SARS-CoV-2 Spike Protein Interaction Space
Abstract
:1. Introduction
2. Results
3. Discussion
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bakhiet, M.; Taurin, S. SARS-CoV-2: Targeted managements and vaccine development. Cytokine Growth Factor Rev. 2021, 58, 16–29. [Google Scholar] [CrossRef]
- De Oliveira Campos, D.M.; Fulco, U.L.; de Oliveira, C.B.S.; Oliveira, J.I.N. SARS-CoV-2 virus infection: Targets and antiviral pharmacological strategies. J. Evid. Based Med. 2020, 13, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281. [Google Scholar] [CrossRef]
- Awadasseid, A.; Wu, Y.; Tanaka, Y.; Zhang, W. SARS-CoV-2 variants evolved during the early stage of the pandemic and the effects of mutations on adaptation in Wuhan populations. Int. J. Biol. Sci. 2021, 17, 97–106. [Google Scholar] [CrossRef]
- Sternberg, A.; Naujokat, C. Structural features of coronavirus SARS-CoV-2 spike protein: Targets for vaccination. Life Sci. 2020, 257, 118056. [Google Scholar] [CrossRef] [PubMed]
- Gestwicki, J.E.; Cairo, C.W.; Strong, L.E.; Oetjen, K.A.; Kiessling, L.L. Influencing receptor-ligand binding mechanisms with multivalent ligand architecture. J. Am. Chem. Soc. 2002, 124, 14922–14933. [Google Scholar] [CrossRef] [PubMed]
- Kapingidza, A.B.; Kowal, K.; Chruszcz, M. Antigen-Antibody Complexes. Subcell Biochem. 2020, 94, 465–497. [Google Scholar] [PubMed]
- Barbeau, E.J. Polynomials; Springer: Berlin/Heidelberg, Germany, 2003; ISBN 978-0-387-40627-5. [Google Scholar]
- Cahen, P.J.; Chabert, J.L. Integer-Valued Polynomials; American Mathematical Society: Providence, RI, USA, 1997; ISBN 978-0-8218-0388-2. [Google Scholar]
- Discriminant. Mathematics. Encyclopedia Britannica. Available online: https://www.britannica.com/science/discriminant (accessed on 26 July 2022).
- Dickenstein, A.; Emiris, I.Z. Solving Polynomial Equations: Foundations, Algorithms, and Applications; Chapter 1; Springer: Berlin/Heidelberg, Germany, 2005; p. 26. ISBN 3-540-24326-7. [Google Scholar]
- Elfand, I.M.; Kapranov, M.M.; Zelevinsky, A.V. Discriminants, Resultants and Multidimensional Determinants; Birkhäuser: Basel, Switzerland, 1994; p. 1. ISBN 3-7643-3660-9. [Google Scholar]
- Hobson, E.W. John Napier and the Invention of Logarithms, 1614, a Lecture; University of California Libraries University Press: Cambridge, MA, USA, 1914. [Google Scholar]
