Dynamic Transcriptional Landscape of Mycobacterium smegmatis under Cold Stress
Abstract
:1. Introduction
2. Results
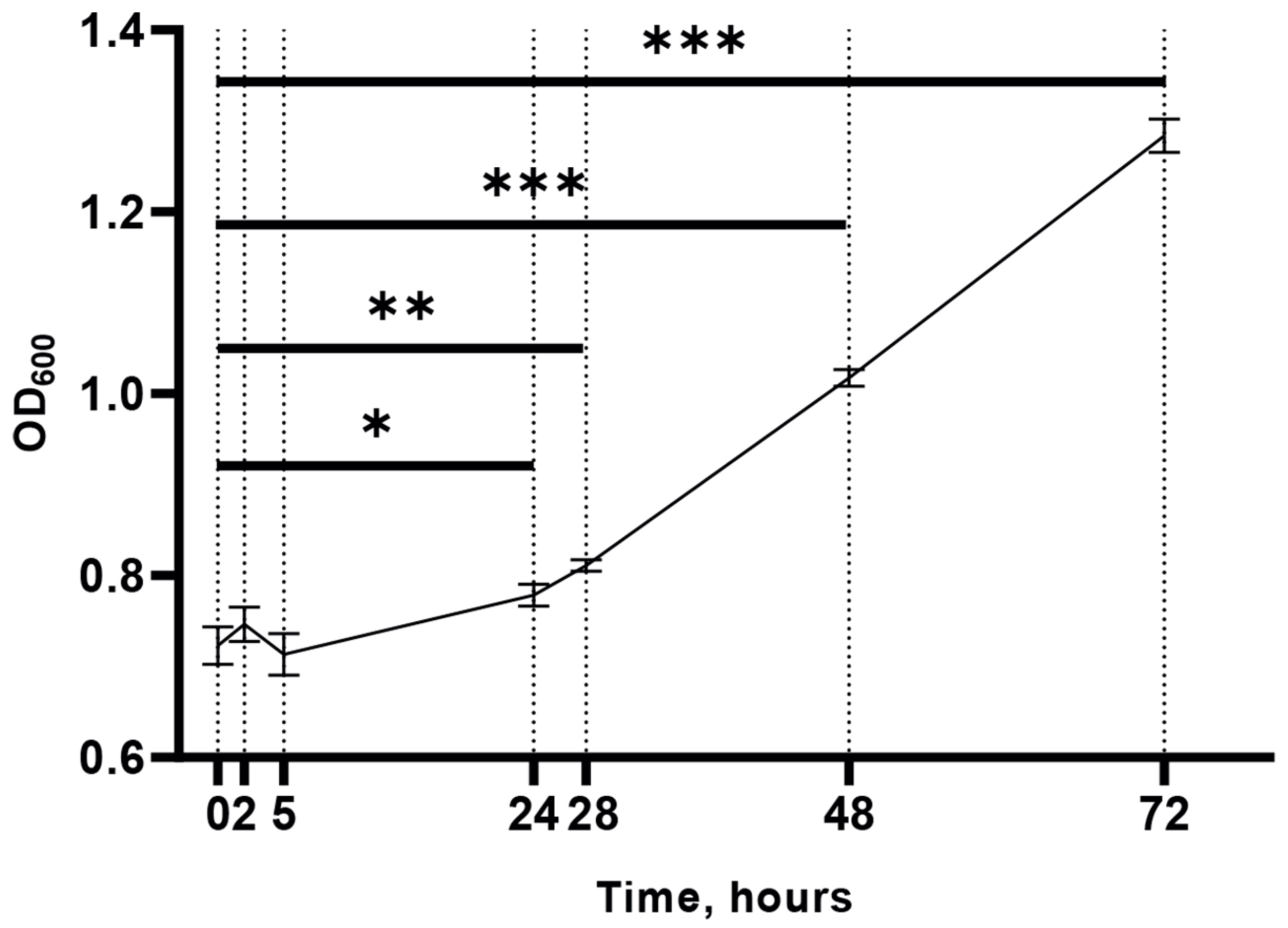
2.1. M. smegmatis Growth under Cold Stress
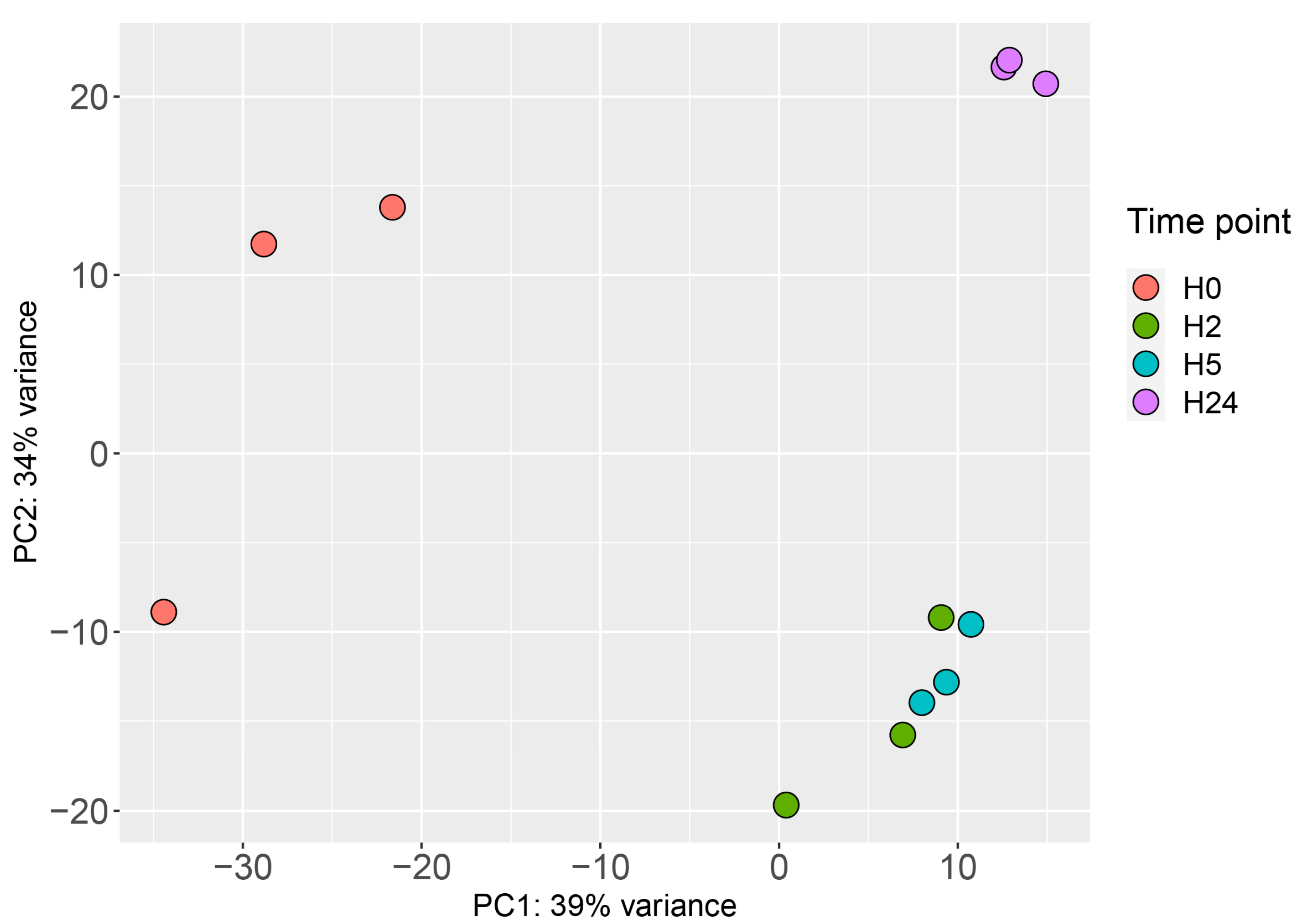
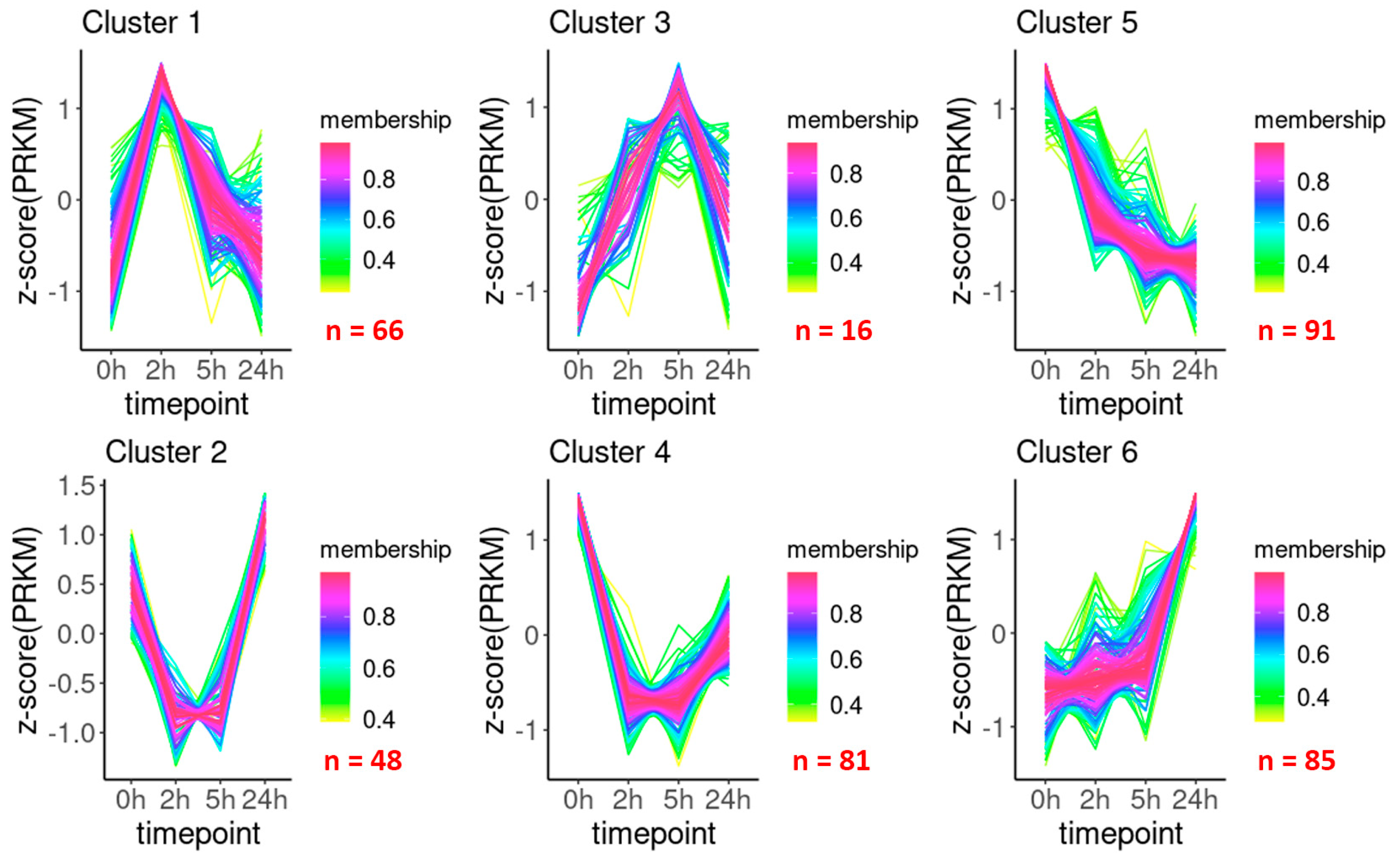
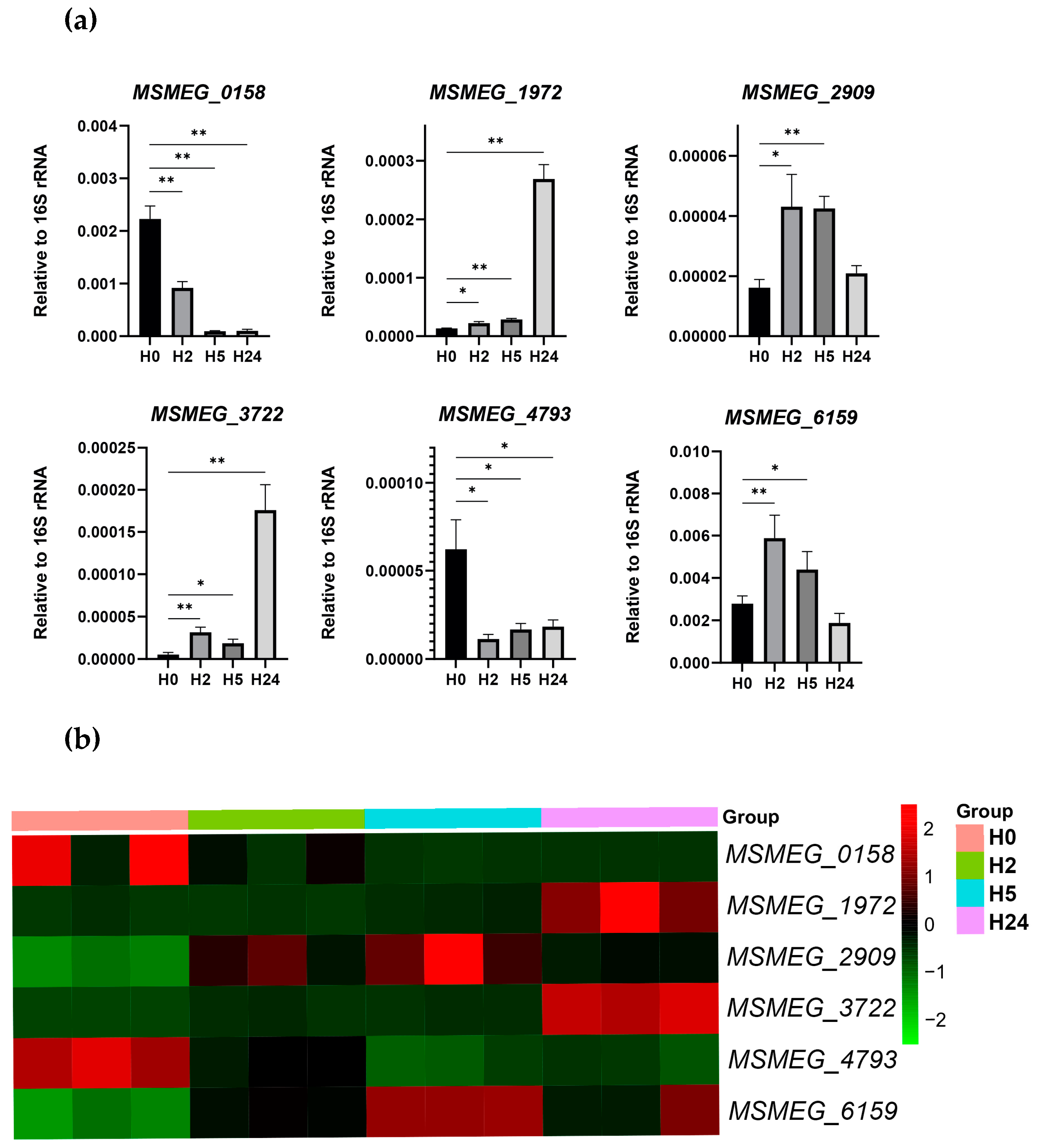
2.2. Transcriptional Landscape
2.3. Small Non-Coding RNAs
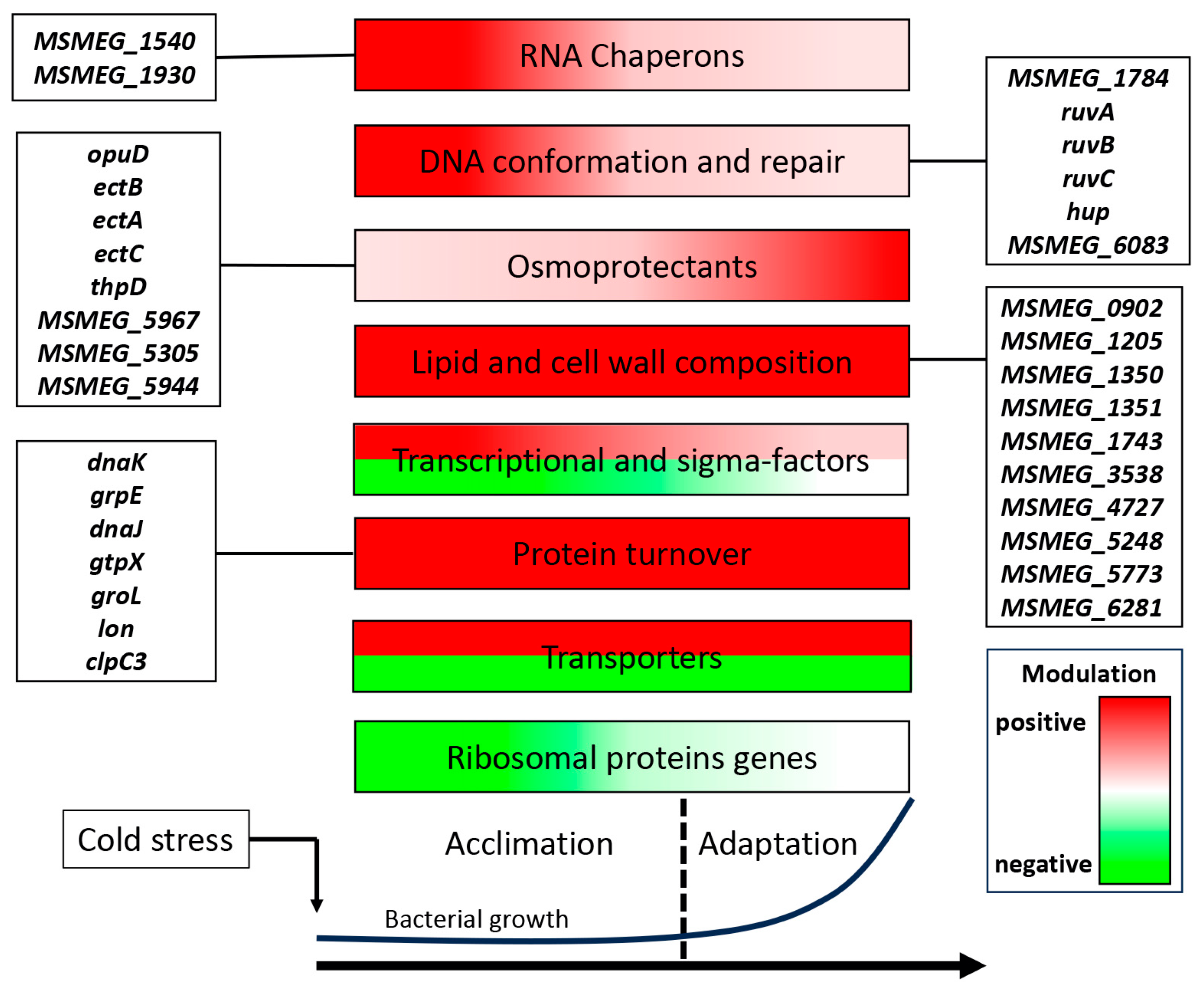
3. Discussion
3.1. RNA Chaperones
3.2. DNA Conformation and Repair Genes
3.3. Osmoprotectant (Compatible Solute) Genes
3.4. Transcriptional Regulator and Sigma Factor Genes
| Gene | Expression Pattern (Fold Change vs. H0) | Regulatory Function | ||
|---|---|---|---|---|
| H2 | H5 | H24 | ||
| MSMEG_6903 | 16.06 | 18.84 | 7.29 | Inositol metabolism [39] |
| MSMEG_0120 | 4.33 | 2.64 | N/A | Lipid metabolism and cellular redox balance [40] |
| MSMEG_5174 | 3.89 | 3.80 | 4.95 | Purine metabolism [41]; the MTB homologue (Rv1152) is strongly expressed in persisting and starved cells [42,43] |
| MSMEG_1747 (sigH7) | 3.81 | N/A | 2.03 | Response to starvation, heat shock and oxidative stress [44]; Interestingly, in a previous study, the response of this sigma factor to cold stress was not detected [44] |
| MSMEG_2694 | 3.78 | 2.17 | N/A | Redox stress; control of σH and σE regulons and Clp protease expression [45] |
| MSMEG_2794 | 2.60 | 2.20 | N/A | Biosynthesis of mycolic acids through repression of the kas-operon [46]; the MTB homolog (Rv0494) is induced by starvation and hypoxia [47,48] |
| MSMEG_6077 (carD) | 2.04 | N/A | N/A | Global transcriptional regulation in M. smegmatis and MTB; direct interaction with RNA polymerase, transcriptional control of over 350 gene; critical for stress response [49] |
| MSMEG_1995 | 2.24 | N/A | N/A | MTB homolog (Rv0165c) represses the mammalian cell entry 1 (mce1) operon [50] |
| MSMEG_0156 (oxyS) | N/A | 0.17 | 0.27 | Response to oxidative stress [51] |
| MSMEG_0937 (regX3) | 0.34 | 0.35 | N/A | Response to phosphate starvation [52] |
| MSMEG_5872 (phoP) | 0.17 | 0.27 | N/A | Response to oxidative stress and hypoxia [53] |
3.5. Translation-Related Genes
3.6. Genes Involved in Protein Turnover
3.7. Genes Related to Cell Wall and Lipid Composition
3.8. Transporter Genes
