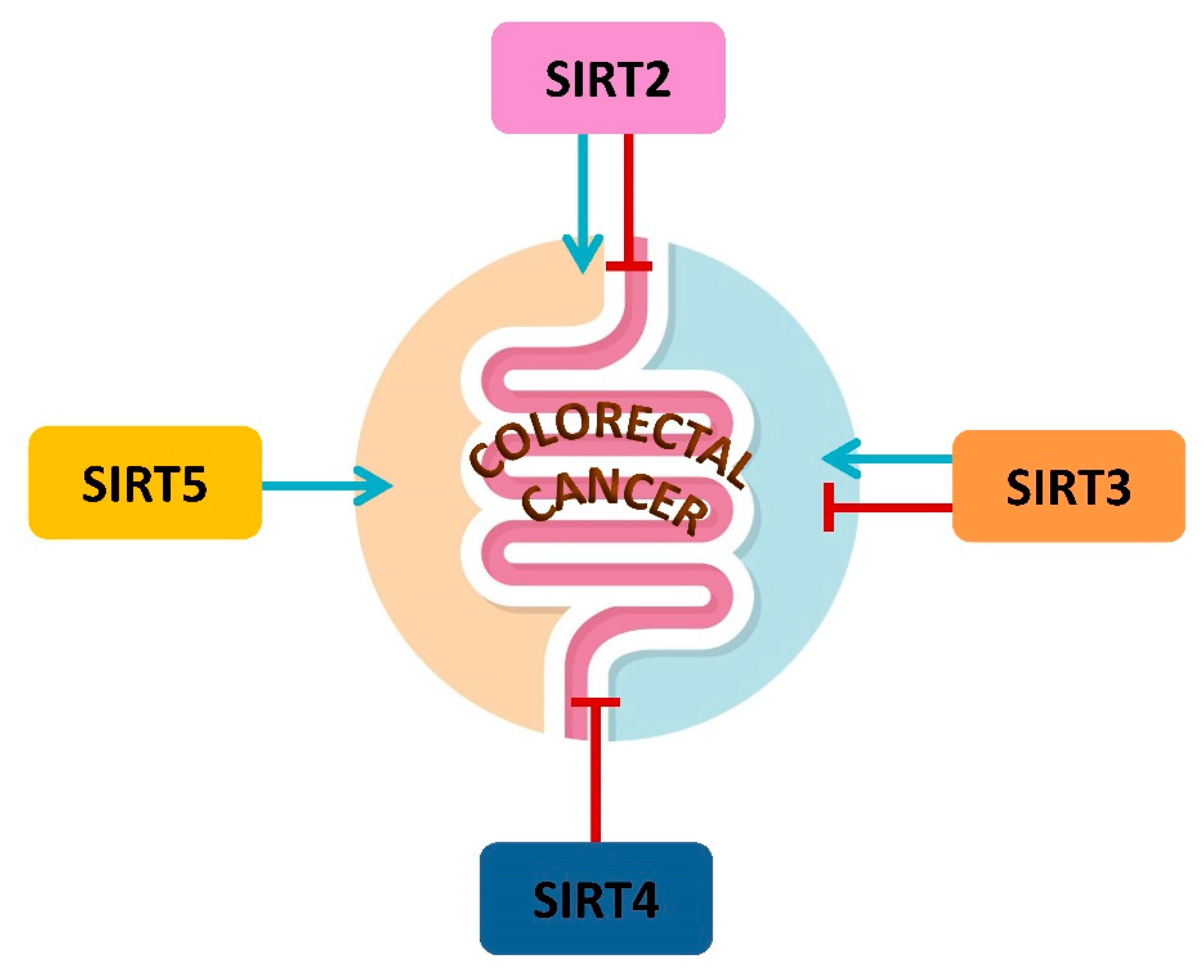
4. Mitochondrial Sirtuins and CRC
4.1. SIRT2 in CRC Metabolism
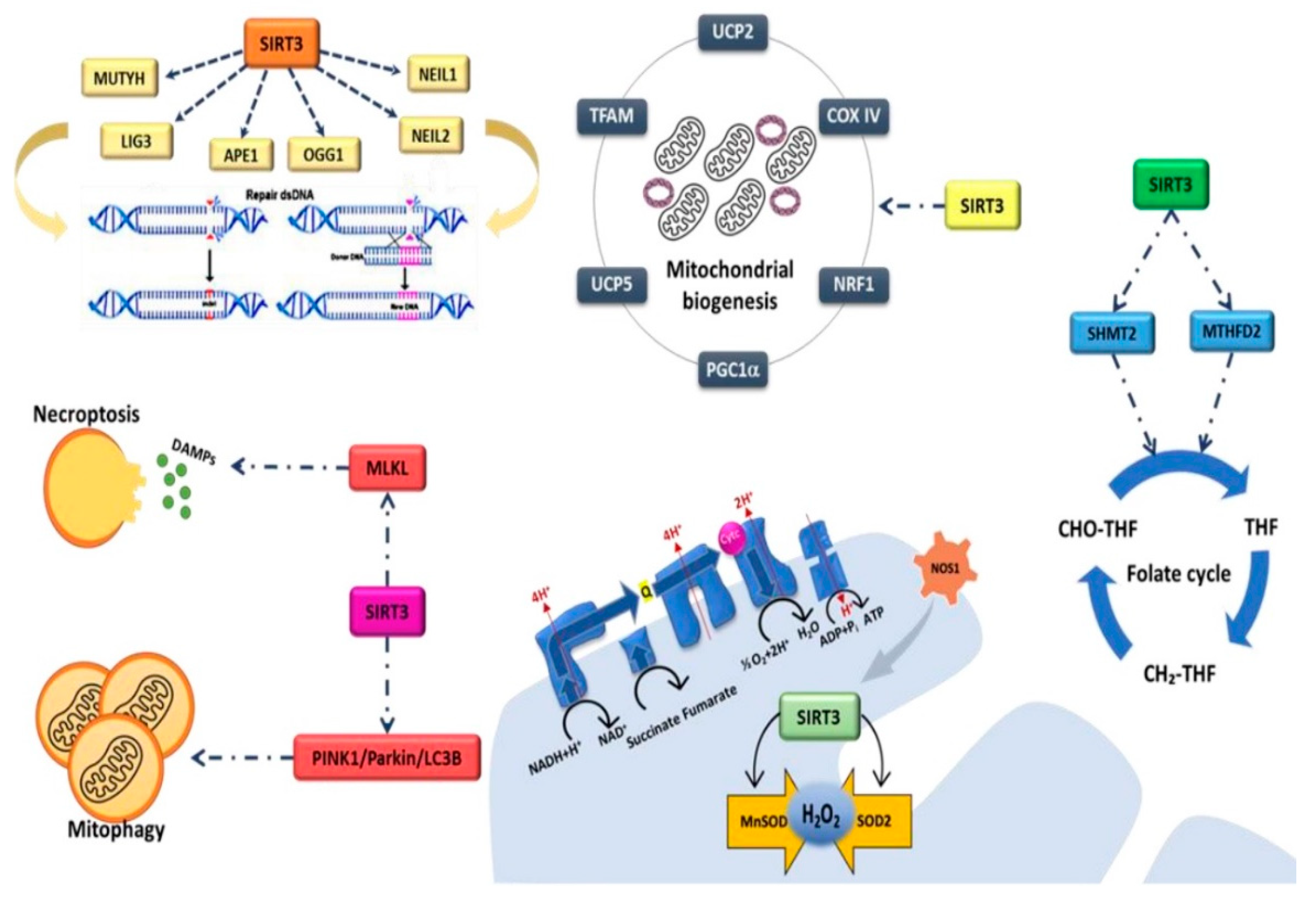
4.2. SIRT3 and CRC Metabolism
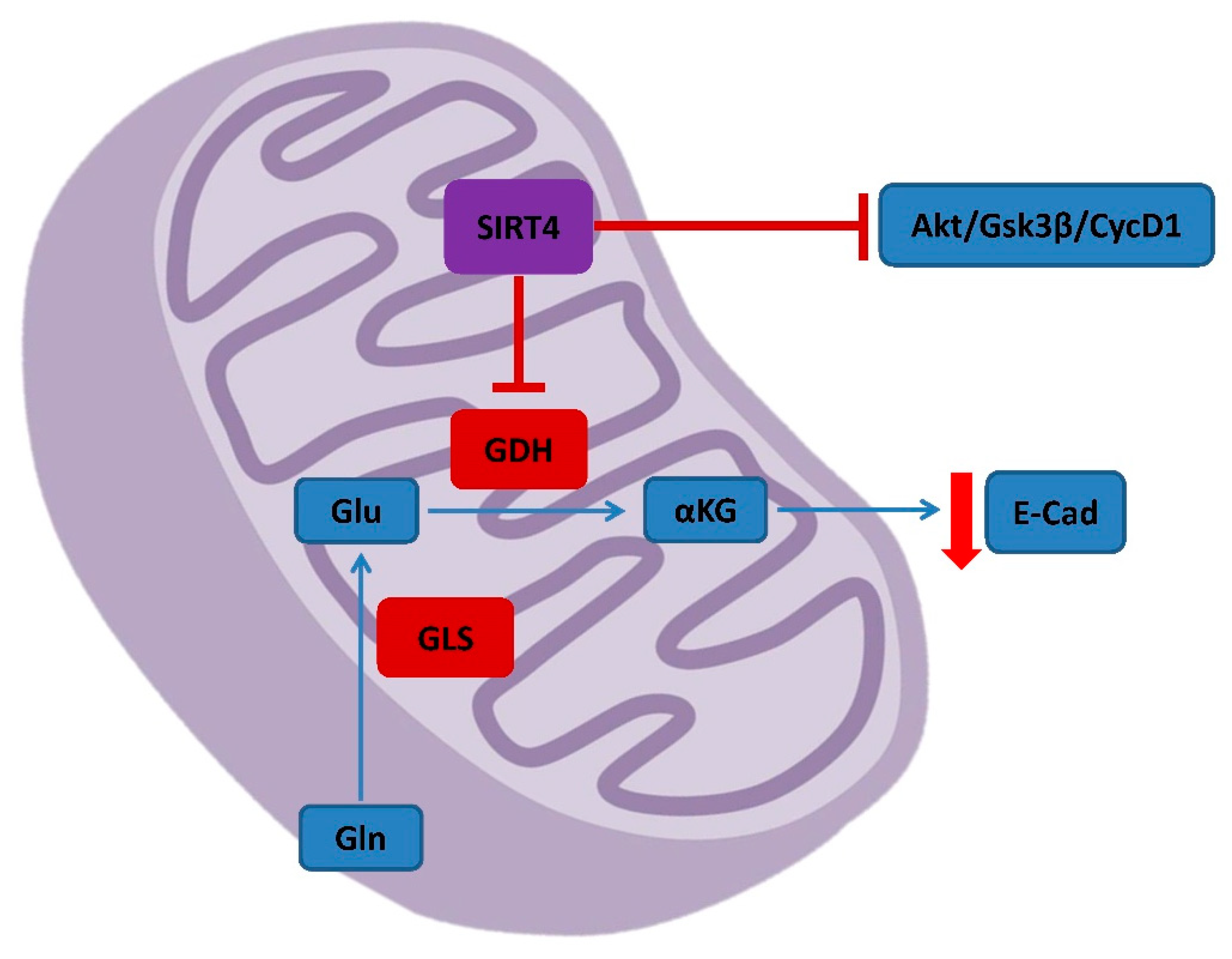
4.3. SIRT4 in CRC Metabolism and Prognosis
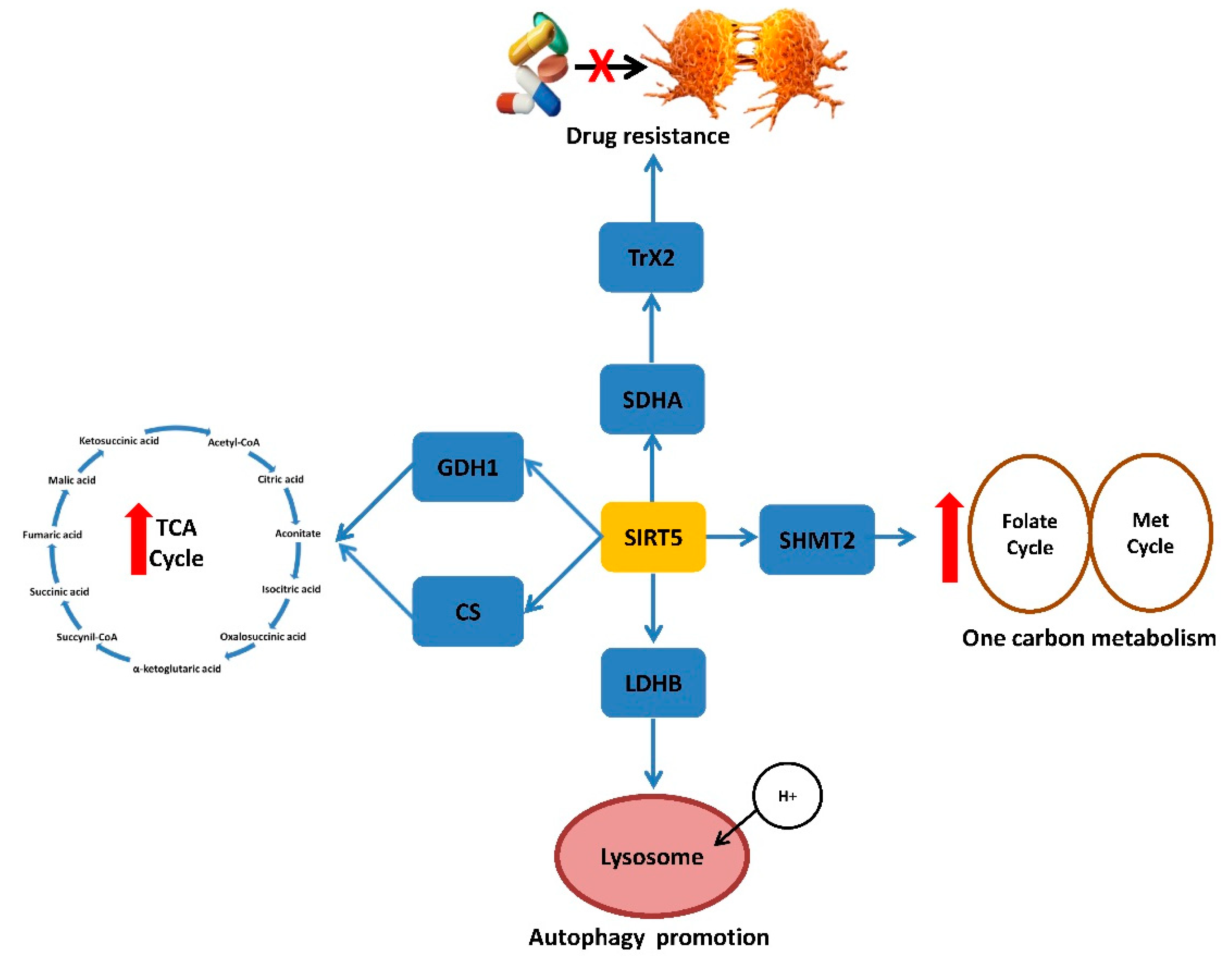
4.4. SIRT5 and CRC Metabolism, Prognosis and Therapy
5. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Teixeira, C.S.S.; Cerqueira, N.M.F.S.A.; Gomes, P.; Sousa, S.F. A molecular perspective on sirtuin activity. Int. J. Mol. Sci. 2020, 21, 8609. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yang, J.; Hong, T.; Chen, X.; Cui, L. SIRT2: Controversy and multiple roles in disease and physiology. Ageing Res. Rev. 2019, 55, 100961. [Google Scholar] [CrossRef]
- Shahgaldi, S.; Kahmini, F.R. A comprehensive review of Sirtuins: With a major focus on redox homeostasis and metabolism. Life Sci. 2021, 282, 119803. [Google Scholar] [CrossRef] [PubMed]
- Zullo, A.; Simone, E.; Grimaldi, M.; Musto, V.; Mancini, F.P. Sirtuins as Mediator of the Anti-Ageing Effects of Calorie Restriction in Skeletal and Cardiac Muscle. Int. J. Mol. Sci. 2018, 19, 928. [Google Scholar] [CrossRef] [Green Version]
- Mei, Z.; Zhang, X.; Yi, J.; Huang, J.; He, J.; Tao, Y. Sirtuins in metabolism, DNA repair and cancer. J. Exp. Clin. Cancer Res. 2016, 35, 182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, L.; Cao, J.; Hu, K.; He, X.; Yun, D.; Tong, T.; Han, L. Sirtuins and their Biological Relevance in Aging and Age-Related Diseases. Aging Dis. 2020, 11, 927–945. [Google Scholar] [CrossRef] [PubMed]
- Kratz, E.M.; Sołkiewicz, K.; Kubis-Kubiak, A.; Piwowar, A. Sirtuins as Important Factors in Pathological States and the Role of Their Molecular Activity Modulators. Int. J. Mol. Sci. 2021, 22, 630. [Google Scholar] [CrossRef]
- Lee, Y.T.; Tan, Y.J.; Oon, C.E. Molecular targeted therapy: Treating cancer with specificity. Eur. J. Pharmacol. 2018, 834, 188–196. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Siegel, R.L.; Jakubowski, C.D.; Fedewa, S.A.; Davis, A.; Azad, N.S. Colorectal Cancer in the Young: Epidemiology, Prevention, Management. Am. Soc. Clin. Oncol. Educ. Book 2020, 40, e75–e88. [Google Scholar] [CrossRef]
- Sánchez-Aragó, M.; Chamorro, M.; Cuezva, J.M. Selection of cancer cells with repressed mitochondria triggers colon cancer progression. Carcinogenesis 2010, 31, 567–576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.W.; Chou, C.T.; Chang, C.C.; Li, Y.J.; Chen, S.T.; Lin, I.C.; Kok, S.H.; Cheng, S.J.; Lee, J.J.; Wu, T.S.; et al. HMGCS2 enhances invasion and metastasis via direct interaction with PPARα to activate Src signaling in colorectal cancer and oral cancer. Oncotarget 2017, 8, 22460–22476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Motawi, T.K.; Shaker, O.G.; Ismail, M.F.; Sayed, N.H. Peroxisome Proliferator-Activated Receptor Gamma in Obesity and Cchenolorectal Cancer: The Role of Epigenetics. Sci. Rep. 2017, 7, 10714. [Google Scholar] [CrossRef] [PubMed]
- Boussios, S.; Ozturk, M.A.; Moschetta, M.; Karathanasi, A.; Zakynthinakis-Kyriakou, N.; Katsanos, K.H.; Christodoulou, D.K.; Pavlidis, N. The Developing Story of Predictive Biomarkers in Colorectal Cancer. J. Pers. Med. 2019, 9, 12. [Google Scholar] [CrossRef] [Green Version]
- Qiu, Z.; Deng, W.; Hong, Y.; Zhao, L.; Li, M.; Guan, Y.; Su, Y.; Chen, C.; Shi, Q.; Yu, J.; et al. Biological Behavior and Lipid Metabolism of Colon Cancer Cells are Regulated by a Combination of Sterol Regulatory Element-Binding Protein 1 and ATP Citrate Lyase. Onco. Targets Ther. 2021, 14, 1531–1542. [Google Scholar] [CrossRef]
- La Vecchia, S.; Sebastián, C. Metabolic pathways regulating colorectal cancer initiation and progression. Semin. Cell Dev. Biol. 2020, 98, 63–70. [Google Scholar] [CrossRef]
- Zuo, L.; Prather, E.R.; Stetskiv, M.; Garrison, D.E.; Meade, J.R.; Peace, T.I.; Zhou, T. Inflammaging and Oxidative Stress in Human Diseases: From Molecular Mechanisms to Novel Treatments. Int. J. Mol. Sci. 2019, 20, 4472. [Google Scholar] [CrossRef] [Green Version]
- Kondo, T.; Nakano, Y.; Adachi, S.; Murohara, T. Effects of Tobacco Smoking on Cardiovascular Disease. Circ. J. 2019, 83, 1980–1985. [Google Scholar] [CrossRef] [Green Version]
- Di Renzo, L.; Gualtieri, P.; Romano, L.; Marrone, G.; Noce, A.; Pujia, A.; Perrone, M.A.; Aiello, V.; Colica, C.; De Lorenzo, A. Role of Personalized Nutrition in Chronic-Degenerative Diseases. Nutrients 2019, 11, 1707. [Google Scholar] [CrossRef] [Green Version]
- Yu, H.; Lin, L.; Zhang, Z.; Zhang, H.; Hu, H. Targeting NF-κB pathway for the therapy of diseases: Mechanism and clinical study. Signal. Transduct. Target Ther. 2020, 5, 209. [Google Scholar] [CrossRef]
- Margină, D.; Ungurianu, A.; Purdel, C.; Tsoukalas, D.; Sarandi, E.; Thanasoula, M.; Tekos, F.; Mesnage, R.; Kouretas, D.; Tsatsakis, A. Chronic Inflammation in the Context of Everyday Life: Dietary Changes as Mitigating Factors. Int. J. Environ. Res. Public Health 2020, 17, 4135. [Google Scholar] [CrossRef] [PubMed]