- Remmert, R. Theory of Complex Functions; Springer: New York, NY, USA, 1991; ISBN 0387971955. [Google Scholar]
- Apostol, T.M. Logarithm. In NIST Handbook of Mathematical Functions; Olver, F.W.J., Lozier, D.M., Boisvert, R.F., Clark, C.W., Eds.; Cambridge University Press: Cambridge, UK, 2010; ISBN 978-0-521-19225-5. [Google Scholar]
- Weisstein, E.W. Domain. Available online: https://mathworld.wolfram.com (accessed on 27 July 2022).
- Freedman, M.H.; Quinn, F. Topology of 4 Manifolds; University Press: Princeton, NJ, USA, 1990; ISBN 0-691-08577-3. [Google Scholar]
- Fulton, W. Intersection Theory; Springer: Berlin/Heidelberg, Germany; New York, NY, USA, 1998; ISBN 978-0-387-98549-7. [Google Scholar]
- Navarro, A.; Navarro, J. On the Riemann-Roch formula without projective hypothesis. arXiv 2017, arXiv:170510769N. [Google Scholar]
- Hirzebruch, H. Topological Methods in Algebraic Geometry; Springer: Berlin, Germany; New York, NY, USA, 1966; ISBN 3-540-58663-6. [Google Scholar]
- Toen, B. Riemann-Roch Theorems for Deligne-Mumford Stacks. arXiv 1998, arXiv:math/9803076. [Google Scholar]
- Barrowcliffe, T.W. Viral inactivation vs. biological activity. Dev. Biol. Stand. 1993, 81, 125–135. [Google Scholar]
- Mansimov, E.; Mahmood, O.; Kang, S.; Cho, K. Molecular Geometry Prediction using a Deep Generative Graph Neural Network. Sci. Rep. 2019, 9, 20381. [Google Scholar] [CrossRef] [Green Version]
- Esfandi, B.; Atabati, M. Sequential Dihedral Angles (SDAs): A Method for Evaluating the 3D Structure of Proteins. Protein J. 2021, 40, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Pechlaner, M.; Dorta, A.P.; Lin, Z.; Rusu, V.H.; van Gunsteren, W.F. A method to apply bond-angle constraints in molecular dynamics simulations. J. Comput. Chem. 2021, 42, 418–434. [Google Scholar] [CrossRef] [PubMed]
- Altis, A.; Nguyen, P.H.; Hegger, R.; Stock, G. Dihedral angle principal component analysis of molecular dynamics simulations. J. Chem. Phys. 2007, 126, 244111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Hu, X.; Feng, Z.; Wang, S.; Sun, K.; Xu, S. Recognizing Ion Ligand-Binding Residues by Random Forest Algorithm Based on Optimized Dihedral Angle. Front. Bioeng. Biotechnol. 2020, 8, 493. [Google Scholar] [CrossRef]
- Helles, G.; Fonseca, R. Predicting dihedral angle probability distributions for protein coil residues from primary sequence using neural networks. BMC Bioinform. 2009, 10, 338. [Google Scholar] [CrossRef] [Green Version]