- Transporters of osmoprotectants betaine and ectoine MSMEG_1883 and MSMEG_5368, respectively, which were upregulated. This process likely reflects another pathway for the accumulation of osmoprotectants in the cell, in addition to the synthesis described earlier;
- Transporters of nutrients (amino acids, sugars, and ions), most of which were downregulated which may reflect a general slowdown in metabolic activity under cold stress—this could be a survival strategy, as slowing down metabolism reduces the energy demand and helps the cell to conserve resources;
- Efflux pumps, some of which were upregulated (multidrug efflux pump gene MSMEG_6225 and daunorubicin export gene MSMEG_6509), which might indicate an increased sensitivity to the toxic stress under cold conditions, and others downregulated, probably representing a strategy for energy conservation;
- Lipid transporters demonstrate various patterns of cold response and distributed among all six clusters, reflecting dynamic changes in M. smegmatis lipid composition during cold stress adaptation. Among them, the genes of mycobacteria-specific mce operons, which encode ABC-like lipid transporters participating in cell wall remodeling [77], were downregulated at the acclimation stage, except for MSMEG_6540, which was upregulated. As described earlier, changes in lipid composition can affect the fluidity and integrity of the cell membrane, which are crucial for maintaining cell function during temperature changes. Alterations in the spectrum of lipid transporters is an integral part of this adaptive process.
3.9. sRNAs
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
4.2. Total RNA Extraction
4.3. Isolation of sRNA Fraction
4.4. Generation of RNA-seq Libraries and Data Analyses
4.5. sRNA Annotation
4.6. Quantitative Real-Time PCR (qRT-PCR)
4.7. Data Presentation and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Barria, C.; Malecki, M.; Arraiano, C.M. Bacterial Adaptation to Cold. Microbiology 2013, 159, 2437–2443. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Gross, C.A. Cold Shock Response in Bacteria. Annu. Rev. Genet. 2021, 55, 377–400. [Google Scholar] [CrossRef]
- Ehrt, S.; Schnappinger, D. Mycobacterial Survival Strategies in the Phagosome: Defence against Host Stresses. Cell. Microbiol. 2009, 11, 1170–1178. [Google Scholar] [CrossRef] [Green Version]