- Mendes, K.L.; Lelis, D.F.; Santos, S.H.S. Nuclear sirtuins and inflammatory signaling pathways. Cytokine Growth Factor Rev. 2017, 38, 98–105. [Google Scholar] [CrossRef] [PubMed]
- van de Ven, R.A.H.; Santos, D.; Haigis, M.C. Mitochondrial Sirtuins and Molecular Mechanisms of Aging. Trends Mol. Med. 2017, 23, 320–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Zhou, M.; Ge, Y.; Wang, X. SIRT1 and aging related signaling pathways. Mech. Ageing Dev. 2020, 187, 111215. [Google Scholar] [CrossRef]
- Lee, S.H.; Lee, J.H.; Lee, H.Y.; Min, K.J. Sirtuin signaling in cellular senescence and aging. BMB Rep. 2019, 52, 24–34. [Google Scholar] [CrossRef] [Green Version]
- Xu, D.; Jiang, X.; He, H.; Liu, D.; Yang, L.; Chen, H.; Wu, L.; Geng, G.; Li, Q. SIRT2 functions in aging, autophagy, and apoptosis in post-maturation bovine oocytes. Life Sci. 2019, 232, 116639. [Google Scholar] [CrossRef]
- He, M.; Chiang, H.H.; Luo, H.; Zheng, Z.; Qiao, Q.; Wang, L.; Tan, M.; Ohkubo, R.; Mu, W.C.; Zhao, S.; et al. An Acetylation Switch of the NLRP3 Inflammasome Regulates Aging-Associated Chronic Inflammation and Insulin Resistance. Cell Metab. 2020, 31, 580–591. [Google Scholar] [CrossRef]
- Li, X.; Liu, L.; Li, T.; Liu, M.; Wang, Y.; Ma, H.; Mu, N.; Wang, H. SIRT6 in Senescence and Aging-Related Cardiovascular Diseases. Front. Cell Dev. Biol. 2021, 9, 641315. [Google Scholar] [CrossRef]
- Bi, S.; Liu, Z.; Wu, Z.; Wang, Z.; Liu, X.; Wang, S.; Ren, J.; Yao, Y.; Zhang, W.; Song, M.; et al. SIRT7 antagonizes human stem cell aging as a heterochromatin stabilizer. Protein Cell 2020, 11, 483–504. [Google Scholar] [CrossRef]
- Luc, K.; Schramm-Luc, A.; Guzik, T.J.; Mikolajczyk, T.P. Oxidative stress and inflammatory markers in prediabetes and diabetes. J. Physiol. Pharmacol. 2019, 70, 809–824. [Google Scholar] [CrossRef]
- D’onofrio, N.; Servillo, L.; Balestrieri, M.L. SIRT1 and SIRT6 Signaling Pathways in Cardiovascular Disease Protection. Antioxid. Redox Signal. 2018, 28, 711–732. [Google Scholar] [CrossRef] [PubMed]
- D’Onofrio, N.; Pieretti, G.; Ciccarelli, F.; Gambardella, A.; Passariello, N.; Rizzo, M.R.; Barbieri, M.; Marfella, R.; Nicoletti, G.; Balestrieri, M.L.; et al. Abdominal Fat SIRT6 Expression and Its Relationship with Inflammatory and Metabolic Pathways in Pre-Diabetic Overweight Patients. Int. J. Mol. Sci. 2019, 20, 1153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sardu, C.; Pieretti, G.; D’Onofrio, N.; Ciccarelli, F.; Paolisso, P.; Passavanti, M.B.; Marfella, R.; Cioffi, M.; Mone, P.; Dalise, A.M.; et al. Inflammatory Cytokines and SIRT1 Levels in Subcutaneous Abdominal Fat: Relationship With Cardiac Performance in Overweight Pre-diabetics Patients. Front. Physiol. 2018, 9, 1030. [Google Scholar] [CrossRef] [PubMed]
- Scisciola, L.; Rizzo, M.R.; Marfella, R.; Cataldo, V.; Fontanella, R.A.; Boccalone, E.; Paolisso, G.; Barbieri, M. New insight in molecular mechanisms regulating SIRT6 expression in diabetes: Hyperglycaemia effects on SIRT6 DNA methylation. J. Cell Physiol. 2021, 236, 4604–4613. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Yang, Q.; Yang, Y.; Gao, Z.; Ma, Y.; Zhang, L.; Liang, W.; Ding, G. Sirt6 Suppresses High Glucose-Induced Mitochondrial Dysfunction and Apoptosis in Podocytes through AMPK Activation. Int. J. Biol. Sci. 2019, 15, 701–713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Dong, Y.; Xiong, Z.; Zhu, Z.; Gao, F.; Wang, T.; Man, W.; Sun, D.; Lin, J.; Li, T.; et al. Sirt6-Mediated Endothelial-to-Mesenchymal Transition Contributes Toward Diabetic Cardiomyopathy via the Notch1 Signaling Pathway. Diabetes Metab. Syndr. Obes. 2020, 13, 4801–4808. [Google Scholar] [CrossRef] [PubMed]
- Alqudah, A.; Eastwood, K.A.; Jerotic, D.; Todd, N.; Hoch, D.; McNally, R.; Obradovic, D.; Dugalic, S.; Hunter, A.J.; Holmes, V.A.; et al. FKBPL and SIRT-1 Are Downregulated by Diabetes in Pregnancy Impacting on Angiogenesis and Endothelial Function. Front. Endocrinol. 2021, 12, 650328. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Sun, W.; Cheng, Y.; Xu, Z.; Cai, L. Role of sirtuin-1 in diabetic nephropathy. J. Mol. Med. 2019, 97, 291–309. [Google Scholar] [CrossRef] [Green Version]
- Ren, H.; Shao, Y.; Wu, C.; Ma, X.; Lv, C.; Wang, Q. Metformin alleviates oxidative stress and enhances autophagy in diabetic kidney disease via AMPK/SIRT1-FoxO1 pathway. Mol. Cell Endocrinol. 2020, 500, 110628. [Google Scholar] [CrossRef]
- Wang, C.H.; Wei, Y.H. Roles of Mitochondrial Sirtuins in Mitochondrial Function, Redox Homeostasis, Insulin Resistance and Type 2 Diabetes. Int. J. Mol. Sci. 2020, 21, 5266. [Google Scholar] [CrossRef]
- Boniakowski, A.M.; denDekker, A.D.; Davis, F.M.; Joshi, A.; Kimball, A.S.; Schaller, M.; Allen, R.; Bermick, J.; Nycz, D.; Skinner, M.E.; et al. SIRT3 Regulates Macrophage-Mediated Inflammation in Diabetic Wound Repair. J. Investig. Dermatol. 2019, 139, 2528–2537. [Google Scholar] [CrossRef] [PubMed]
