Atmospheric and Room Temperature Plasma (ARTP) Mutagenesis Improved the Anti-MRSA Activity of Brevibacillus sp. SPR20
Abstract
:1. Introduction
2. Results
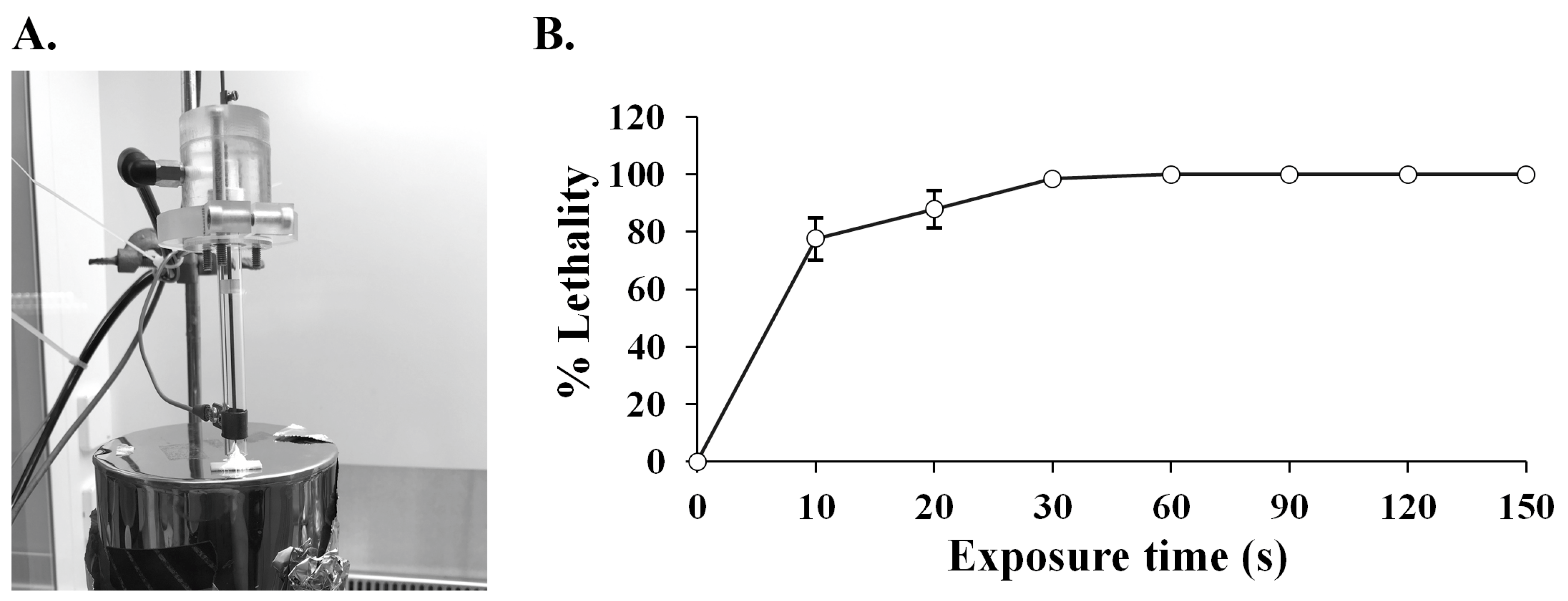
2.1. ARTP Mutagenesis on SPR20
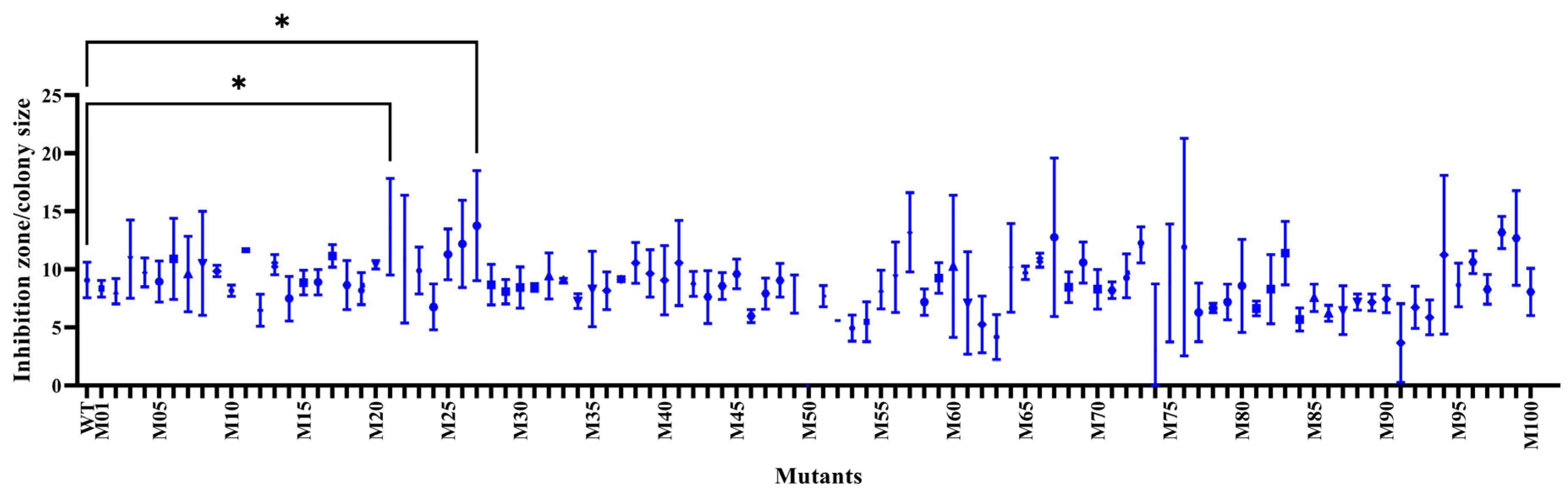
2.2. Screening for the Positive Mutations
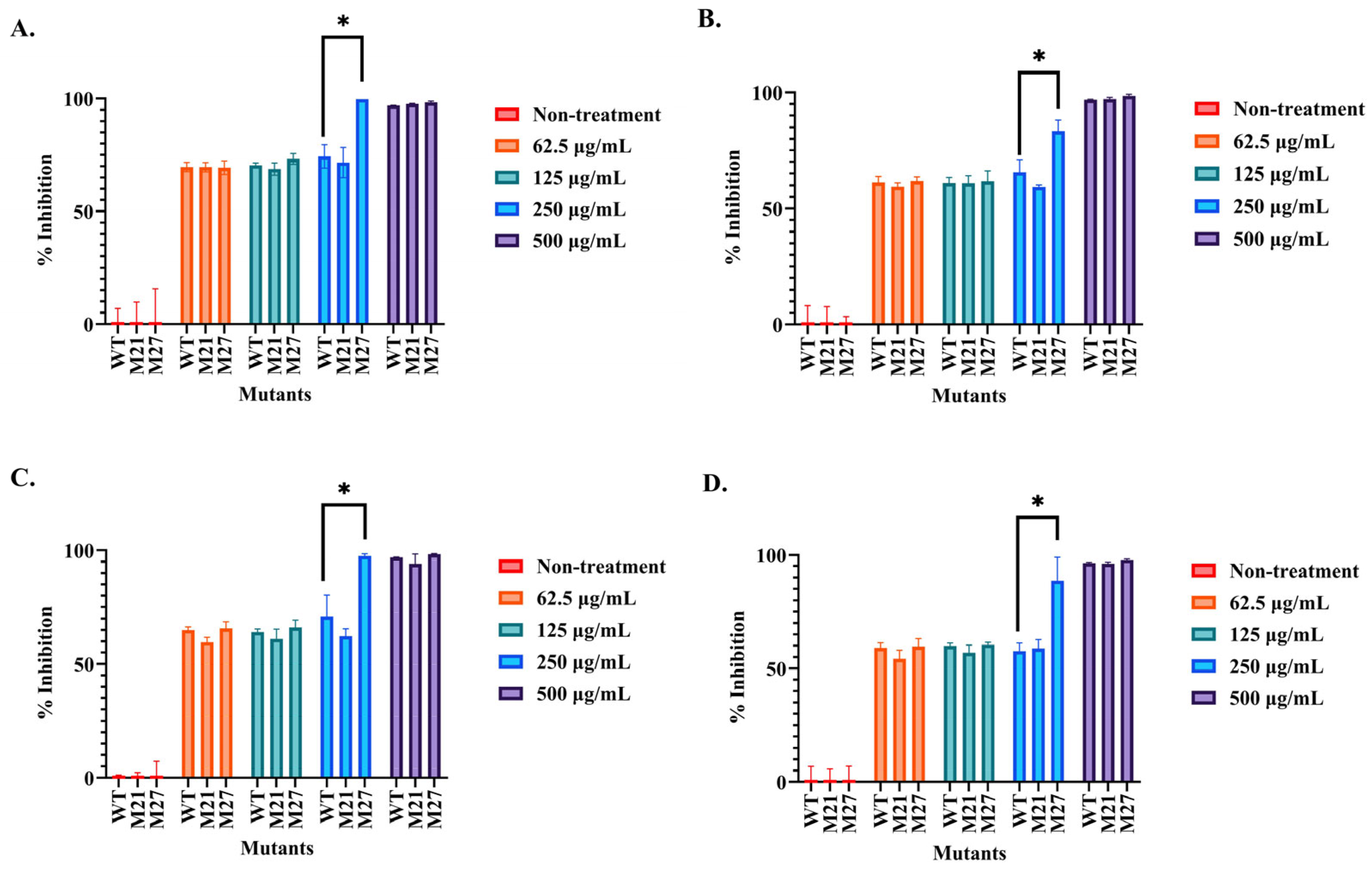
2.3. Determination of the Antibacterial Activity of the Selected SPR20 Mutants
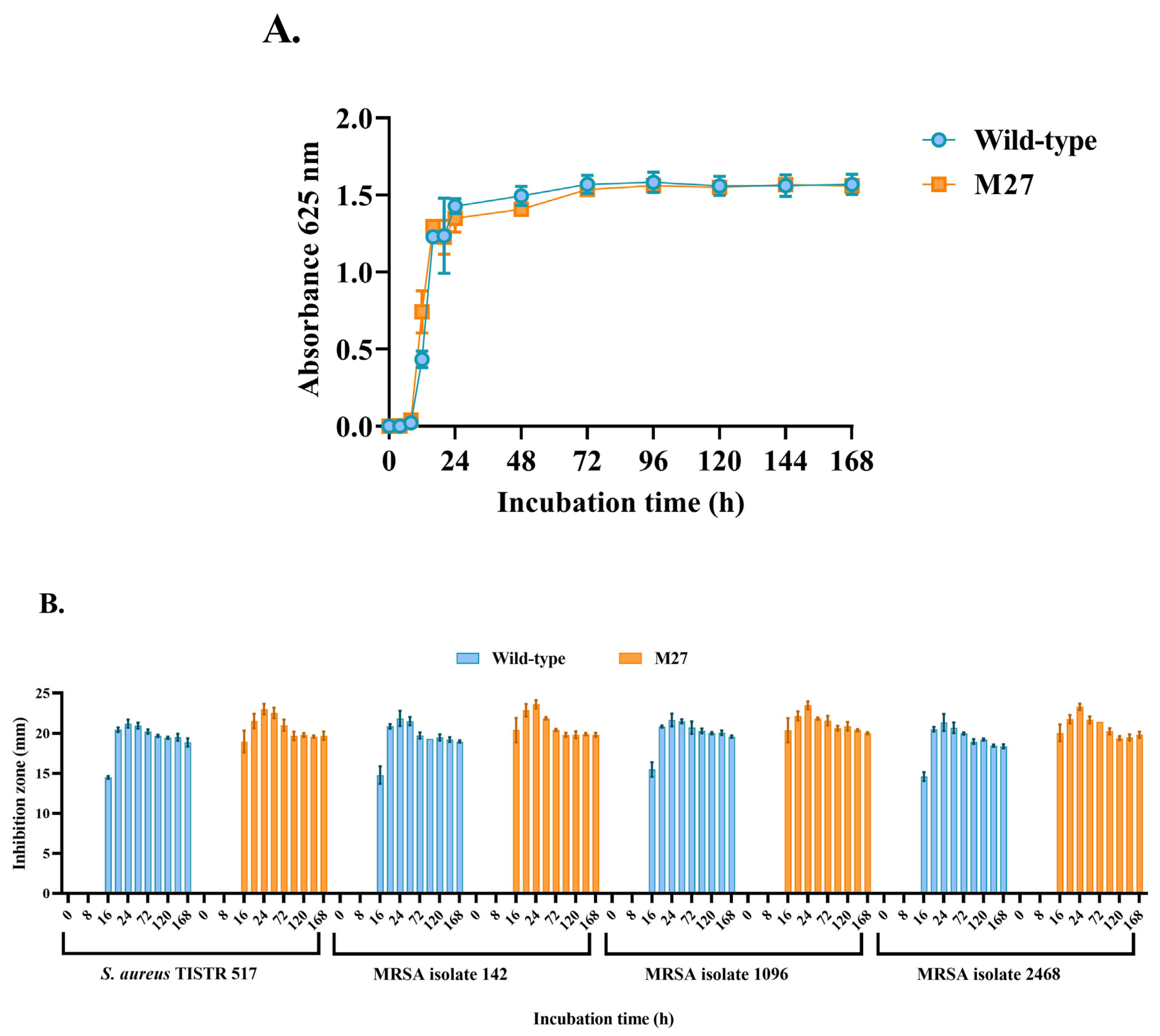
2.4. Comparison of Growth Curve and Antibacterial Activity
2.5. Strain Stability of the Selected Mutant
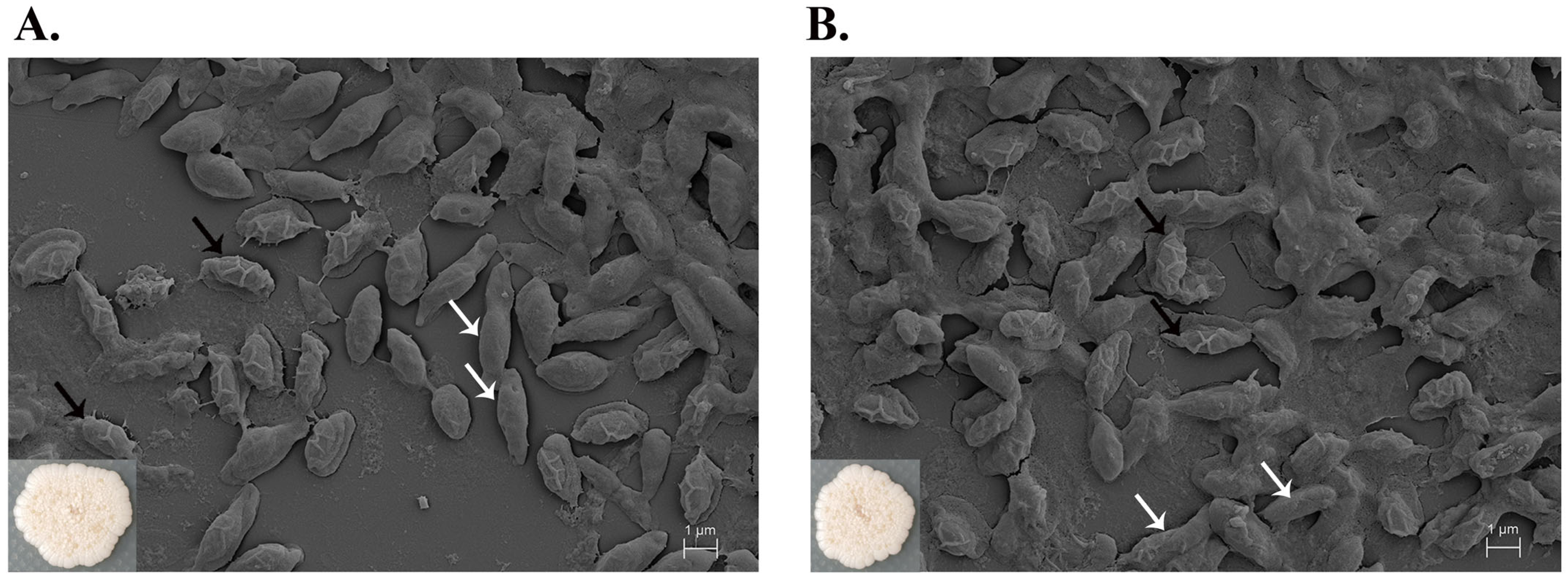
2.6. Cell Morphology by Scanning Electron Microscopy (SEM)
2.7. Antibiotic Susceptibility Assay
2.8. Proteomic Variability by Principal Component Analysis
2.9. Comparative Analysis of Proteomics
3. Discussion
4. Materials and Methods
4.1. Microorganisms and Culture Conditions
4.2. Atmospheric and Room Temperature Plasma (ARTP) Mutagenesis
4.3. Mutant Screening by Agar Overlay Method
4.4. Antibacterial Testing by Broth Microdilution Assay
4.5. Growth Curve and Agar Well Diffusion Assay
4.6. Stability of the Mutant Strains
4.7. Cell Morphology by Scanning Electron Microscope (SEM)
4.8. Antibiotic Susceptibility Test
4.9. Label-Free Quantitated Proteomics Analysis
4.10. Proteomics Data Analysis and Bioinformatics Analysis
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bloom, D.E.; Cadarette, D. Infectious disease threats in the twenty-first century: Strengthening the global response. Front. Immunol. 2019, 10, 549. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Huang, W.E.; Yang, Q. Clinical perspective of antimicrobial resistance in bacteria. Infect. Drug Resist. 2022, 15, 735–746. [Google Scholar] [CrossRef]
- Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Munita, J.M.; Arias, C.A. Mechanisms of antibiotic resistance. Microbiol. Spectr. 2016, 4, VMBF-0016-2015. [Google Scholar] [CrossRef] [Green Version]
- Patel, J.; Harant, A.; Fernandes, G.; Mwamelo, A.J.; Hein, W.; Dekker, D.; Sridhar, D. Measuring the global response to antimicrobial resistance, 2020–2021: A systematic governance analysis of 114 countries. Lancet Infect. Dis. 2023, 23, 706–718. [Google Scholar] [CrossRef]