- Mavi, P.S.; Singh, S.; Kumar, A. Reductive Stress: New Insights in Physiology and Drug Tolerance of Mycobacterium. Antioxid. Redox Signal. 2020, 32, 1348–1366. [Google Scholar] [CrossRef]
- Saviola, B. Mycobacterium Tuberculosis Adaptation to Survival in a Human Host. In Tuberculosis—Current Issues in Diagnosis and Management; IntechOpen: London, UK, 2013; ISBN 978-953-51-1049-1. [Google Scholar]
- Shires, K.; Steyn, L. The Cold-Shock Stress Response in Mycobacterium Smegmatis Induces the Expression of a Histone-like Protein. Mol. Microbiol. 2001, 39, 994–1009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rustad, T.R.; Roberts, D.M.; Liao, R.P.; Sherman, D.R. Isolation of Mycobacterial RNA. In Mycobacteria Protocols, 2nd ed.; Parish, T., Brown, A.C., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2009; pp. 13–22. ISBN 978-1-59745-207-6. [Google Scholar]
- Hruska, K.; Kaevska, M. Mycobacteria in Water, Soil, Plants and Air: A Review. Vet. Med. 2012, 57, 623–679. [Google Scholar] [CrossRef] [Green Version]
- Sparks, I.L.; Derbyshire, K.M.; Jacobs, W.R.; Morita, Y.S. Mycobacterium Smegmatis: The Vanguard of Mycobacterial Research. J. Bacteriol. 2023, 205, e00337-22. [Google Scholar] [CrossRef] [PubMed]
- Gray, A.N.; Koo, B.-M.; Shiver, A.L.; Peters, J.M.; Osadnik, H.; Gross, C.A. High-Throughput Bacterial Functional Genomics in the Sequencing Era. Curr. Opin Microbiol. 2015, 27, 86–95. [Google Scholar] [CrossRef] [Green Version]
- Kobras, C.M.; Fenton, A.K.; Sheppard, S.K. Next-Generation Microbiology: From Comparative Genomics to Gene Function. Genome Biol. 2021, 22, 123. [Google Scholar] [CrossRef]
- Salina, E.G.; Grigorov, A.S.; Bychenko, O.S.; Skvortsova, Y.V.; Mamedov, I.Z.; Azhikina, T.L.; Kaprelyants, A.S. Resuscitation of Dormant “Non-Culturable” Mycobacterium Tuberculosis Is Characterized by Immediate Transcriptional Burst. Front. Cell. Infect. Microbiol. 2019, 9, 272. [Google Scholar] [CrossRef] [Green Version]
- Li, S.-K.; Ng, P.K.-S.; Qin, H.; Lau, J.K.-Y.; Lau, J.P.-Y.; Tsui, S.K.-W.; Chan, T.-F.; Lau, T.C.-K. Identification of Small RNAs in Mycobacterium Smegmatis Using Heterologous Hfq. RNA 2013, 19, 74–84. [Google Scholar] [CrossRef] [Green Version]
- Tsai, C.-H.; Baranowski, C.; Livny, J.; McDonough, K.A.; Wade, J.T.; Contreras, L.M. Identification of Novel SRNAs in Mycobacterial Species. PLoS ONE 2013, 8, e79411. [Google Scholar] [CrossRef] [PubMed]