- Saravanan, K.; Krishnaswamy, S. Analysis of dihedral angle preferences for alanine and glycine residues in alpha and beta transmembrane regions. J. Biomol. Struct. Dyn. 2014, 33, 552–562. [Google Scholar] [CrossRef] [PubMed]
- Dayalan, S.; Gooneratne, N.D.; Bevinakoppa, S.; Schroder, H. Dihedral angle and secondary structure database of short amino acid fragments. Bioinformation 2006, 1, 78–80. [Google Scholar] [CrossRef]
- Talipov, M.R.; Navale, T.S.; Hossain, M.M.; Shukla, R.; Ivanov, M.V.; Rathore, R. Dihedral-Angle-Controlled Crossover from Static Hole Delocalization to Dynamic Hopping in Biaryl Cation Radicals. Angew. Chem. Int. Ed. Engl. 2017, 56, 266–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, H.; Singh, S.; Raghava, G.P.S. Evaluation of Protein Dihedral Angle Prediction Methods. PLoS ONE 2014, 9, e105667. [Google Scholar] [CrossRef]
- Middlemiss, R.R.; Marks, J.L. Smart. 15-4. Surfaces of Revolution. In Analytic Geometry, 3rd ed.; McGraw-Hill Book Company: New York, NY, USA, 1968; p. 378. [Google Scholar]
- Edwards, R.D.; Magee, J. “14”Technical Analysis of Stock Trends; Stock Trend Service: Springfield, MA, USA, 1948; p. 505. ISBN 1-880408-00-7. [Google Scholar]
- Cannon, K.; Ochoa, J.M.; Yeates, T. High-symmetry protein assemblies: Patterns and emerging applications. Curr. Opin. Struct. Biol. 2019, 55, 77–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balaji, S. Internal symmetry in protein structures: Prevalence, functional relevance and evolution. Curr. Opin. Struct. Biol. 2015, 32, 156–166. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Bhardwaj, V.K.; Purohit, R. Inhibition of nonstructural protein 15 of SARS CoV 2 by golden spice: A cmputational insight. Cell Biochem. Funct. 2022, 40, 926–934. [Google Scholar] [CrossRef] [PubMed]
- Rahul, S.; Vijay, K.B.; Rituraj, P. Potential of turmric-derived compounds against dependent RNA polymerase of SARs CoV 2. Comput. Biol. Med. 2021, 139, 104965. [Google Scholar]
- Baig, A.; Srinivasan, H. SARS-CoV-2 Inhibitors from Nigella Sativa. Appl. Biochem. Biotechnol. 2022, 194, 1051–1090. [Google Scholar] [CrossRef]
- Singh, S.P.; Pritam, M.; Pandey, B.; Yadav, T.P. Microstructure, pathophysiology, and potential therapeutics of COVID-19: A comprehensive review. J. Med. Virol. 2021, 93, 275–299. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Hutchinson, E.G.; Michie, A.D.; Wallace, A.C.; Jones, M.L.; Thornton, J.M. PDBsum: A Web-based database of summaries and analyses of all PDB structures. Trends Biochem. Sci. 1997, 22, 488–490. [Google Scholar] [CrossRef]