- Mancuso, G.; Midiri, A.; Gerace, E.; Biondo, C. Bacterial antibiotic resistance: The most critical pathogens. Pathogens 2021, 10, 1310. [Google Scholar] [CrossRef]
- Antimicrobial Resistance Division. 2019 Antibacterial Agents in Clinical Development: An Analysis of the Antibacterial Clinical Development Pipeline, 1st ed.; World Health Organization: Geneva, Switzerland, 2019; pp. 1–5. [Google Scholar]
- Terreni, M.; Taccani, M.; Pregnolato, M. New antibiotics for multidrug-resistant bacterial strains: Latest research developments and future perspectives. Molecules 2021, 26, 2671. [Google Scholar] [CrossRef] [PubMed]
- Sharrar, A.M.; Crits-Christoph, A.; Méheust, R.; Diamond, S.; Starr, E.P.; Banfield, J.F. Bacterial secondary metabolite biosynthetic potential in soil varies with phylum, depth, and vegetation type. mBio 2020, 11, e00416-20. [Google Scholar] [CrossRef] [PubMed]
- Songnaka, N.; Lertcanawanichakul, M.; Atipairin, A. Promising anti-MRSA activity of Brevibacillus sp. isolated from soil and strain improvement by UV mutagenesis. Sci. Pharm. 2021, 89, 1. [Google Scholar] [CrossRef]
- Songnaka, N.; Lertcanawanichakul, M.; Hutapea, A.M.; Krobthong, S.; Yingchutrakul, Y.; Atipairin, A. Purification and characterization of novel anti-MRSA peptides produced by Brevibacillus sp. SPR-20. Molecules 2022, 27, 8452. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.P.; Sun, J.; Ma, Y. Biomanufacturing: History and perspective. J. Ind. Microbiol. Biotechnol. 2017, 44, 773–784. [Google Scholar] [CrossRef]
- Parekh, S.; Vinci, V.A.; Strobel, R.J. Improvement of microbial strains and fermentation processes. Appl. Microbiol. Biotechnol. 2000, 54, 287–301. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Wang, Y.; Liu, X.; Hu, Z.; Qian, J.; Shi, T.; Wang, Y.; Ye, C. Metabolic analysis of Schizochytrium sp. mutants with high EPA content achieved with ARTP mutagenesis screening. Bioprocess Biosyst. Eng. 2023, 46, 893–901. [Google Scholar] [CrossRef]