- Poznyak, A.; Grechko, A.V.; Poggio, P.; Myasoedova, V.A.; Alfieri, V.; Orekhov, A.N. The Diabetes Mellitus-Atherosclerosis Connection: The Role of Lipid and Glucose Metabolism and Chronic Inflammation. Int. J. Mol. Sci. 2020, 21, 1835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balestrieri, M.L.; Rizzo, M.R.; Barbieri, M.; Paolisso, P.; D’Onofrio, N.; Giovane, A.; Siniscalchi, M.; Minicucci, F.; Sardu, C.; D’Andrea, D.; et al. Sirtuin 6 expression and inflammatory activity in diabetic atherosclerotic plaques: Effects of incretin treatment. Diabetes 2015, 64, 1395–1406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, T.; Sun, C.; Hu, L.; Gao, E.; Li, C.; Wang, H.; Sun, D. Sirt6 stabilizes atherosclerosis plaques by promoting macrophage autophagy and reducing contact with endothelial cells. Biochem. Cell Biol. 2020, 98, 120–129. [Google Scholar] [CrossRef]
- Sasaki, Y.; Ikeda, Y.; Miyauchi, T.; Uchikado, Y.; Akasaki, Y.; Ohishi, M. Estrogen-SIRT1 Axis Plays a Pivotal Role in Protecting Arteries Against Menopause-Induced Senescence and Atherosclerosis. J. Atheroscler. Thromb. 2020, 27, 47–59. [Google Scholar] [CrossRef] [Green Version]
- Yuan, P.; Hu, Q.; He, X.; Long, Y.; Song, X.; Wu, F.; He, Y.; Zhou, X. Laminar flow inhibits the Hippo/YAP pathway via autophagy and SIRT1-mediated deacetylation against atherosclerosis. Cell Death Dis. 2020, 11, 141. [Google Scholar] [CrossRef] [Green Version]
- Agita, A.; Alsagaff, M.T. Inflammation, Immunity, and Hypertension. Acta Med. Indones. 2017, 49, 158–165. [Google Scholar]
- Dikalova, A.E.; Pandey, A.; Xiao, L.; Arslanbaeva, L.; Sidorova, T.; Lopez, M.G.; Billings, F.T., IV; Verdin, E.; Auwerx, J.; Harrison, D.G.; et al. Mitochondrial Deacetylase Sirt3 Reduces Vascular Dysfunction and Hypertension While Sirt3 Depletion in Essential Hypertension Is Linked to Vascular Inflammation and Oxidative Stress. Circ. Res. 2020, 126, 439–452. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.; Wang, Z.; Wu, J.; Liu, M.; Li, M.; Sun, Y.; Huang, W.; Li, Y.; Zhang, Y.; Tang, W.; et al. Endothelial SIRT6 Is Vital to Prevent Hypertension and Associated Cardiorenal Injury Through Targeting Nkx3.2-GATA5 Signaling. Circ. Res. 2019, 124, 1448–1461. [Google Scholar] [CrossRef]
- Yeong, K.Y.; Berdigaliyev, N.; Chang, Y. Sirtuins and Their Implications in Neurodegenerative Diseases from a Drug Discovery Perspective. ACS Chem. Neurosci. 2020, 11, 4073–4091. [Google Scholar] [CrossRef]
- Chandramowlishwaran, P.; Vijay, A.; Abraham, D.; Li, G.; Mwangi, S.M.; Srinivasan, S. Role of Sirtuins in Modulating Neurodegeneration of the Enteric Nervous System and Central Nervous System. Front. Neurosci. 2020, 14, 614331. [Google Scholar] [CrossRef]
- Anamika; Khanna, A.; Acharjee, P.; Acharjee, A.; Trigun, S.K. Mitochondrial SIRT3 and neurodegenerative brain disorders. J. Chem. Neuroanat. 2019, 95, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Pratiwi, R.; Nantasenamat, C.; Ruankham, W.; Suwanjang, W.; Prachayasittikul, V.; Prachayasittikul, S.; Phopin, K. Mechanisms and Neuroprotective Activities of Stigmasterol Against Oxidative Stress-Induced Neuronal Cell Death via Sirtuin Family. Front. Nutr. 2021, 8, 648995. [Google Scholar] [CrossRef] [PubMed]
- Almeida, M.; Porter, R.M. Sirtuins and FoxOs in osteoporosis and osteoarthritis. Bone 2019, 121, 284–292. [Google Scholar] [CrossRef]
- Zhang, G.Z.; Deng, Y.J.; Xie, Q.Q.; Ren, E.H.; Ma, Z.J.; He, X.G.; Gao, Y.C.; Kang, X.W. Sirtuins and intervertebral disc degeneration: Roles in inflammation, oxidative stress, and mitochondrial function. Clin. Chim. Acta 2020, 508, 33–42. [Google Scholar] [CrossRef]
- Singh, N.; Baby, D.; Rajguru, J.P.; Patil, P.B.; Thakkannavar, S.S.; Pujari, V.B. Inflammation and cancer. Ann. Afr. Med. 2019, 18, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Dong, Z.; Ke, X.; Hou, J.; Zhao, E.; Zhang, K.; Wang, F.; Yang, L.; Xiang, Z.; Cui, H. The roles of sirtuins family in cell metabolism during tumor development. Semin. Cancer Biol. 2019, 57, 59–71. [Google Scholar] [CrossRef]
- Xia, C.; Dong, X.; Li, H.; Cao, M.; Sun, D.; He, S.; Yang, F.; Yan, X.; Zhang, S.; Li, N.; et al. Cancer statistics in China and United States 2022: Profiles, trends and determinants. Chin. Med. J. 2022, 135, 584–590. [Google Scholar] [CrossRef]
- Greene, K.S.; Lukey, M.J.; Wang, X.; Blank, B.; Druso, J.E.; Lin, M.J.; Stalnecker, C.A.; Zhang, C.; Negrón Abril, Y.; Erickson, J.W.; et al. SIRT5 stabilizes mitochondrial glutaminase and supports breast cancer tumorigenesis. Proc. Natl. Acad. Sci. USA 2019, 116, 26625–26632. [Google Scholar] [CrossRef]
- Tang, X.; Shi, L.; Xie, N.; Liu, Z.; Qian, M.; Meng, F.; Xu, Q.; Zhou, M.; Cao, X.; Zhu, W.G.; et al. SIRT7 antagonizes TGF-β signaling and inhibits breast cancer metastasis. Nat. Commun. 2017, 8, 318. [Google Scholar] [CrossRef] [Green Version]