- Eren, E.; Watts, N.R.; Dearborn, A.D.; Palmer, I.W.; Kaufman, J.D.; Steven, A.C.; Wingfield, P.T. Structures of Hepatitis B Virus Core- and e-Antigen Immune Complexes Suggest Multi-point Inhibition. Structure 2018, 26, 1314. [Google Scholar] [CrossRef] [Green Version]
- Baggen, J.; Liu, Y.; Lyoo, H.; van Vliet, A.L.W.; Wahedi, M.; de Bruin, J.W.; Roberts, R.W.; Overduin, P.; Meijer, A.; Rossmann, M.G.; et al. Bypassing pan-enterovirus host factor PLA2G16. Nat. Commun. 2019, 10, 3171. [Google Scholar] [CrossRef] [Green Version]
- Singh, H.; Jakhar, R.; Sehrawat, N. Designing spike protein (S-Protein) based multi-epitope peptide vaccine against SARS COVID-19 by immunoinformatics. Heliyon 2020, 6, e05528. [Google Scholar] [CrossRef]
- Ong, E.; Wong, M.U.; Huffman, A.; He, Y. COVID-19 Coronavirus Vaccine Design Using Reverse Vaccinology and Machine Learning. Front. Immunol. 2020, 11, 1581. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, M.E. SARS-CoV-2 Proteins: Are They Useful as Targets for COVID-19 Drugs and Vaccines? Curr. Mol. Med. 2022, 22, 50–66. [Google Scholar] [CrossRef] [PubMed]
- Zierep, P.F.; Vita, R.; Blazeska, N.; Moumbock, A.F.A.; Greenbaum, J.A.; Peters, B.; Günther, S. Towards the prediction of non-peptidic epitopes. PLoS Comput. Biol. 2022, 18, e1009151. [Google Scholar] [CrossRef]
- Trolle, T.; Metushi, I.G.; Greenbaum, J.A.; Kim, Y.; Sidney, J.; Lund, O.; Sette, A.; Peters, B.; Nielsen, M. Automated benchmarking of peptide-MHC class I binding predictions. Bioinformatics 2015, 31, 2174–2181. [Google Scholar] [CrossRef] [Green Version]
- Kunik, V.; Ashkenazi, S.; Ofran, Y. Paratome: An online tool for systematic identification of antigen-binding regions in antibodies based on sequence or structure. Nucleic Acids Res. 2012, 40, W521–W524. [Google Scholar] [CrossRef] [Green Version]
- Liberis, E.; Veličković, P.; Sormanni, P.; Vendruscolo, M.; Liò, P. Parapred: Antibody paratope prediction using convolutional and recurrent neural networks. Bioinformatics 2018, 34, 2944–2950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murakami, Y.; Mizuguchi, K. Homology-based prediction of interactions between proteins using Averaged One-Dependence Estimators. BMC Bioinform. 2014, 15, 213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Garcia, J.; Valls-Comamala, V.; Guney, E.; Andreu, D.; Muñoz, F.J.; Fernandez-Fuentes, N.; Oliva, B. iFrag: A Protein–Protein Interface Prediction Server Based on Sequence Fragments. J. Mol. Biol. 2017, 429, 382–389. [Google Scholar] [CrossRef]