- Xu, H.; Dai, C.; Tang, Y.; Xu, X.; Umego, E.C.; He, R.; Ma, H. The selective breeding and mutagenesis mechanism of high-yielding surfactin Bacillus subtilis strains with atmospheric and room temperature plasma. J. Sci. Food Agric. 2021, 102, 1851–1861. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.Y.; Huang, Z.L.; Li, G.; Zhao, H.X.; Xing, X.H.; Sun, W.T.; Li, H.P.; Gou, Z.X.; Bao, C.Y. Novel mutation breeding method for Streptomyces avermitilis using an atmospheric pressure glow discharge plasma. J. Appl. Microbiol. 2010, 108, 851–858. [Google Scholar] [CrossRef] [PubMed]
- Ottenheim, C.; Nawrath, M.; Wu, J.C. Microbial mutagenesis by atmospheric and room-temperature plasma (ARTP): The latest development. Bioresour. Bioprocess. 2018, 5, 12. [Google Scholar] [CrossRef] [Green Version]
- Yu, F.; Zhang, M.; Sun, J.; Wang, F.; Li, X.; Liu, Y.; Wang, Z.; Zhao, X.; Li, J.; Chen, J.; et al. Improved neomycin sulfate potency in Streptomyces fradiae using atmospheric and room temperature plasma (ARTP) mutagenesis and fermentation medium optimization. Microorganisms 2022, 10, 94. [Google Scholar] [CrossRef]
- Schulze, W.X.; Mann, M. A novel proteomic screen for peptide-protein interactions. J. Biol. Chem. 2004, 279, 10756–10764. [Google Scholar] [CrossRef] [Green Version]
- Lasch, P.; Schneider, A.; Blumenscheit, C.; Doellinger, J. Identification of microorganisms by liquid chromatography-mass spectrometry (LC-MS) and in silico peptide mass libraries. Mol. Cell. Proteom. 2020, 19, 2125–2139. [Google Scholar] [CrossRef]
- Gorshkov, V.; Hotta, S.Y.; Verano-Braga, T.; Kjeldsen, F. Peptide de novo sequencing of mixture tandem mass spectra. Proteomics 2016, 16, 2470–2479. [Google Scholar] [CrossRef] [Green Version]
- Wei, R.; Wang, J.; Su, M.; Jia, E.; Chen, S.; Chen, T.; Ni, Y. Missing value imputation approach for mass spectrometry-based metabolomics data. Sci. Rep. 2018, 8, 663. [Google Scholar] [CrossRef] [Green Version]
- Kurita, H.; Haruta, N.; Uchihashi, Y.; Seto, T.; Takashima, K. Strand breaks and chemical modification of intracellular DNA induced by cold atmospheric pressure plasma irradiation. PLoS ONE 2020, 15, e0232724. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Xia, H.; Yang, M.; Geng, W.; Zuo, J.; Ostrikov, K. Insights into generation of OH radicals in plasma jets with constant power: The effects of driving voltage and frequency. Vacuum 2022, 198, 110901. [Google Scholar] [CrossRef]