- Aljada, A.; Saleh, A.M.; Al Suwaidan, S. Modulation of insulin/IGFs pathways by sirtuin-7 inhibition in drug-induced chemoreistance. Diagn. Pathol. 2014, 9, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, L.; Liu, X.; Ren, Y.; Li, J.; Li, P.; Jiao, Q.; Meng, P.; Wang, F.; Wang, Y.; Wang, Y.S.; et al. Loss of SIRT4 promotes the self-renewal of Breast Cancer Stem Cells. Theranostics 2020, 10, 9458–9476. [Google Scholar] [CrossRef] [PubMed]
- Bijoux, W.; Cordina-Duverger, E.; Balbolia, S.; Lamy, P.J.; Rebillard, X.; Tretarre, B.; Cenee, S.; Menegaux, F. Occupation and prostate Cancer risk: Results from the epidemiological study of prostate cancer (EPICAP). J. Occup. Med. Toxicol. 2022, 17, 5. [Google Scholar] [CrossRef]
- Li, T.; Li, Y.; Liu, T.; Hu, B.; Li, J.; Liu, C.; Liu, T.; Li, F. Mitochondrial PAK6 inhibits prostate cancer cell apoptosis via the PAK6-SIRT4-ANT2 complex. Theranostics 2020, 10, 2571–2586. [Google Scholar] [CrossRef] [PubMed]
- Guan, J.; Jiang, X.; Gai, J.; Sun, X.; Zhao, J.; Li, J.; Li, Y.; Cheng, M.; Du, T.; Fu, L.; et al. Sirtuin 5 regulates the proliferation, invasion and migration of prostate cancer cells through acetyl-CoA acetyltransferase 1. J. Cell Mol. Med. 2020, 24, 14039–14049. [Google Scholar] [CrossRef]
- Ghose, A.; Moschetta, M.; Pappas-Gogos, G.; Sheriff, M.; Boussios, S. Genetic Aberrations of DNA Repair Pathways in Prostate Cancer: Translation to the Clinic. Int. J. Mol. Sci. 2021, 22, 9783. [Google Scholar] [CrossRef]
- Ding, M.; Jiang, C.Y.; Zhang, Y.; Zhao, J.; Han, B.M.; Xia, S.J. SIRT7 depletion inhibits cell proliferation and androgen-induced autophagy by suppressing the AR signaling in prostate cancer. J. Exp. Clin. Cancer Res. 2020, 39, 28. [Google Scholar] [CrossRef]
- Sawant Dessai, A.; Dominguez, M.P.; Chen, U.I.; Hasper, J.; Prechtl, C.; Yu, C.; Katsuta, E.; Dai, T.; Zhu, B.; Jung, S.Y.; et al. Transcriptional Repression of SIRT3 Potentiates Mitochondrial Aconitase Activation to Drive Aggressive Prostate Cancer to the Bone. Cancer Res. 2021, 81, 50–63. [Google Scholar] [CrossRef]
- Fu, W.; Li, H.; Fu, H.; Zhao, S.; Shi, W.; Sun, M.; Li, Y. The SIRT3 and SIRT6 Promote Prostate Cancer Progression by Inhibiting Necroptosis-Mediated Innate Immune Response. J. Immunol. Res. 2020, 2020, 8820355. [Google Scholar] [CrossRef]
- Garcia-Peterson, L.M.; Li, X. Trending topics of SIRT1 in tumorigenicity. Biochim. Biophys. Acta Gen. Subj. 2021, 1865, 129952. [Google Scholar] [CrossRef]
- Cao, Y.; Zhao, R.; Guo, K.; Ren, S.; Zhang, Y.; Lu, Z.; Tian, L.; Li, T.; Chen, X.; Wang, Z. Potential Metabolite Biomarkers for Early Detection of Stage-I Pancreatic Ductal Adenocarcinoma. Front. Oncol. 2022, 11, 744667. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.; Shukla, S.K.; Vernucci, E.; He, C.; Wang, D.; King, R.J.; Jha, K.; Siddhanta, K.; Mullen, N.J.; Attri, K.S.; et al. Metabolic Rewiring by Loss of Sirt5 Promotes Kras-Induced Pancreatic Cancer Progression. Gastroenterology 2021, 161, 1584–1600. [Google Scholar] [CrossRef]
- Tsai, Y.C.; Chen, S.L.; Peng, S.L.; Tsai, Y.L.; Chang, Z.M.; Chang, V.H.; Ch’ang, H.J. Upregulating sirtuin 6 ameliorates glycolysis, EMT and distant metastasis of pancreatic adenocarcinoma with krüppel-like factor 10 deficiency. Exp. Mol. Med. 2021, 53, 1623–1635. [Google Scholar] [CrossRef] [PubMed]
- Abati, S.; Bramati, C.; Bondi, S.; Lissoni, A.; Trimarchi, M. Oral Cancer and Precancer: A Narrative Review on the Relevance of Early Diagnosis. Int. J. Environ. Res. Public Health 2020, 17, 9160. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Zhang, S.; Wu, S.; Zhu, Q.; Li, W. MiR-770 promotes oral squamous cell carcinoma migration and invasion by regulating the Sirt7/Smad4 pathway. IUBMB Life 2021, 73, 264–272. [Google Scholar] [CrossRef]
- D’Onofrio, N.; Mele, L.; Martino, E.; Salzano, A.; Restucci, B.; Cautela, D.; Tatullo, M.; Balestrieri, M.L.; Campanile, G. Synergistic Effect of Dietary Betaines on SIRT1-Mediated Apoptosis in Human Oral Squamous Cell Carcinoma Cal 27. Cancers 2020, 12, 2468. [Google Scholar] [CrossRef]
- Islam, S.; Abiko, Y.; Uehara, O.; Chiba, I. Sirtuin 1 and oral cancer. Oncol. Lett. 2019, 17, 729–738. [Google Scholar] [CrossRef] [Green Version]
- Penrose, H.; Heller, S.; Cable, C.; Makboul, R.; Chadalawada, G.; Chen, Y.; Crawford, S.E.; Savkovic, S.D. Epidermal growth factor receptor mediated proliferation depends on increased lipid droplet density regulated via a negative regulatory loop with FOXO3/Sirtuin6. Biochem. Biophys. Res. Commun. 2016, 469, 370–376. [Google Scholar] [CrossRef] [Green Version]
- D’Onofrio, N.; Cacciola, N.A.; Martino, E.; Borrelli, F.; Fiorino, F.; Lombardi, A.; Neglia, G.; Balestrieri, M.L.; Campanile, G. ROS-Mediated Apoptotic Cell Death of Human Colon Cancer LoVo Cells by Milk δ-Valerobetaine. Sci. Rep. 2020, 10, 8978. [Google Scholar] [CrossRef]