- Lamichhane, G.; Arnvig, K.B.; McDonough, K.A. Definition and Annotation of (Myco)Bacterial Non-Coding RNA. Tuberculosis (Edinb) 2013, 93, 26–29. [Google Scholar] [CrossRef]
- Pánek, J.; Krásný, L.; Bobek, J.; Ježková, E.; Korelusová, J.; Vohradský, J. The Suboptimal Structures Find the Optimal RNAs: Homology Search for Bacterial Non-Coding RNAs Using Suboptimal RNA Structures. Nucleic Acids Res. 2011, 39, 3418–3426. [Google Scholar] [CrossRef]
- Šiková, M.; Janoušková, M.; Ramaniuk, O.; Páleníková, P.; Pospíšil, J.; Bartl, P.; Suder, A.; Pajer, P.; Kubičková, P.; Pavliš, O.; et al. Ms1 RNA Increases the Amount of RNA Polymerase in Mycobacterium Smegmatis. Mol. Microbiol. 2019, 111, 354–372. [Google Scholar] [CrossRef]
- Jones, P.G.; VanBogelen, R.A.; Neidhardt, F.C. Induction of Proteins in Response to Low Temperature in Escherichia Coli. J. Bacteriol. 1987, 169, 2092–2095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Czapski, T.R.; Trun, N. Expression of Csp Genes in E. Coli K-12 in Defined Rich and Defined Minimal Media during Normal Growth, and after Cold-Shock. Gene 2014, 547, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Gualerzi, C.O.; Maria Giuliodori, A.; Pon, C.L. Transcriptional and Post-Transcriptional Control of Cold-Shock Genes. J. Mol. Biol. 2003, 331, 527–539. [Google Scholar] [CrossRef] [PubMed]
- D’Auria, G.; Esposito, C.; Falcigno, L.; Calvanese, L.; Iaccarino, E.; Ruggiero, A.; Pedone, C.; Pedone, E.; Berisio, R. Dynamical Properties of Cold Shock Protein A from Mycobacterium Tuberculosis. Biochem. Biophys. Res. Commun. 2010, 402, 693–698. [Google Scholar] [CrossRef]
- Brandi, A.; Spurio, R.; Gualerzi, C.O.; Pon, C.L. Massive Presence of the Escherichia Coli ‘Majorcold-Shock Protein’ CspA under Non-Stress Conditions. EMBO J. 1999, 18, 1653–1659. [Google Scholar] [CrossRef]
- Schwenk, S.; Arnvig, K.B. Regulatory RNA in Mycobacterium Tuberculosis, Back to Basics. Pathog. Dis. 2018, 76, fty035. [Google Scholar] [CrossRef] [PubMed]