- Zhang, C.; Qin, J.; Dai, Y.; Mu, W.; Zhang, T. Atmospheric and room temperature plasma (ARTP) mutagenesis enables xylitol over-production with yeast Candida tropicalis. J. Biotechnol. 2019, 296, 7–13. [Google Scholar] [CrossRef]
- Peng, Q.; Xiao, Y.; Zhang, S.; Zhou, C.; Xie, A.; Li, Z.; Tan, A.; Zhou, L.; Xie, Y.; Zhao, J.; et al. Mutation breeding of Aspergillus niger by atmospheric room temperature plasma to enhance phosphorus solubilization ability. PeerJ 2022, 10, e13076. [Google Scholar] [CrossRef]
- Wende, K.; Bekeschus, S.; Schmidt, A.; Jatsch, L.; Hasse, S.; Weltmann, K.D.; Masur, K.; von Woedtke, T. Risk assessment of a cold argon plasma jet in respect to its mutagenicity. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2016, 798–799, 48–54. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, L.; Zhang, X.; Su, N.; Li, H.; Oda, Y.; Xing, X. Quantitative evaluation of DNA damage caused by atmospheric and room-temperature plasma (ARTP) and other mutagenesis methods using a rapid umu-microplate test protocol for microbial mutation breeding. Chin. J. Chem. Eng. 2021, 39, 205–210. [Google Scholar] [CrossRef]
- Songnaka, N.; Nisoa, M.; Atipairin, A.; Wanganuttara, T.; Chinnawong, T. Enhanced antibacterial activity of Brevibacillus sp. SPR19 by atmospheric and room temperature plasma mutagenesis (ARTP). Sci. Pharm. 2022, 90, 23. [Google Scholar] [CrossRef]
- Šimončicová, J.; Kryštofová, S.; Medvecká, V.; Ďurišová, K.; Kaliňáková, B. Technical applications of plasma treatments: Current state and perspectives. Appl. Microbiol. Biotechnol. 2019, 103, 5117–5129. [Google Scholar] [CrossRef]
- Niedźwiedź, I.; Juzwa, W.; Skrzypiec, K.; Skrzypek, T.; Waśko, A.; Kwiatkowski, M.; Pawłat, J.; Polak-Berecka, M. Morphological and physiological changes in Lentilactobacillus hilgardii cells after cold plasma treatment. Sci. Rep. 2020, 10, 18882. [Google Scholar] [CrossRef]
- Xu, Y.; Jing, Y.; Zhang, Q.; Xiu, J.; Tian, M.; Cui, Q.; Ma, Y.; Yi, L.; Han, L.; Qian, Y.; et al. Improving rhamnolipids biosynthesis in Pseudomonas sp. L01 through atmospheric and room-temperature plasma (ARTP) mutagenesis. Microorganisms 2023, 11, 1182. [Google Scholar] [CrossRef]