- Jung, J.; Lee, Y.H.; Fang, X.; Kim, S.J.; Kim, S.H.; Kim, D.H.; Song, N.Y.; Na, H.K.; Baek, J.H.; Surh, Y.J. IL-1β induces expression of proinflammatory cytokines and migration of human colon cancer cells through upregulation of SIRT1. Arch. Biochem. Biophys. 2021, 703, 108847. [Google Scholar] [CrossRef]
- Yu, L.; Dong, L.; Li, H.; Liu, Z.; Luo, Z.; Duan, G.; Dai, X.; Lin, Z. Ubiquitination-mediated degradation of SIRT1 by SMURF2 suppresses CRC cell proliferation and tumorigenesis. Oncogene 2020, 39, 4450–4464. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Liu, Q.; Dai, M.; Peng, R.; Li, X.; Zuo, W.; Gou, J.; Zhou, F.; Yu, S.; Liu, H.; et al. FOXQ1-mediated SIRT1 upregulation enhances stemness and radio-resistance of colorectal cancer cells and restores intestinal microbiota function by promoting β-catenin nuclear translocation. J. Exp. Clin. Cancer Res. 2022, 41, 70. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.H.; Kim, S.J.; Fang, X.; Song, N.Y.; Kim, D.H.; Suh, J.; Na, H.K.; Kim, K.O.; Baek, J.H.; Surh, Y.J. JNK-mediated Ser27 phosphorylation and stabilization of SIRT1 promote growth and progression of colon cancer through deacetylation-dependent activation of Snail. Mol. Oncol. 2021; Online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Aventaggiato, M.; Vernucci, E.; Barreca, F.; Russo, M.A.; Tafani, M. Sirtuins’ control of autophagy and mitophagy in cancer. Pharmacol. Ther. 2021, 221, 107748. [Google Scholar] [CrossRef]
- Warburg, O.; Minami, S. Versuche an Überlebendem Carcinom-gewebe. Klin. Wochenschr. 1923, 2, 776–777. [Google Scholar] [CrossRef]
- Pascale, R.M.; Calvisi, D.F.; Simile, M.M.; Feo, C.F.; Feo, F. The Warburg Effect 97 Years after Its Discovery. Cancers 2020, 12, 2819. [Google Scholar] [CrossRef]
- Pinho, A.V.; Mawson, A.; Gill, A.; Arshi, M.; Warmerdam, M.; Giry-Laterriere, M.; Eling, N.; Lie, T.; Kuster, E.; Camargo, S.; et al. Sirtuin 1 stimulates the proliferation and the expression of glycolysis genes in pancreatic neoplastic lesions. Oncotarget 2016, 7, 74768–74778. [Google Scholar] [CrossRef] [Green Version]
- Tambay, V.; Raymond, V.A.; Bilodeau, M. MYC Rules: Leading Glutamine Metabolism toward a Distinct Cancer Cell Phenotype. Cancers 2021, 13, 4484. [Google Scholar] [CrossRef]
- Bensard, C.L.; Wisidagama, D.R.; Olson, K.A.; Berg, J.A.; Krah, N.M.; Schell, J.C.; Nowinski, S.M.; Fogarty, S.; Bott, A.J.; Wei, P.; et al. Regulation of Tumor Initiation by the Mitochondrial Pyruvate Carrier. Cell Metab. 2020, 31, 284.e7–300.e7. [Google Scholar] [CrossRef]
- Kłos, P.; Dabravolski, S.A. The Role of Mitochondria Dysfunction in Inflammatory Bowel Diseases and Colorectal Cancer. Int. J. Mol. Sci. 2021, 22, 11673. [Google Scholar] [CrossRef]
- Liu, G.; Park, S.H.; Imbesi, M.; Nathan, W.J.; Zou, X.; Zhu, Y.; Jiang, H.; Parisiadou, L.; Gius, D. Loss of NAD-Dependent Protein Deacetylase Sirtuin-2 Alters Mitochondrial Protein Acetylation and Dysregulates Mitophagy. Antioxid. Redox Signal. 2017, 26, 849–863. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Ye, Y.; Yang, X.; Liu, B.; Wang, Z.; Chen, S.; Jiang, K.; Zhang, W.; Jiang, H.; Mustonen, H.; et al. SIRT2-dependent IDH1 deacetylation inhibits colorectal cancer and liver metastases. EMBO Rep. 2020, 21, e48183. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhou, Y.; Kim, J.T.; Sengoku, T.; Alstott, M.C.; Weiss, H.L.; Wang, Q.; Evers, B.M. Regulation of SIRT2 by Wnt/β-catenin signaling pathway in colorectal cancer cells. Biochim. Biophys. Acta Mol. Cell Res. 2021, 1868, 118966. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Sun, X.; Li, G.; Wu, Q.; Chen, Y.; Yang, X.; Luo, X.; Hu, J.; Wang, G. Inhibition of SIRT2 limits tumour angiogenesis via inactivation of the STAT3/VEGFA signalling pathway. Cell Death Dis. 2018, 10, 9. [Google Scholar] [CrossRef] [PubMed]
- Cheon, M.G.; Kim, W.; Choi, M.; Kim, J.E. AK-1 a specific SIRT2 inhibitor.; induces cell cycle arrest by downregulating Snail in HCT116 human colon carcinoma cells. Cancer Lett. 2015, 356, 637–645. [Google Scholar] [CrossRef]
- Hou, J.Y.; Cao, J.; Gao, L.J.; Zhang, F.P.; Shen, J.; Zhou, L.; Shi, J.Y.; Feng, Y.L.; Yan, Z.; Wang, D.P.; et al. Upregulation of α enolase (ENO1) crotonylation in colorectal cancer and its promoting effect on cancer cell metastasis. Biochem. Biophys. Res. Commun. 2021, 578, 77–83. [Google Scholar] [CrossRef]
- Yang, Y.; Ding, J.; Gao, Z.G.; Wang, Z.J. A variant in SIRT2 gene 3′-UTR is associated with susceptibility to colorectal cancer. Oncotarget 2017, 8, 41021–41025. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Xiang, H.; Liu, J.; Chen, Y.; He, R.R.; Liu, B. Mitochondrial Sirtuin 3: New emerging biological function and therapeutic target. Theranostics 2020, 10, 8315–8342. [Google Scholar] [CrossRef]
- Torrens-Mas, M.; Hernández-López, R.; Pons, D.G.; Roca, P.; Oliver, J.; Sastre-Serra, J. Sirtuin 3 silencing impairs mitochondrial biogenesis and metabolism in colon cancer cells. Am. J. Physiol. Cell Physiol. 2019, 317, C398–C404. [Google Scholar] [CrossRef]