- Biswas, S.; Chatterjee, S.; Dey, T.; Dey, S.; Manna, S.; Nandy, A.; Basak, S.C. Insilico Aproach for peptide Vaccine Design for COVID 19. MOL2NET 2020, 6. [Google Scholar]
- Basak, S.C.; Majumdar, S.; Vracko, M.; Nandy, A.; Bhattacharjee, A. A Generic Computer Assisted Four -Pronged Approach for the Management of Emerging Global Phatogens: Some Comments on COVID 19 CCADD. Curr. Comput.-Aided Drug Des. 2020, 16, 351–353. [Google Scholar] [CrossRef]
- Zhang, Y.-F.; Ho, M. Humanization of rabbit monoclonal antibodies via grafting combined Kabat/IMGT/Paratome complementarity-determining regions: Rationale and examples. Mabs 2017, 9, 419–429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, G.B. 6.7: Area of a Surface of Revolution; 6.11: The Theorems of Pappus. In Calculus, 3rd ed.; Addison-Wesley Pub. Co.: Boston, MA, USA, 1969; pp. 206–209, 217–219. [Google Scholar]
- Bhachoo, J.; Beuming, T. Investigating Protein–Peptide Interactions Using the Schrödinger Computational Suite. Methods Mol. Biol. 2017, 1561, 235–254. [Google Scholar] [PubMed]
- Vilar, S.; Cozza, G.; Moro, S. Medicinal chemistry and the molecular operating environment (MOE): Application of QSAR and molecular docking to drug discovery. Curr. Top. Med. Chem. 2008, 8, 1555–1572. [Google Scholar] [CrossRef] [PubMed]
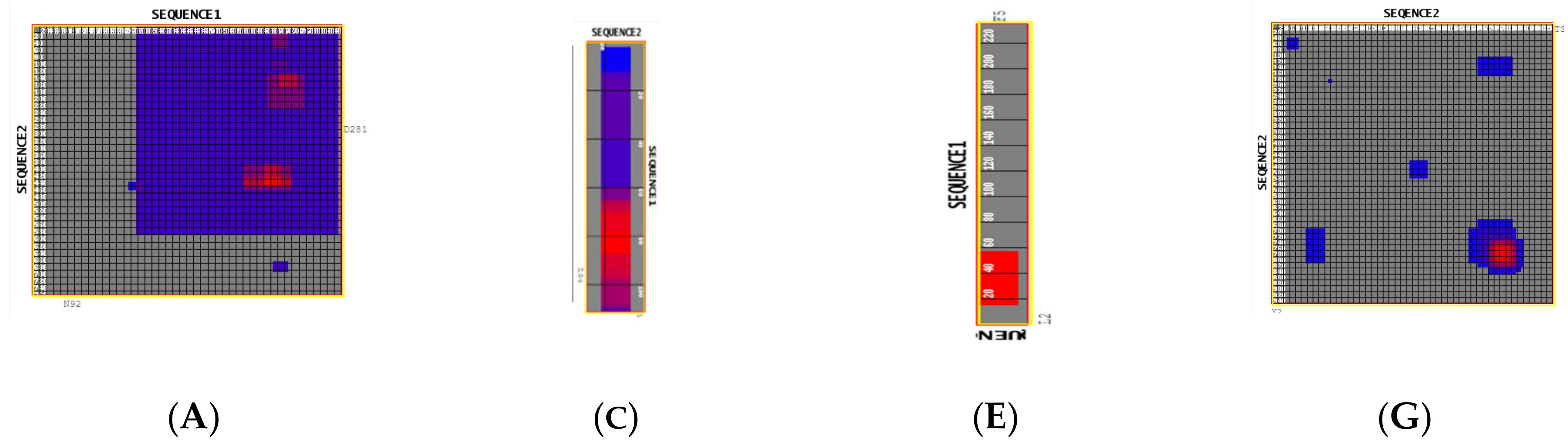
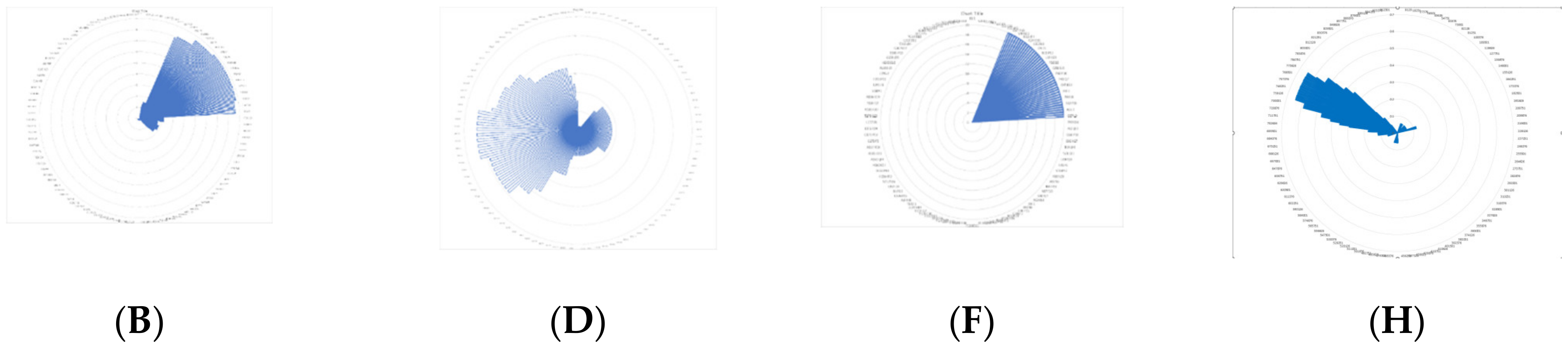

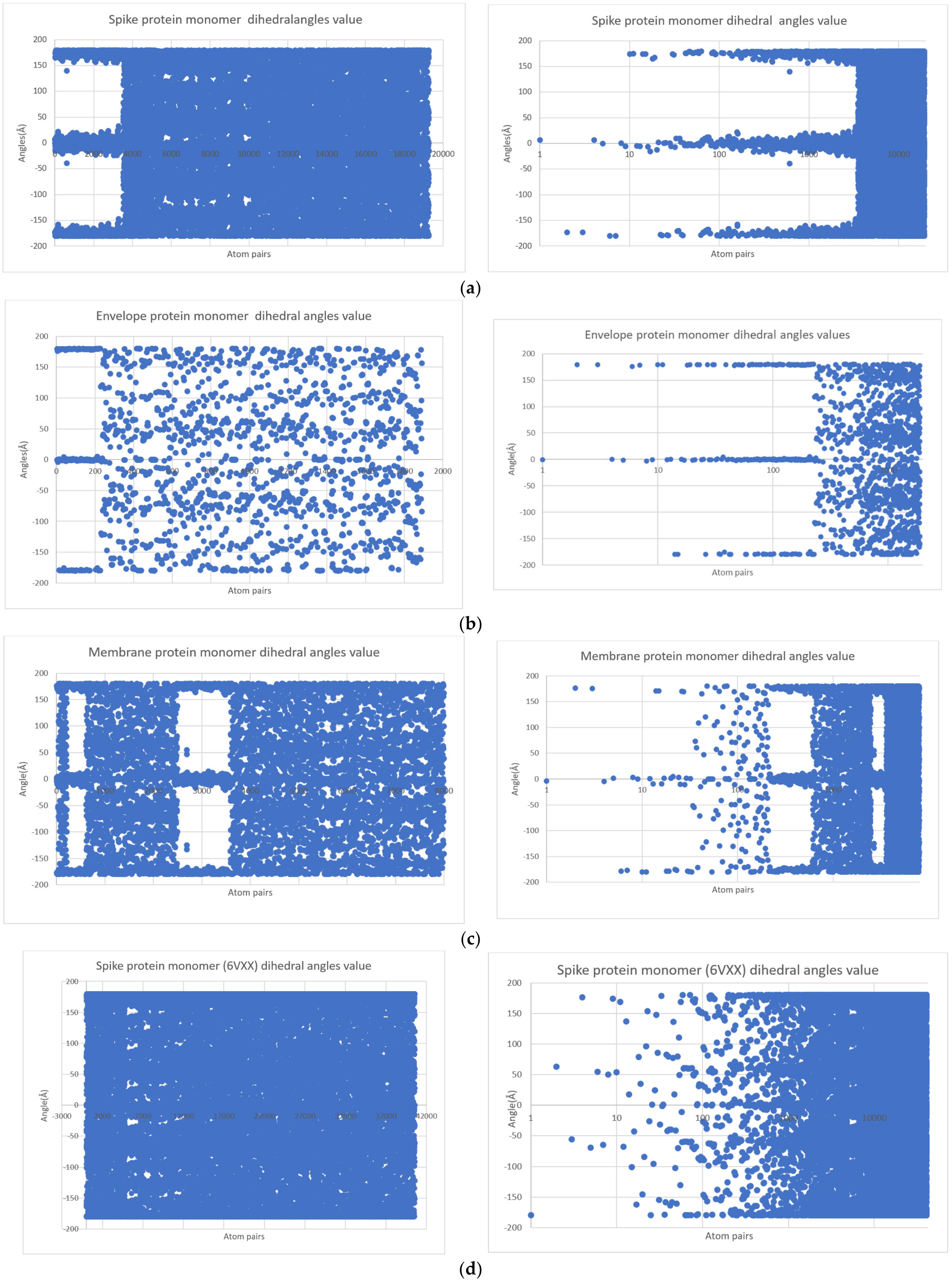
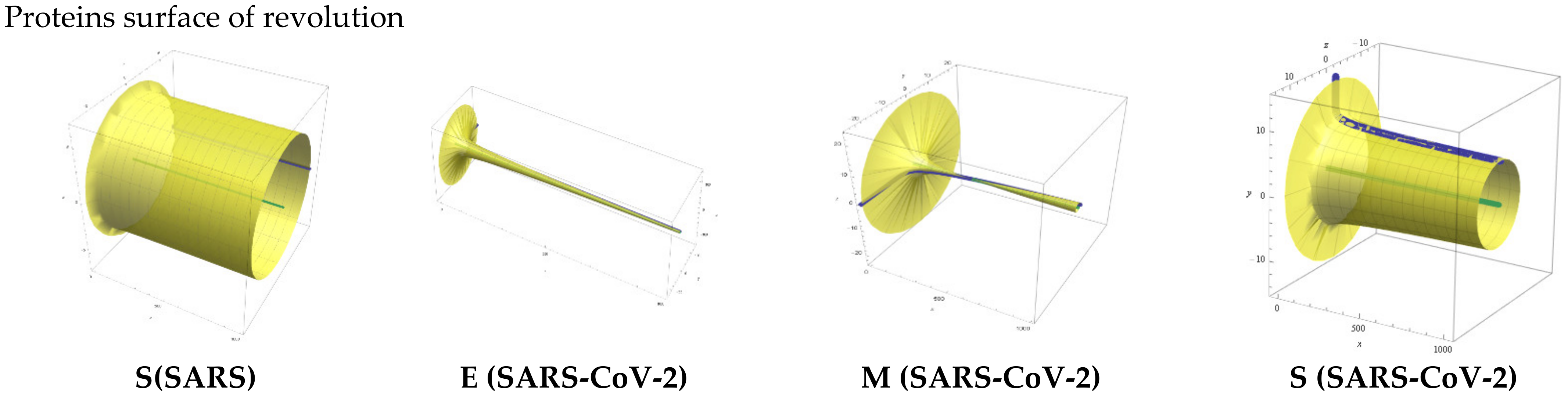

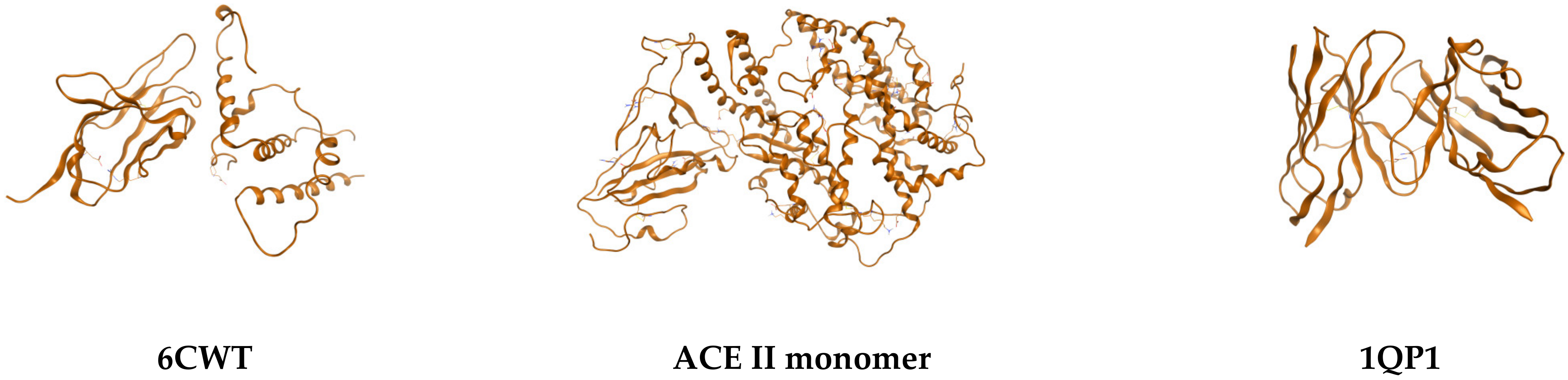
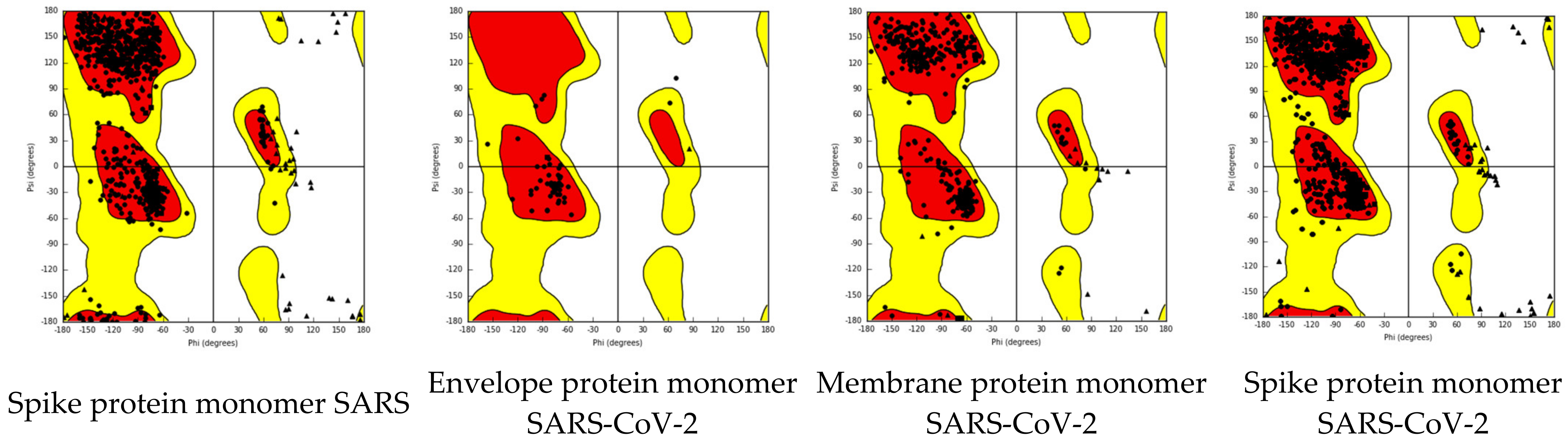
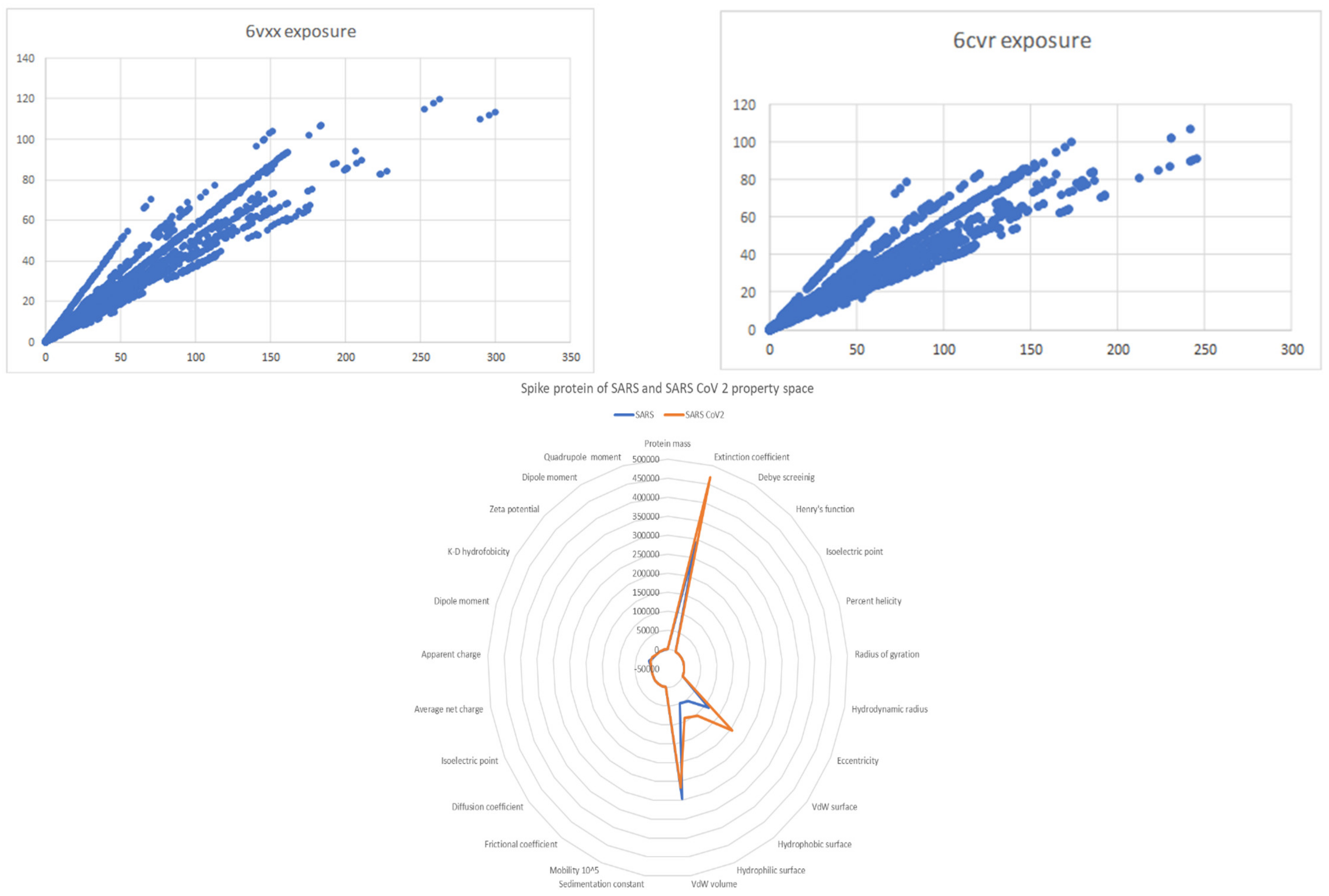
| Nr | Structure | Polynomial Eq. | Logarithmic Eq. | Polynomial Discriminant Eq. Solution | Logarithmic Eq. Solution |
|---|---|---|---|---|---|
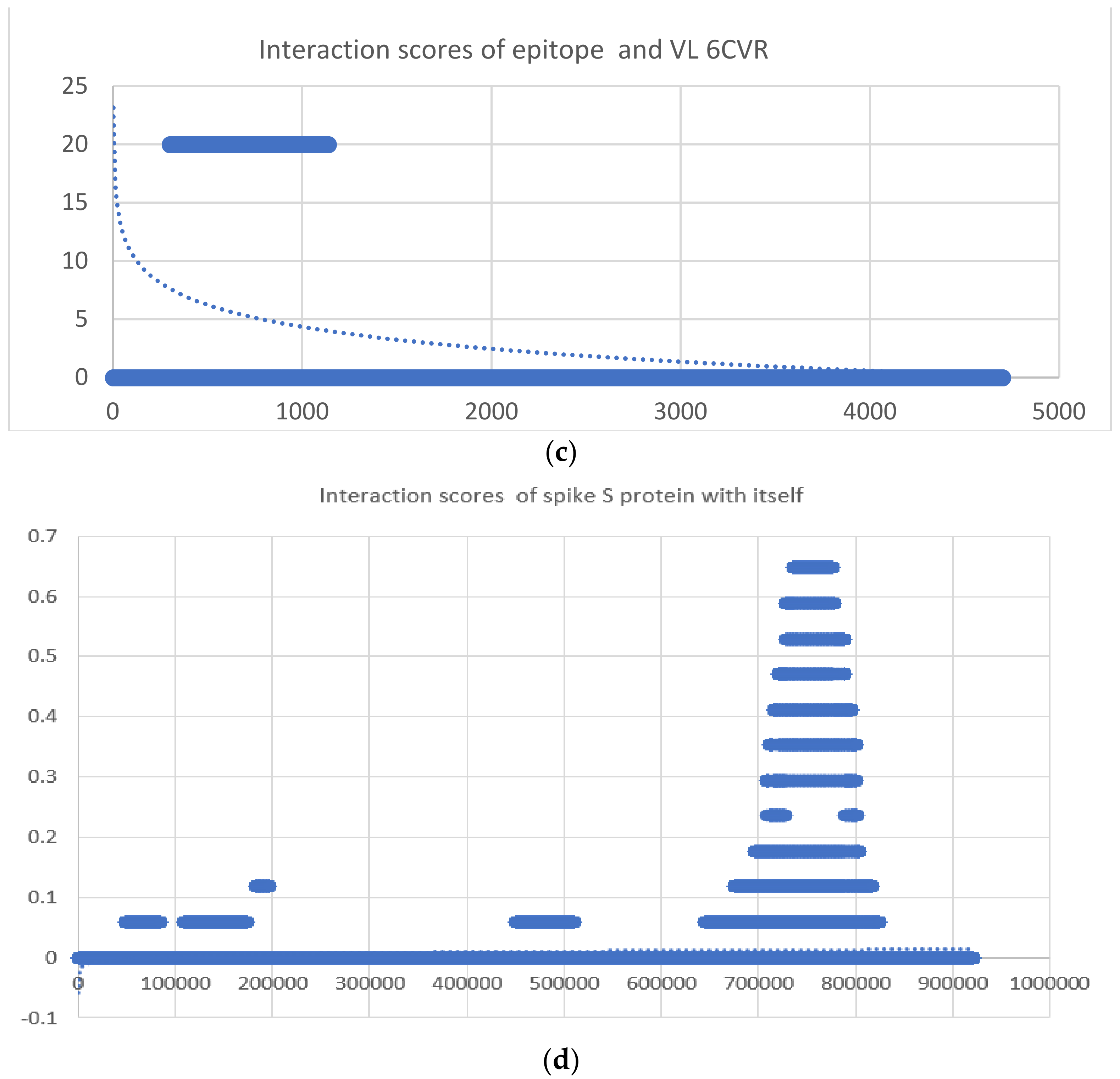
| 1 | 6cvr | y = −21x6 − 16x5 − 12x4 − 0.9x3 − 0.5x2 + 0.0226x − 2.6132 | y = −0.964 ln(x) + 8.6756 | −1.25027 × 1013 | 8099.72 |
| 2 | 5M76 | y = −19x6 − 15x5 − 12x4 − 0.8x3 − 0.6x2 − 0.0007x + 0.0118 | y = 0.2959 ln(x) − 1.43 | 1.75267 × 109 | 125.551 |