- Gao, X.; Liu, E.; Yin, Y.; Yang, L.; Huang, Q.; Chen, S.; Ho, C.T. Enhancing activities of salt-tolerant proteases secreted by Aspergillus oryzae Using atmospheric and room-temperature plasma mutagenesis. J. Agric. Food. Chem. 2020, 68, 2757–2764. [Google Scholar] [CrossRef] [PubMed]
- Qian, X.; Xin, K.; Zhang, L.; Zhou, J.; Xu, A.; Dong, W.; Jiang, M. Integration of ARTP mutation and adaptive laboratory evolution to reveal 1,4-butanediol degradation in Pseudomonas putida KT2440. Microbiol. Spectr. 2023, 11, e0498822. [Google Scholar] [CrossRef] [PubMed]
- Laroussi, M.; Mendis, D.A.; Rosenberg, M. Plasma interaction with microbes. New J. Phys. 2003, 5, 41.1–41.10. [Google Scholar] [CrossRef]
- Mouz, N.; Di Guilmi, A.M.; Gordon, E.; Hakenbeck, R.; Dideberg, O.; Vernet, T. Mutations in the active site of penicillin-binding protein PBP2x from Streptococcus pneumonia: Role in the specificity for beta-lactam antibiotics. J. Biol. Chem. 1999, 274, 19175–19180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, K.; Zhao, H. Quinolone antibiotics: Resistance and therapy. Infect. Drug Resist. 2023, 16, 811–820. [Google Scholar] [CrossRef]
- Lopatkin, A.J.; Bening, S.C.; Manson, A.L.; Stokes, J.M.; Kohanski, M.A.; Badran, A.H.; Earl, A.M.; Cheney, N.J.; Yang, J.H.; Collins, J.J. Clinically relevant mutations in core metabolic genes confer antibiotic resistance. Science 2021, 371, eaba0862. [Google Scholar] [CrossRef]
- Richts, B.; Rosenberg, J.; Commichau, F.M. A Survey of pyridoxal 5’-phosphate-dependent proteins in the Gram-positive model bacterium Bacillus subtilis. Front. Mol. Biosci. 2019, 6, 32. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.K.; Kuo, H.C.; Lai, C.H.; Chou, C.C. Single amino acid utilization for bacterial categorization. Sci. Rep. 2020, 10, 12686. [Google Scholar] [CrossRef]
- Romero, R.M.; Roberts, M.F.; Phillipson, J.D. Anthranilate synthase in microorganisms and plants. Phytochemistry 1995, 39, 263–276. [Google Scholar] [CrossRef]
- Li, X.H.; Kim, S.K.; Lee, J.H. Anti-biofilm effects of anthranilate on a broad range of bacteria. Sci. Rep. 2017, 7, 8604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mishra, A.K.; Choi, J.; Moon, E.; Baek, K.H. Tryptophan-rich and proline-rich antimicrobial peptides. Molecules 2018, 23, 815. [Google Scholar] [CrossRef] [Green Version]