- Beckering, C.L.; Steil, L.; Weber, M.H.W.; Völker, U.; Marahiel, M.A. Genomewide Transcriptional Analysis of the Cold Shock Response in Bacillus Subtilis. J. Bacteriol. 2002, 184, 6395–6402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, P.G.; Mitta, M.; Kim, Y.; Jiang, W.; Inouye, M. Cold Shock Induces a Major Ribosomal-Associated Protein That Unwinds Double-Stranded RNA in Escherichia Coli. Proc. Natl. Acad. Sci. USA 1996, 93, 76–80. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, T.; Kataoka, K.; Ogata, Y.; Inoue, R.; Sekimizu, K. Increase in Negative Supercoiling of Plasmid DNA in Escherichia Coli Exposed to Cold Shock. Mol. Microbiol. 1997, 23, 381–386. [Google Scholar] [CrossRef]
- Los, D.A. The Effect of Low-Temperature-Induced DNA Supercoiling on the Expression of the Desaturase Genes in Synechocystis. Cell Mol. Biol. (Noisy-le-grand) 2004, 50, 605–612. [Google Scholar]
- Panoff, J.-M.; Thammavongs, B.; Guéguen, M.; Boutibonnes, P. Cold Stress Responses in Mesophilic Bacteria. Cryobiology 1998, 36, 75–83. [Google Scholar] [CrossRef]
- Duru, I.C.; Ylinen, A.; Belanov, S.; Pulido, A.A.; Paulin, L.; Auvinen, P. Transcriptomic Time-Series Analysis of Cold- and Heat-Shock Response in Psychrotrophic Lactic Acid Bacteria. BMC Genom. 2021, 22, 28. [Google Scholar] [CrossRef]
- Donaldson, J.R.; Courcelle, C.T.; Courcelle, J. RuvABC Is Required to Resolve Holliday Junctions That Accumulate Following Replication on Damaged Templates in Escherichia Coli. J. Biol. Chem. 2006, 281, 28811–28821. [Google Scholar] [CrossRef] [Green Version]
- Kurthkoti, K.; Srinath, T.; Kumar, P.; Malshetty, V.S.; Sang, P.B.; Jain, R.; Manjunath, R.; Varshney, U. A Distinct Physiological Role of MutY in Mutation Prevention in Mycobacteria. Microbiology 2010, 156, 88–93. [Google Scholar] [CrossRef] [Green Version]
- Ofer, N.; Wishkautzan, M.; Meijler, M.; Wang, Y.; Speer, A.; Niederweis, M.; Gur, E. Ectoine Biosynthesis in Mycobacterium Smegmatis. Appl. Environ. Microbiol. 2012, 78, 7483–7486. [Google Scholar] [CrossRef] [Green Version]
- Jebbar, M.; Talibart, R.; Gloux, K.; Bernard, T.; Blanco, C. Osmoprotection of Escherichia Coli by Ectoine: Uptake and Accumulation Characteristics. J. Bacteriol. 1992, 174, 5027–5035. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Wang, Q.; Xu, W.; Liu, X.; Gao, X.; Zhang, Y. Stationary Phase-Dependent Accumulation of Ectoine Is an Efficient Adaptation Strategy in Vibrio Anguillarum against Cold Stress. Microbiol. Res. 2017, 205, 8–18. [Google Scholar] [CrossRef]