| 5M6A | y = −19x6 − 15x5 − 11x4 − 0.8x3 − 0.5x2 − 0.001x + 0.0703 | y = 0.1133 ln(x) − 0.0932 | 3.13803 × 109 | 2.2764 | |
| 4 | 5C9K | y =−19x6 − 15x5 − 12x4 − 0.9x3 − 0.6x2 + 0.0028x − 0.1209 | y = 0.1457 ln(x) − 0.2645 | −1.74887 × 1010 | 6.14337 |
| 5 | 5ACM | y = −19x6 − 16x5 − 12x4 − 0.9x3 − 0.6x2 + 0.0023x − 0.0865 | y = 0.1653 ln(x) − 0.7071 | −1.20161 × 1010 | 72.0728 |
| 6 | 4X4Z | y = −19x6 − 15x5 − 11x4 − 0.8x3 − 0.6x2 + 0.0021x + 0.4072 | y = −0.039 ln(x) + 1.4726 | 7.63376 × 1010 | 25,033,075,340,170,460 |
| 7 | 4N1C | y = −19x6 − 15x5 − 12x4 − 0.9x3 − 0.7x2 + 0.0015x + 0.0317 | y = −0.022 ln(x) + 0.4792 | 1.26141 × 1010 | 2.8822 × 109 |
| 8 | 4LVE | y = −19x6 − 15x5 − 12x4 − 0.9x3 − 0.6x2 − 0.0017x + 0.2568 | y = 0.1423 ln(x) − 0.33 | 3.61698 × 1010 | 10.166 |
| 9 | 4K07 | y = −20x6 − 16x5 − 13x4 − 1.0x3 − 0.6x2 + 0.0028x + 0.4214 | y = −0.105 ln(x) + 1.5204 | 5.92218 × 1010 | 1.9435 × 106 |
| 10 | 4K3G | y = −19×6 − 15x5 − 11x4 − 0.8x3 − 0.5x2 − 0.0022x + 0.1699 | y = 0.2815 ln(x) − 1.1125 | 7.39881 × 109 | 52.0416 |
| 11 | 4BJL | y = −19x6 − 15x5 − 12x4 − 0.8x3 − 0.5x2 − 0.0021x + 0.1734 | y = 0.0611 ln(x) − 0.0185 | 7.84012 × 109 | 1.35362 |
| 12 | 3UPA | y = −19x6 − 15x5 − 12x4 − 0.9x3 − 0.6x2 + 0.0047x + 0.3328 | y = −0.067 ln(x) + 1.6866 | 4.86836 × 1010 | 8.56157 × 1010 |
| 13 | 3T0W | y = −19x6 − 15x5 − 11x4 − 0.8x3 − 0.6x2 + 0.001x − 0.009 | y = 0.1872 ln(x) − 0.4948 | −1.39331 × 109 | 14.0576 |
| 14 | 3MCG | y = −20x6 − 17x5 − 12x4 − 0.9x3 − 0.6x2 + 0.0018x − 0.0765 | y = 0.1462 ln(x) − 0.641 | −1.241 × 1010 | 80.1905 |
| 15 | 3CDC | y = −20x6 − 16x5 − 13x4 − 0.9x3 − 0.6x2 + 0.0037x + 0.4392 | y = −0.119 ln(x) + 1.6773 | 8.86941 × 1010 | 1.3224 × 106 |
| 16 | 3BJL | y = −19x6 − 15x5 − 11x4 − 0.8x3 − 0.5x2 − 0.0045x + 0.3602 | y = 0.1276 ln(x) − 0.0371 | 1.71228 × 1010 | 1.33743 |
| 17 | 3BDX | y = −19x6 − 15x5 − 11x4 − 0.8x3 − 0.5x2 − 0.0016x + 0.1355 | y = 0.3303 ln(x) − 1.4623 | 5.93645 × 109 | 83.6957 |
| 18 | 3BD3 | y = −19x6 − 15x5 − 11x4 − 0.8x3 − 0.5x2 + 0.0059x − 0.1529 | y = 0.2444 ln(x) − 0.7981 | −7.37006 × 109 | 26.1945 |
| 19 | 3B5G | y = −19x6 − 15x5 − 11x4 − 0.8x3 − 0.6x2 − 0.0014x + 0.1304 | y = 0.3172 ln(x) − 1.4154 | 2.09459 × 1010 | 86.6753 |
| 20 | 2Q2O | y = −20x6 − 16x5 − 12x4 − 0.9x3 − 0.5x2 + 0.0062x + 0.3398 | y = −0.093 ln(x) + 1.6544 | 1.34625 × 1010 | 5.31829 × 107 |
| 21 | 2OMB | y = −19x6 − 15x5 − 12x4 − 0.8x3 − 0.6x2 − 0.5x + 0.0285 | y = 0.1881 ln(x) − 0.6922 | 2.85107 × 1012 | 39.6447 |