- Miljkovic, M.; Jovanovic, S.; O’Connor, P.M.; Mirkovic, N.; Jovcic, B.; Filipic, B.; Dinic, M.; Studholme, D.J.; Fira, D.; Cotter, P.D.; et al. Brevibacillus laterosporus strains BGSP7, BGSP9 and BGSP11 isolated from silage produce broad spectrum multi-antimicrobials. PLoS ONE 2019, 14, e0216773. [Google Scholar] [CrossRef]
- Theodore, C.M.; Stamps, B.W.; King, J.B.; Price, L.S.; Powell, D.R.; Stevenson, B.S.; Cichewicz, R.H. Genomic and metabolomic insights into the natural product biosynthetic diversity of a feral-hog-associated Brevibacillus laterosporus strain. PLoS ONE 2014, 9, e90124. [Google Scholar] [CrossRef]
- Hoskisson, P.A.; Seipke, R.F. Cryptic or silent? The known unknowns, unknown knowns, and unknown unknowns of secondary metabolism. mBio 2022, 11, e02642-20. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Huang, Z.; Gui, X.; Xiang, W.; Jin, Y.; Chen, J.; Zhao, J. Multi-omics comparative analysis of streptomyces mutants obtained by iterative atmosphere and room-temperature plasma mutagenesis. Front. Microbiol. 2021, 11, 630309. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, 11th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018; pp. 15–52. [Google Scholar]
- Zhang, B.; Zhang, H.D.; Zhou, Y.T.; Huang, K.; Liu, Z.Q.; Zheng, Y.G. Improvement of amphotericin B production by a newly isolated Streptomyces nodosus mutant. Biotechnol. Appl. Biochem. 2018, 65, 188. [Google Scholar] [CrossRef]
- Benkova, M.; Soukup, O.; Marek, J. Antimicrobial susceptibility testing: Currently used methods and devices and the near future in clinical practice. J. Appl. Microbiol. 2020, 129, 806–822. [Google Scholar] [CrossRef]
- Jorgensen, J.H.; Ferraro, M.J. Antimicrobial susceptibility testing: A review of general principles and contemporary practices. Clin. Infect. Dis. 2009, 49, 1749–1755. [Google Scholar] [CrossRef]