- Ballista-Hernández, J.; Martínez-Ferrer, M.; Vélez, R.; Climent, C.; Sánchez-Vázquez, M.M.; Torres, C.; Rodríguez-Muñoz, A.; Ayala-Peña, S.; Torres-Ramos, C.A. Mitochondrial DNA Integrity Is Maintained by APE1 in Carcinogen-Induced Colorectal Cancer. Mol. Cancer Res. 2017, 15, 831–841. [Google Scholar] [CrossRef] [Green Version]
- Errichiello, E.; Venesio, T. Mitochondrial DNA variants in colorectal carcinogenesis: Drivers or passengers? J. Cancer Res. Clin. Oncol. 2017, 143, 1905–1914. [Google Scholar] [CrossRef] [PubMed]
- Kleist, B.; Meurer, T.; Poetsch, M. Mitochondrial DNA alteration in primary and metastatic colorectal cancer: Different frequency and association with selected clinicopathological and molecular markers. Tumour Biol. 2017, 39, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ansari, A.; Rahman, M.S.; Saha, S.K.; Saikot, F.K.; Deep, A.; Kim, K.H. Function of the SIRT3 mitochondrial deacetylase in cellular physiology, cancer, and neurodegenerative disease. Aging Cell 2017, 16, 4–16. [Google Scholar] [CrossRef] [PubMed]
- Kabziński, J.; Walczak, A.; Mik, M.; Majsterek, I. Sirt3 regulates the level of mitochondrial DNA repair activity through deacetylation of NEIL1, NEIL2, OGG1, MUTYH, APE1 and LIG3 in colorectal cancer. Pol. J. Surg. 2019, 92, 1–4. [Google Scholar] [CrossRef]
- Parodi-Rullán, R.M.; Chapa-Dubocq, X.R.; Javadov, S. Acetylation of Mitochondrial Proteins in the Heart: The Role of SIRT3. Front. Physiol. 2018, 9, 1094. [Google Scholar] [CrossRef] [Green Version]
- Newman, A.C.; Maddocks, O.D.K. One-carbon metabolism in cancer. Br. J. Cancer 2017, 116, 1499–1504. [Google Scholar] [CrossRef] [Green Version]
- Ducker, G.S.; Rabinowitz, J.D. One-Carbon Metabolism in Health and Disease. Cell Metab. 2017, 25, 27–42. [Google Scholar] [CrossRef] [Green Version]
- Ducker, G.S.; Chen, L.; Morscher, R.J.; Ghergurovich, J.M.; Esposito, M.; Teng, X.; Kang, Y.; Rabinowitz, J.D. Reversal of Cytosolic One-Carbon Flux Compensates for Loss of the Mitochondrial Folate Pathway. Cell Metab. 2016, 23, 1140–1153. [Google Scholar] [CrossRef] [Green Version]
- Miyo, M.; Konno, M.; Colvin, H.; Nishida, N.; Koseki, J.; Kawamoto, K.; Tsunekuni, K.; Nishimura, J.; Hata, T.; Takemasa, I.; et al. The importance of mitochondrial folate enzymes in human colorectal cancer. Oncol. Rep. 2017, 37, 417–425. [Google Scholar] [CrossRef] [Green Version]
- Wei, Z.; Song, J.; Wang, G.; Cui, X.; Zheng, J.; Tang, Y.; Chen, X.; Li, J.; Cui, L.; Liu, C.Y.; et al. Deacetylation of serine hydroxymethyl-transferase 2 by SIRT3 promotes colorectal carcinogenesis. Nat. Commun. 2018, 9, 4468. [Google Scholar] [CrossRef] [Green Version]
- Wan, X.; Wang, C.; Huang, Z.; Zhou, D.; Xiang, S.; Qi, Q.; Chen, X.; Arbely, E.; Liu, C.Y.; Du, P.; et al. Cisplatin inhibits SIRT3-deacetylation MTHFD2 to disturb cellular redox balance in colorectal cancer cell. Cell Death Dis. 2020, 11, 649. [Google Scholar] [CrossRef] [PubMed]
- Mandal, P. Insight of nitric oxide signaling: A potential biomarker with multifaceted complex mechanism in colorectal carcinogenesis. Biochem. Biophys. Res. Commun. 2018, 495, 1766–1768. [Google Scholar] [CrossRef]
- Wang, Q.; Ye, S.; Chen, X.; Xu, P.; Li, K.; Zeng, S.; Huang, M.; Gao, W.; Chen, J.; Zhang, Q.; et al. Mitochondrial NOS1 suppresses apoptosis in colon cancer cells through increasing SIRT3 activity. Biochem. Biophys. Res. Commun. 2019, 515, 517–523. [Google Scholar] [CrossRef] [PubMed]
- Torrens-Mas, M.; Hernández-López, R.; Oliver, J.; Roca, P.; Sastre-Serra, J. Sirtuin 3 silencing improves oxaliplatin efficacy through acetylation of MnSOD in colon cancer. J. Cell Physiol. 2018, 233, 6067–6076. [Google Scholar] [CrossRef] [PubMed]
- Gaya-Bover, A.; Hernández-López, R.; Alorda-Clara, M.; Ibarra de la Rosa, J.M.; Falcó, E.; Fernández, T.; Company, M.M.; Torrens-Mas, M.; Roca, P.; Oliver, J.; et al. Antioxidant enzymes change in different non-metastatic stages in tumoral and peritumoral tissues of colorectal cancer. Int. J. Biochem. Cell Biol. 2020, 120, 105698. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, X.L.; Zhou, M.; Kang, C.; Lang, H.D.; Chen, M.T.; Hui, S.C.; Wang, B.; Mi, M.T. Crosstalk between gut microbiota and Sirtuin-3 in colonic inflammation and tumorigenesis. Exp. Mol. Med. 2018, 50, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Paku, M.; Haraguchi, N.; Takeda, M.; Fujino, S.; Ogino, T.; Takahashi, H.; Miyoshi, N.; Uemura, M.; Mizushima, T.; Yamamoto, H.; et al. SIRT3-Mediated SOD2 and PGC-1α Contribute to Chemoresistance in Colorectal Cancer Cells. Ann. Surg. Oncol. 2021, 28, 4720–4732. [Google Scholar] [CrossRef]
- D’Onofrio, N.; Martino, E.; Mele, L.; Colloca, A.; Maione, M.; Cautela, D.; Castaldo, D.; Balestrieri, M.L. Colorectal Cancer Apoptosis Induced by Dietary δ-Valerobetaine Involves PINK1/Parkin Dependent-Mitophagy and SIRT3. Int. J. Mol. Sci. 2021, 22, 8117. [Google Scholar] [CrossRef] [PubMed]