- Kuhlmann, A.U.; Bursy, J.; Gimpel, S.; Hoffmann, T.; Bremer, E. Synthesis of the Compatible Solute Ectoine in Virgibacillus Pantothenticus Is Triggered by High Salinity and Low Growth Temperature. Appl. Environ. Microbiol. 2008, 74, 4560–4563. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, T.; Bremer, E. Protection of Bacillus Subtilis against Cold Stress via Compatible-Solute Acquisition. J. Bacteriol. 2011, 193, 1552–1562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Landfald, B.; Strøm, A.R. Choline-Glycine Betaine Pathway Confers a High Level of Osmotic Tolerance in Escherichia Coli. J. Bacteriol. 1986, 165, 849–855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wargo, M.J. Homeostasis and Catabolism of Choline and Glycine Betaine: Lessons from Pseudomonas Aeruginosa. Appl. Environ. Microbiol. 2013, 79, 2112–2120. [Google Scholar] [CrossRef] [Green Version]
- Patel, N.; O’Malley, T.; Zhang, Y.-K.; Xia, Y.; Sunde, B.; Flint, L.; Korkegian, A.; Ioerger, T.R.; Sacchettini, J.; Alley, M.R.K.; et al. A Novel 6-Benzyl Ether Benzoxaborole Is Active against Mycobacterium TuberculosisIn Vitro. Antimicrob. Agents Chemother. 2017, 61, e01205-17. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.; Shee, S.; Tv, R.; Mohanraj, K.; Nagarajan, S.; Das, M.; Malhotra, N.; Sinha, V.; Thakur, C.; Rajmani, R.; et al. Biosensor-Integrated Transposon Mutagenesis Reveals Rv0158 as a Coordinator of Metabolism-Linked Redox Homeostasis in Mycobacterium Tuberculosis. 2022; in review. [Google Scholar] [CrossRef]
- Deng, W.; Zheng, Z.; Chen, Y.; Yang, M.; Yan, J.; Li, W.; Zeng, J.; Xie, J.; Gong, S.; Zeng, H. Deficiency of GntR Family Regulator MSMEG_5174 Promotes Mycobacterium Smegmatis Resistance to Aminoglycosides via Manipulating Purine Metabolism. Front. Microbiol. 2022, 13, 919538. [Google Scholar] [CrossRef] [PubMed]
- Keren, I.; Minami, S.; Rubin, E.; Lewis, K. Characterization and Transcriptome Analysis of Mycobacterium Tuberculosis Persisters. mBio 2011, 2, e00100-11. [Google Scholar] [CrossRef] [Green Version]
- Betts, J.C.; Lukey, P.T.; Robb, L.C.; McAdam, R.A.; Duncan, K. Evaluation of a Nutrient Starvation Model of Mycobacterium Tuberculosis Persistence by Gene and Protein Expression Profiling. Mol. Microbiol. 2002, 43, 717–731. [Google Scholar] [CrossRef]