- Krobthong, S.; Choowongkomon, K.; Suphakun, P.; Kuaprasert, B.; Samutrtai, P.; Yingchutrakul, Y. The anti-oxidative effect of Lingzhi protein hydrolysates on lipopolysaccharide-stimulated A549 cells. Food Biosci. 2021, 41, 101093. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; García-Seisdedos, D.; Hewapathirana, S.; Kamatchinathan, S.; Kundu, D.J.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022, 50, D543–D552. [Google Scholar] [CrossRef] [PubMed]
- UniProt Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef] [PubMed]




| Antibiotics | Inhibition Zone (mm ± SD) | |
|---|---|---|
| M27 | Wild-Type | |
| Cefoxitin (FOX; 30 µg) | 33.02 ± 0.51 | 34.04 ± 0.00 |
| Ceftriaxone (CRO; 30 µg) | 32.07 ± 1.34 | 32.91 ± 0.78 |
| Ciprofloxacin (CIP; 5 µg) | 35.56 ± 1.02 | 35.56 ± 0.51 |
| Doxycycline (DO; 30 µg) | 40.47 ± 0.29 | 39.79 ± 0.78 |
| Erythromycin (E; 15 µg) | 36.91 ± 0.64 | 37.76 ± 0.39 |
| Gentamicin (CN; 10 µg) | 26.66 ± 0.15 | 26.49 ± 0.73 |
| Imipenem (IPM; 10 µg) | 35.05 ± 1.02 | 34.71 ± 0.29 |
| Vancomycin (VA; 30 µg) | 21.08 ± 0.44 | 20.91 ± 0.96 |
| Piperacillin + Tazobactam (TZP; 110 µg) | 35.56 ± 0.25 | 36.41 ± 0.53 |
| No. | Gene Name | Protein Name | Biological Process | Molecular Function |
|---|---|---|---|---|
| 1 | pcrB | Heptaprenylglyceryl phosphate synthase | ||
| 2 | A0A075RAN9 | Putative copper-importing P-type ATPase A | transport | catalytic activity; metal ion binding; nucleotide binding; transporter activity |
| 3 | A0A2S5H3D0 | Multifunctional 2′,3′-cyclic-nucleotide 2′-phosphodiesterase/5′-nucleotidase/3′-nucleotidase | ||
| 4 | A0A385TD34 | Threonine synthase | ||
| 5 | A0A177XSD9 | Urocanate reductase | metabolic process | catalytic activity; nucleotide binding |
| 6 | A0A075R7M4 | Flagellin | cellular component movement | structural molecule activity |
| 7 | A0A0K9YU01 | Anthranilate synthase component II | metabolic process | catalytic activity |
| 8 | A0A3M8C1C7 | Lipoprotein | ||
| 9 | glpX | Fructose-1,6-bisphosphatase | ||
| 10 | J3AIB6 | Uncharacterized protein | ||
| 11 | rocD | Ornithine aminotransferase | ||
| 12 | A0A4Y3PDX0 | Probable glycine dehydrogenase (decarboxylating) subunit 1 | ||
| 13 | dut | Deoxyuridine 5′-triphosphate nucleotidohydrolase | ||
| 14 | A0A177XIL4 | Polyketide synthase | metabolic process | catalytic activity |
| 15 | ribH | 6,7-dimethyl-8-ribityllumazine synthase | ||