- D’Onofrio, N.; Martino, E.; Balestrieri, A.; Mele, L.; Neglia, G.; Balestrieri, M.L.; Campanile, G. SIRT3 and Metabolic Reprogramming Mediate the Antiproliferative Effects of Whey in Human Colon Cancer Cells. Cancers 2021, 13, 5196. [Google Scholar] [CrossRef]
- D’Onofrio, N.; Martino, E.; Balestrieri, A.; Mele, L.; Cautela, D.; Castaldo, D.; Balestrieri, M.L. Diet-derived ergothioneine induces necroptosis in colorectal cancer cells by activating the SIRT3/MLKL pathway. FEBS Lett. 2022. [Google Scholar] [CrossRef]
- Bai, Y.; Yang, J.; Cui, Y.; Yao, Y.; Wu, F.; Liu, C.; Fan, X.; Zhang, Y. Research Progress of Sirtuin4 in Cancer. Front. Oncol. 2021, 10, 562950. [Google Scholar] [CrossRef] [PubMed]
- Smith, H.Q.; Li, C.; Stanley, C.A.; Smith, T.J. Glutamate Dehydrogenase, a Complex Enzyme at a Crucial Metabolic Branch Point. Neurochem. Res. 2019, 44, 117–132. [Google Scholar] [CrossRef]
- Miyo, M.; Yamamoto, H.; Konno, M.; Colvin, H.; Nishida, N.; Koseki, J.; Kawamoto, K.; Ogawa, H.; Hamabe, A.; Uemura, M.; et al. Tumour-suppressive function of SIRT4 inhuman colorectal cancer. Br. J. Cancer 2015, 113, 492–499. [Google Scholar] [CrossRef] [Green Version]
- Huang, G.; Cheng, J.; Yu, F.; Liu, X.; Yuan, C.; Liu, C.; Chen, X.; Peng, Z. Clinical and therapeuticsignificance of sirtuin-4 expression in colorectal cancer. Oncol. Rep. 2016, 35, 2801–2810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- AlShamaileh, H.; Wang, T.; Xiang, D.; Yin, W.; Tran, P.H.; Barrero, R.A.; Zhang, P.Z.; Li, Y.; Kong, L.; Liu, K.; et al. Aptamer-mediated survivin RNAi enables 5-fluorouracil to eliminate colorectal cancer stem cells. Sci. Rep. 2017, 7, 5898. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Wang, G.; Li, X.; Wang, T.; Weng, M.; Zhang, Y. Knockout of SIRT4 decreases chemosensitivity to 5-FU in colorectal cancer cells. Oncol. Lett. 2018, 16, 1675–1681. [Google Scholar] [CrossRef]
- Cui, Y.; Bai, Y.; Yang, J.; Yao, Y.; Zhang, C.; Liu, C.; Shi, J.; Li, Q.; Zhang, J.; Lu, X.; et al. SIRT4 is the molecular switch mediating cellular proliferation in colorectal cancer through GLS mediated activation of AKT/GSK3β/CyclinD1 pathway. Carcinogenesis 2021, 42, 481–492. [Google Scholar] [CrossRef]
- Kumar, S.; Lombard, D.B. Mitochondrial sirtuins and their relationships with metabolic disease and cancer. Antioxid. Redox Sign. 2015, 22, 1060–1077. [Google Scholar] [CrossRef]
- Ryter, S.W.; Cloonan, S.M.; Choi, A.M. Autophagy: A critical regulator of cellular metabolism and homeostasis. Mol. Cells 2013, 36, 7–16. [Google Scholar] [CrossRef] [Green Version]
- Shi, L.; Yan, H.; An, S.; Shen, M.; Jia, W.; Zhang, R.; Zhao, L.; Huang, G.; Liu, J. SIRT5-mediated deacetylation of LDHB promotes autophagy and tumorigenesis in colorectal cancer. Mol. Oncol. 2019, 13, 358–375. [Google Scholar] [CrossRef]
- Wang, Y.Q.; Wang, H.L.; Xu, J.; Tan, J.; Fu, L.N.; Wang, J.L.; Zou, T.H.; Sun, D.F.; Gao, Q.Y.; Chen, Y.X.; et al. Sirtuin5 contributes to colorectal carcinogenesis by enhancing glutaminolysis in a deglutarylation-dependent manner. Nat. Commun. 2018, 9, 545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, M.; Yang, X.; Bie, J.; Wang, Z.; Liu, M.; Li, Y.; Shao, G.; Luo, J. Citrate synthase desuccinylation by SIRT5 promotes colon cancer cell proliferation and migration. Biol. Chem. 2020, 401, 1031–1039. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wang, Z.; Li, X.; Liu, B.; Liu, M.; Liu, L.; Chen, S.; Ren, M.; Wang, Y.; Yu, M.; et al. SHMT2 Desuccinylation by SIRT5 Drives Cancer Cell Proliferation. Cancer Res. 2018, 78, 372–386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, Z.; Liu, X.; Chen, T.; Gao, W.; Wu, Z.; Hu, Z.; Wei, D.; Gao, C.; Li, Q. Targeting a Sirt5-Positive Subpopulation Overcomes Multidrug Resistance in Wild-Type Kras Colorectal Carcinomas. Cell Rep. 2018, 23, 3975–3978. [Google Scholar] [CrossRef] [PubMed]
- Ekremoglu, O.; Koc, A. The role of SIRT5 and p53 proteins in the sensitivity of colon cancer cells to chemotherapeutic agent 5-Fluorouracil. Mol. Biol. Rep. 2021, 48, 5485–5495. [Google Scholar] [CrossRef] [PubMed]
- Khodaei, F.; Hosseini, S.M.; Omidi, M.; Hosseini, S.F.; Rezaei, M. Cytotoxicity of metformin against HT29 colon cancer cells contributes to mitochondrial Sirt3 upregulation. J. Biochem. Mol. Toxicol. 2021, 35, e22662. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Li, X.P.; Jiang, R.; Chen, Y.; Lv, X.T.; Guo, X.X.; Tian, K.; Yuan, D.Z.; Lv, Y.W.; Ran, J.H.; et al. Evodiamine inhibits migration and invasion by Sirt1-mediated post-translational modulations in colorectal cancer. Anticancer Drugs 2019, 30, 611–617. [Google Scholar] [CrossRef]
- Tan, Y.J.; Lee, Y.T.; Mancera, R.L.; Oon, C.E. BZD9L1 sirtuin inhibitor: Identification of key molecular targets and their biological functions in HCT 116 colorectal cancer cells. Life Sci. 2021, 284, 119747. [Google Scholar] [CrossRef]