- Singh, A.K.; Singh, B.N. Differential Expression of SigH Paralogs during Growth and under Different Stress Conditions in Mycobacterium Smegmatis. J. Bacteriol. 2009, 191, 2888–2893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGillivray, A.; Golden, N.A.; Gautam, U.S.; Mehra, S.; Kaushal, D. The Mycobacterium Tuberculosis Rv2745c Plays an Important Role in Responding to Redox Stress. PLoS ONE 2014, 9, e93604. [Google Scholar] [CrossRef] [PubMed]
- Biswas, R.K.; Dutta, D.; Tripathi, A.; Feng, Y.; Banerjee, M.; Singh, B.N. Identification and Characterization of Rv0494: A Fatty Acid-Responsive Protein of the GntR/FadR Family from Mycobacterium Tuberculosis. Microbiology 2013, 159, 913–923. [Google Scholar] [CrossRef] [Green Version]
- Galagan, J.E.; Minch, K.; Peterson, M.; Lyubetskaya, A.; Azizi, E.; Sweet, L.; Gomes, A.; Rustad, T.; Dolganov, G.; Glotova, I.; et al. The Mycobacterium Tuberculosis Regulatory Network and Hypoxia. Nature 2013, 499, 178–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yousuf, S.; Angara, R.; Vindal, V.; Ranjan, A. Rv0494 Is a Starvation-Inducible, Auto-Regulatory FadR-like Regulator from Mycobacterium Tuberculosis. Microbiology 2015, 161, 463–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stallings, C.L.; Stephanou, N.C.; Chu, L.; Hochschild, A.; Nickels, B.E.; Glickman, M.S. CarD Is an Essential Regulator of RRNA Transcription Required for Mycobacterium Tuberculosis Persistence. Cell 2009, 138, 146–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uchida, Y.; Casali, N.; White, A.; Morici, L.; Kendall, L.V.; Riley, L.W. Accelerated Immunopathological Response of Mice Infected with Mycobacterium Tuberculosis Disrupted in the Mce1 Operon Negative Transcriptional Regulator. Cell. Microbiol. 2007, 9, 1275–1283. [Google Scholar] [CrossRef]
- Daugherty, A.; Powers, K.M.; Standley, M.S.; Kim, C.S.; Purdy, G.E. Mycobacterium Smegmatis RoxY Is a Repressor of OxyS and Contributes to Resistance to Oxidative Stress and Bactericidal Ubiquitin-Derived Peptides. J. Bacteriol. 2011, 193, 6824–6833. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; You, D.; Ye, B.-C. RegX3 Controls Glyoxylate Shunt and Mycobacteria Survival by Directly Regulating the Transcription of Isocitrate Lyase Gene in Mycobacterium Smegmatis. ACS Infect. Dis. 2021, 7, 927–936. [Google Scholar] [CrossRef]