| 22 | 2MCG | y = −20x6 − 17x5 − 12x4 − 0.9x3 − 0.6x2 + 0.0018x − 0.0765 | y = 0.1462 ln(x) − 0.641 | −1.241 × 1010 | 80.1905 |
| 23 | 2KQN | y = − 19x6 − 15x5 − 12x4 − 0.8x3 − 0.5x2 + 0.0063x + 0.2908 | y = −0.131 ln(x) + 1.8406 | 1.4796 × 1010 | 1.26475 × 106 |
| 24 | 2KQM | y = − 20x6 − 16x5 − 13x4 − 1.1x3 − 0.6x2 + 0.001x + 0.1356 | y = −0.034 ln(x) + 0.4816 | 1.60522 × 1010 | 1.41793 × 106 |
| 25 | 1REI | y = −19x6 − 15x5 − 12x4 − 0.9x3 − 0.6x2 + 0.0047x + 0.0759 | y = −0.099 ln(x) + 1.7129 | 1.07351 × 1010 | 3.26717 × 107 |
| 26 | 1QP1 | y = −20x6 − 16x5 − 12x4 − 0.9x3 − 0.6x2 + 0.0013x + 0.161 | y = −0.053 ln(x) + 0.6762 | 2.62376 × 1010 | 347,490 |
| 27 | 1QAC | y = −20x6 − 16x5 − 12x4 − 0.9x3 − 0.6x2 − 0.0003x + 0.0601 | y = 0.0241 ln(x) + 0.0217 | 9.87188 × 109 | 0.406401 |
| 28 | 1MCW | y = −20x6 − 16x5 − 12x4 − 0.9x3 − 0.6x2 + 0.002x − 0.0665 | y = −0.006 ln(x) + 0.2463 | −1.11613 × 1010 | 672,640,970,952,404,352 |
| 29 | 1MCS | y = −19x6 − 16x5 − 13x4 − 0.9x3 − 0.6x2 + 0.0014x − 0.0533 | y = 0.1255 ln(x) − 0.5198 | −6.94971 × 109 | 62.918 |
| 30 | 1MCJ | y = −19x6 − 16x5 − 12x4 − 1.0x3 − 0.6x2 + 0.0012x − 0.0402 | y = 0.1337 ln(x) − 0.5691 | −5.368 × 109 | 70.5657 |
| 31 | 1MCI | y = −19x6 − 16x5 − 13x4 − 1.0x3 − 0.6x2 + 0.0013x − 0.0482 | y = 0.1293 ln(x) − 0.5426 | −6.26192 × 109 | 66.4495 |
| 32 | 1MCD | y = −19x6 − 16x5 − 12x4 − 1.0x3 − 0.6x2 + 0.0012x − 0.0402 | y = 0.1337 ln(x) − 0.5691 | −5.368 × 109 | 70.5657 |
| 33 | 1MCC | y = −19x6 − 16x5 − 13x4 − 0.9x3 − 0.6x2 + 0.0014x − 0.0533 | y = 0.1255 ln(x) − 0.5198 | −6.94971 × 109 | 62.918 |
| 34 | 1MAJ | y = −19x6 − 15x5 − 12x4 − 0.9x3 − 0.6x2 − 0.0005x + 0.126 | y = 0.0665 ln(x) − 0.1725 | 1.75923 × 1010 | 13.383 |
| 35 | 1LGV | y = −19x6 − 15x5 − 11x4 − 0.8x3 − 0.5x2 − 0.0018x + 0.0735 | y = 0.1481 ln(x) − 0.6409 | 3.27612 × 109 | 75.7533 |
| 36 | 1IVL | y = −20x6 − 16x5 − 12x4 − 0.9x3 − 0.7x2 + 0.0015x + 0.092 | y = 0.0212 ln(x) + 0.333 | 4.36258 × 1010 | 1.50764 × 10−7 |
| 37 | 1EK3 | y = −19x6 − 15x5 − 12x4 − 0.9x3 − 0.6x2 + 0.0002x + 0.1087 | y = 0.0677 ln(x) − 0.0314 | 1.52104 × 1010 | 1.59012 |
| 38 | 1EEU | y = −19x6 − 15x5 − 12x4 − 0.8x3 − 0.6x2 − 0.0013x + 0.1335 | y = 0.0796 ln(x) − 0.1689 | 2.13078 × 1010 | 8.34664 |
| 39 | 1EEQ | y = −19x6 − 15x5 − 12x4 − 0.8x3 − 0.6x2 − 0.0013x + 0.1344 | y = 0.098 ln(x) − 0.2921 | 2.14642 × 1010 | 19.6999 |