- Zarkavelis, G.; Boussios, S.; Papadaki, A.; Katsanos, K.H.; Christodoulou, D.K.; Pentheroudakis, G. Current and future biomarkers in colorectal cancer. Ann. Gastroenterol. 2017, 30, 613–621. [Google Scholar] [CrossRef]
- Kang, Y.J.; Jang, J.Y.; Kwon, Y.H.; Lee, J.H.; Lee, S.; Park, Y.; Jung, Y.S.; Im, E.; Moon, H.R.; Chung, H.Y.; et al. MHY2245, a Sirtuin Inhibitor, Induces Cell Cycle Arrest and Apoptosis in HCT116 Human Colorectal Cancer Cells. Int. J. Mol. Sci. 2022, 23, 1590. [Google Scholar] [CrossRef]
- Kim, M.J.; Kang, Y.J.; Sung, B.; Jang, J.Y.; Ahn, Y.R.; Oh, H.J.; Choi, H.; Choi, I.; Im, E.; Moon, H.R.; et al. Novel SIRT Inhibitor, MHY2256, Induces Cell Cycle Arrest, Apoptosis and Autophagic Cell Death in HCT116 Human Colorectal Cancer Cells. Biomol. Ther. 2020, 28, 561–568. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Zhu, Z.; Chen, Y.; Song, J.; Huang, Y.; Song, K.; Zhong, J.; Xu, X.; Wei, J.; Wang, C.; et al. Small-molecule activating SIRT6 elicits therapeutic effects and synergistically promotes anti-tumor activity of vitamin D3 in colorectal cancer. Theranostics 2020, 10, 5845–5864. [Google Scholar] [CrossRef]
- Iachettini, S.; Trisciuoglio, D.; Rotili, D.; Lucidi, A.; Salvati, E.; Zizza, P.; Di Leo, L.; Del Bufalo, D.; Ciriolo, M.R.; Leonetti, C.; et al. Pharmacological activation of SIRT6 triggers lethal autophagy in human cancer cells. Cell Death Dis. 2018, 9, 996. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Wong, G.; Naffouje, S.; Felder, S.; Sanchez, J.; Dineen, S.; Powers, B.D.; Dessureault, S.; Gurd, E.; Castillo, D.; et al. A Novel Nomogram for Early Identification and Intervention in Colorectal Cancer Patients at Risk for Malnutrition. Am. Surg. 2021, 2021, 31348211058620. [Google Scholar] [CrossRef] [PubMed]
- Gessani, S.; Van Duijnhoven, F.J.; Moreno-Aliaga, M.J. Editorial: Diet, Inflammation and Colorectal Cancer. Front. Immunol. 2019, 10, 2598. [Google Scholar] [CrossRef] [PubMed]
- Serrano, D.; Pozzi, C.; Guglietta, S.; Fosso, B.; Suppa, M.; Gnagnarella, P.; Corso, F.; Bellerba, F.; Macis, D.; Aristarco, V.; et al. Microbiome as Mediator of Diet on Colorectal Cancer Risk: The Role of Vitamin D.; Markers of Inflammation and Adipokines. Nutrients 2021, 13, 363. [Google Scholar] [CrossRef] [PubMed]
- Collins, N.; Belkaid, Y. Control of immunity via nutritional interventions. Immunity 2022, 55, 210–223. [Google Scholar] [CrossRef]
- Santilli, A.; Stefanopoulos, S.; Cresci, G.A.M. The gut barrier and chronic diseases. Curr. Opin. Clin. Nutr. Metab. Care 2022. [Google Scholar] [CrossRef]
- Li, Z.; Wang, K.; Shivappa, N.; Hébert, J.R.; Chen, H.; Liu, H.; Jiang, X. Inflammatory potential of diet and colorectal carcinogenesis: A prospective longitudinal cohort. Br. J. Cancer 2022. [Google Scholar] [CrossRef]
- Rodriguez-Gonzalez, J.C.; Hernández-Balmaseda, I.; Declerck, K.; Pérez-Novo, C.; Logie, E.; Theys, C.; Jakubek, P.; Quiñones-Maza, O.L.; Dantas-Cassali, G.; Carlos Dos Reis, D.; et al. Antiproliferative Antiangiogenic and Antimetastatic Therapy Response by Mangiferin in a Syngeneic Immunocompetent Colorectal Cancer Mouse Model Involves Changes in Mitochondrial Energy Metabolism. Front. Pharmacol. 2021, 12, 670167. [Google Scholar] [CrossRef]
- Buhrmann, C.; Shayan, P.; Brockmueller, A.; Shakibaei, M. Resveratrol Suppresses Cross-Talk between Colorectal Cancer Cells and Stromal Cells in Multicellular Tumor Microenvironment: A Bridge between In Vitro and In Vivo Tumor Microenvironment Study. Molecules 2020, 25, 4292. [Google Scholar] [CrossRef] [PubMed]
- Visioli, F.; Ingram, A.; Beckman, J.S.; Magnusson, K.R.; Hagen, T.M. Strategies to protect against age-related mitochondrial decay: Do natural products and their derivatives help? Free Radic. Biol. Med. 2022, 178, 330–346. [Google Scholar] [CrossRef] [PubMed]
- Mautone, N.; Zwergel, C.; Mai, A.; Rotili, D. Sirtuin modulators: Where are we now? A review of patents from 2015 to 2019. Expert Opin. Ther. Pat. 2020, 6, 389–407. [Google Scholar] [CrossRef] [PubMed]
- Karaman Mayack, B.; Sippl, W.; Ntie-Kang, F. Natural Products as Modulators of Sirtuins. Molecules 2020, 14, 3287. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Colloca, A.; Balestrieri, A.; Anastasio, C.; Balestrieri, M.L.; D’Onofrio, N. Mitochondrial Sirtuins in Chronic Degenerative Diseases: New Metabolic Targets in Colorectal Cancer. Int. J. Mol. Sci. 2022, 23, 3212. https://doi.org/10.3390/ijms23063212
Colloca A, Balestrieri A, Anastasio C, Balestrieri ML, D’Onofrio N. Mitochondrial Sirtuins in Chronic Degenerative Diseases: New Metabolic Targets in Colorectal Cancer. International Journal of Molecular Sciences. 2022; 23(6):3212. https://doi.org/10.3390/ijms23063212
Chicago/Turabian StyleColloca, Antonino, Anna Balestrieri, Camilla Anastasio, Maria Luisa Balestrieri, and Nunzia D’Onofrio. 2022. "Mitochondrial Sirtuins in Chronic Degenerative Diseases: New Metabolic Targets in Colorectal Cancer" International Journal of Molecular Sciences 23, no. 6: 3212. https://doi.org/10.3390/ijms23063212