| 16 | A0A502H7N5 | 3-ketoacyl-ACP reductase | ||
| 17 | A0A177XS18 | Carbonic anhydrase | catalytic activity; metal ion binding | |
| 18 | gatB | Aspartyl/glutamyl-tRNA(Asn/Gln)-amidotransferase subunit B | ||
| 19 | deoD | Purine nucleoside phosphorylase DeoD-type | ||
| 20 | A0A2S5HRX3 | Nickel ABC transporter substrate-binding protein | ||
| 21 | alaC | Aminotransferase | ||
| 22 | A0A3M8D599 | Chromosome segregation protein SMC | ||
| 23 | pgk | Phosphoglycerate kinase | ||
| 24 | accC | Biotin carboxylase | ||
| 25 | yvdD | Cytokinin riboside 5′-monophosphate phosphoribohydrolase | ||
| 26 | fusA | Elongation factor G | ||
| 27 | rnr | Ribonuclease R | ||
| 28 | A0A220MEP0 | Bacillithiol biosynthesis deacetylase BshB1 | metabolic process | catalytic activity |
| 29 | A0A075R422 | Dihydrolipoamide acetyltransferase component of pyruvate dehydrogenase complex | metabolic process | catalytic activity |
| 30 | A0A075RA12 | General stress protein 14 | metabolic process | catalytic activity |
| 31 | A0A177XNX2 | ABC transporter ATP-binding protein | catalytic activity; nucleotide binding |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Songnaka, N.; Lertcanawanichakul, M.; Hutapea, A.M.; Nisoa, M.; Krobthong, S.; Yingchutrakul, Y.; Atipairin, A. Atmospheric and Room Temperature Plasma (ARTP) Mutagenesis Improved the Anti-MRSA Activity of Brevibacillus sp. SPR20. Int. J. Mol. Sci. 2023, 24, 12016. https://doi.org/10.3390/ijms241512016
Songnaka N, Lertcanawanichakul M, Hutapea AM, Nisoa M, Krobthong S, Yingchutrakul Y, Atipairin A. Atmospheric and Room Temperature Plasma (ARTP) Mutagenesis Improved the Anti-MRSA Activity of Brevibacillus sp. SPR20. International Journal of Molecular Sciences. 2023; 24(15):12016. https://doi.org/10.3390/ijms241512016
Chicago/Turabian StyleSongnaka, Nuttapon, Monthon Lertcanawanichakul, Albert Manggading Hutapea, Mudtorlep Nisoa, Sucheewin Krobthong, Yodying Yingchutrakul, and Apichart Atipairin. 2023. "Atmospheric and Room Temperature Plasma (ARTP) Mutagenesis Improved the Anti-MRSA Activity of Brevibacillus sp. SPR20" International Journal of Molecular Sciences 24, no. 15: 12016. https://doi.org/10.3390/ijms241512016