| 40 | 1DCL | y = −20x6 − 16x5 − 13x4 − 0.9x3 − 0.6x2 + 0.002x − 0.0963 | y = 0.1157 ln(x) − 0.4593 | −1.51605 × 1010 | 52.9713 |
| 41 | 1BWW | y = −20x6 − 16x5 − 13x4 − 1.0x3 − 0.6x2 + 0.0013x + 0.0516 | y = −0.03 ln(x) + 0.4975 | 7.65811 × 109 | 1.59239 × 107 |
| 42 | 1BJM | y = −18x6 − 14x5 − 10x4 − 0.7x3 + 0.0002x2 − 0.0464x + 2.9016 | y = −0.03 ln(x) + 1.5043 | 1.58331 × 1013 | 59,837,435,299 × 10−11 |
| 43 | 1B6D | y = −19x6 − 15x5 − 12x4 − 0.9x3 − 0.7x2 + 0.0014x + 0.1079 | y = −0.057 ln(x) + 0.7972 | 4.37707 × 1010 | 1.18584 × 106 |
| 44 | 1BOW | y = −21x6 − 17x5 − 13x4 − 1.0x3 − 0.6x2 + 0.0016x + 0.2476 | y = −0.074 ln(x) + 1.0069 | 3.80249 × 1010 | 811.595 |
| 45 | 1A8J | y = 19x6 − 16x5 − 12x4 − 0.9x3 − 0.6x2 + 0.0023x − 0.0865 | y = 0.1653 ln(x) − 0.7071 | 1.41204 × 108 | 72.0728 |
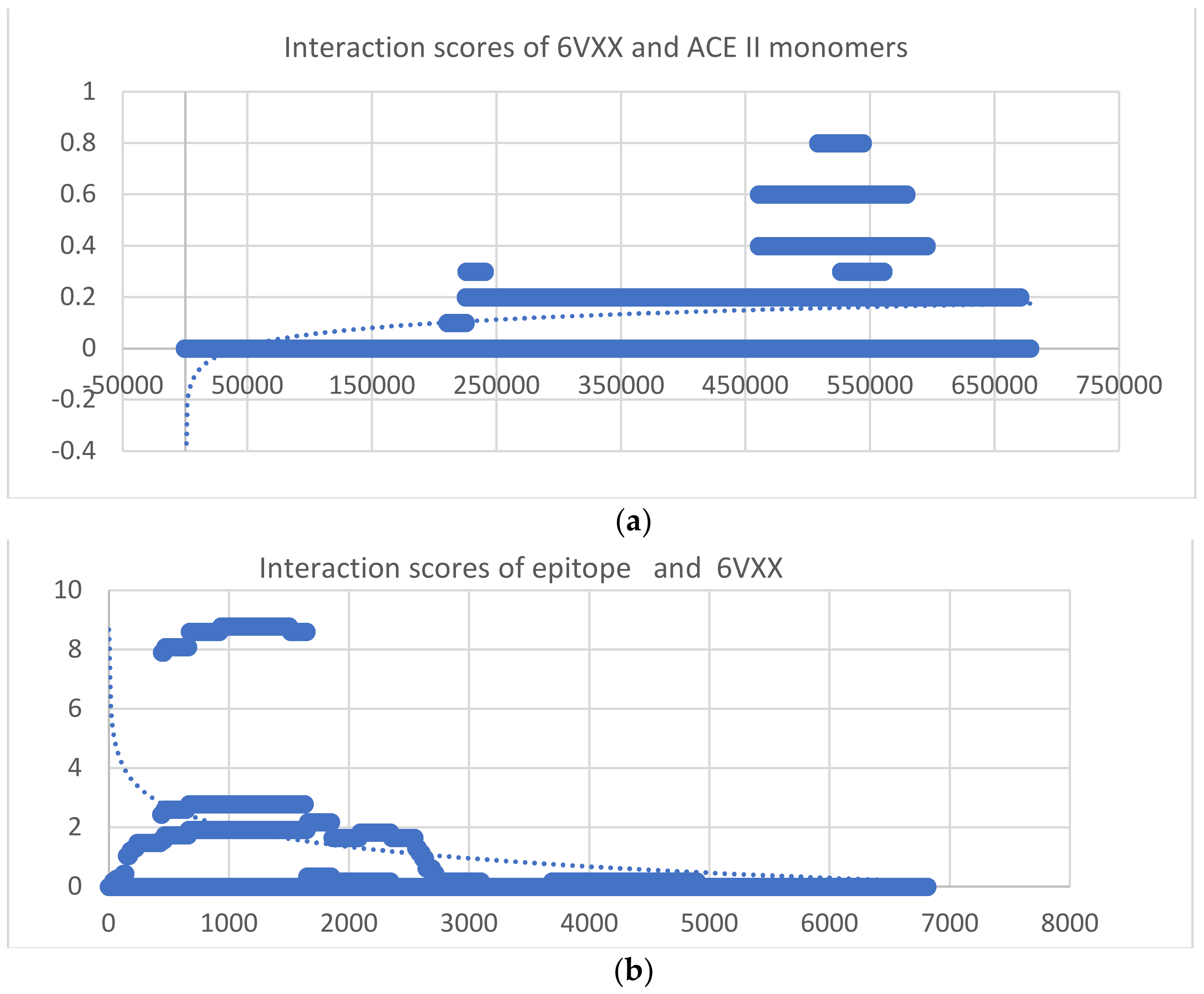
| 46 | 6VXX | y = −19x6 − 15x5 − 11x4 − 0.7x3 − 0.0002x2 + 0.119x − 10.705 | y = −2.722 ln(x) + 23.157 | −1.72453 × 1016 | 4951.01 |
| 47 | ACEII 6VXX | y = −16x6 − 12x5 − 9x4 − 0.6x3 − 0.0001x2 + 0.1x − 9.857 | y = 0.0628 ln(x)−0.668 | −4.79692 × 1015 | 41,645.3 |
| 48 | 6VXX 6VXX | y = −27x6 − 20x5 − 15x4 − 2.4x3 − 0.0001x2 +0.1x − 17.1143 | y = 0.0053 ln(x) − 0.0593 | −6.91299 × 1017 | 723,07.2 |
| ABR L1: QSIGNALA (27–34) |
| ABR L2: LLISAGSNLAS (46–56) |
| ABR L3: LGTYSAIDRA (89–98) |
| Protein Monomer | Equations |
|---|---|
| S(SARS) | y = −0.059 ln(x) + 6.1779 |
| E(SARS-CoV-2) | y = −8.42 ln(x) + 62.544 |
| M(SARTS-CoV-2) | y = 1.5101 ln(x) − 8.9329 |
| S (SARS-CoV-2) | y = −0.443 ln(x) + 9.7581 |
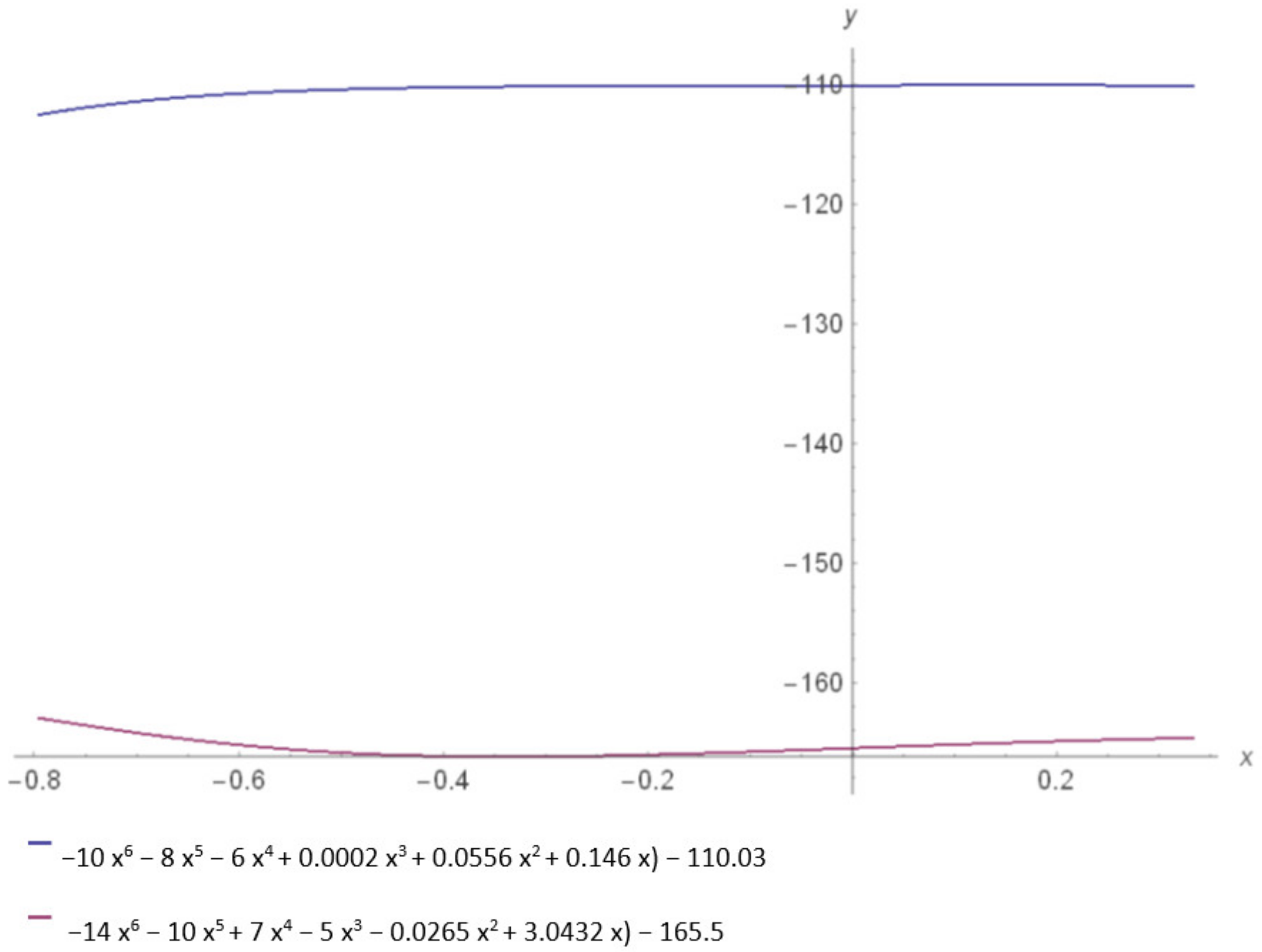
| Protein Monomer Energetically Allowed Regions | Equations |
|---|---|
| S (SARS) | y = −14x6 − 10x5 − 7x4 − 5x3 − 0.0265x2 + 3.0432x − 165.5 |
| S (SARS-CoV-2) | y= −10x6 − 8x5 − 6x4 + 0.0002x3 + 0.0556x2 + 0.146x − 110.03 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lungu, C.N.; Putz, M.V. SARS-CoV-2 Spike Protein Interaction Space. Int. J. Mol. Sci. 2023, 24, 12058. https://doi.org/10.3390/ijms241512058
Lungu CN, Putz MV. SARS-CoV-2 Spike Protein Interaction Space. International Journal of Molecular Sciences. 2023; 24(15):12058. https://doi.org/10.3390/ijms241512058
Chicago/Turabian StyleLungu, Claudiu N., and Mihai V. Putz. 2023. "SARS-CoV-2 Spike Protein Interaction Space" International Journal of Molecular Sciences 24, no. 15: 12058. https://doi.org/10.3390/ijms241512058