- Asensio, J.G.; Maia, C.; Ferrer, N.L.; Barilone, N.; Laval, F.; Soto, C.Y.; Winter, N.; Daffé, M.; Gicquel, B.; Martín, C.; et al. The Virulence-Associated Two-Component PhoP-PhoR System Controls the Biosynthesis of Polyketide-Derived Lipids in Mycobacterium Tuberculosis*. J. Biol. Chem. 2006, 281, 1313–1316. [Google Scholar] [CrossRef] [Green Version]
- Hümpel, A.; Gebhard, S.; Cook, G.M.; Berney, M. The SigF Regulon in Mycobacterium Smegmatis Reveals Roles in Adaptation to Stationary Phase, Heat, and Oxidative Stress. J. Bacteriol. 2010, 192, 2491–2502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, A.K.; Dutta, D.; Singh, V.; Srivastava, V.; Biswas, R.K.; Singh, B.N. Characterization of Mycobacterium Smegmatis SigF Mutant and Its Regulon: Overexpression of SigF Antagonist (MSMEG_1803) in M. Smegmatis Mimics SigF Mutant Phenotype, Loss of Pigmentation, and Sensitivity to Oxidative Stress. Microbiologyopen 2015, 4, 896–916. [Google Scholar] [CrossRef] [Green Version]
- Xia, B.; Ke, H.; Shinde, U.; Inouye, M. The Role of RbfA in 16S RRNA Processing and Cell Growth at Low Temperature in Escherichia Coli. J. Mol. Biol. 2003, 332, 575–584. [Google Scholar] [CrossRef] [PubMed]
- Charollais, J.; Dreyfus, M.; Iost, I. CsdA, a Cold-Shock RNA Helicase from Escherichia Coli, Is Involved in the Biogenesis of 50S Ribosomal Subunit. Nucleic Acids Res. 2004, 32, 2751–2759. [Google Scholar] [CrossRef] [PubMed]
- Giuliodori, A.M.; Fabbretti, A.; Gualerzi, C. Cold-Responsive Regions of Paradigm Cold-Shock and Non-Cold-Shock MRNAs Responsible for Cold Shock Translational Bias. Int. J. Mol. Sci. 2019, 20, 457. [Google Scholar] [CrossRef] [PubMed]
- Giuliodori, A.M.; Brandi, A.; Gualerzi, C.O.; Pon, C.L. Preferential Translation of Cold-Shock MRNAs during Cold Adaptation. RNA 2004, 10, 265–276. [Google Scholar] [CrossRef] [Green Version]
- Brandi, A.; Piersimoni, L.; Feto, N.A.; Spurio, R.; Alix, J.-H.; Schmidt, F.; Gualerzi, C.O. Translation Initiation Factor IF2 Contributes to Ribosome Assembly and Maturation during Cold Adaptation. Nucleic Acids Res. 2019, 47, 4652–4662. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Sharma, M.R.; Koripella, R.K.; Yang, Y.; Kaushal, P.S.; Lin, Q.; Wade, J.T.; Gray, T.A.; Derbyshire, K.M.; Agrawal, R.K.; et al. Zinc Depletion Induces Ribosome Hibernation in Mycobacteria. Proc. Natl. Acad. Sci. USA 2018, 115, 8191–8196. [Google Scholar] [CrossRef] [Green Version]
- Trauner, A.; Lougheed, K.E.A.; Bennett, M.H.; Hingley-Wilson, S.M.; Williams, H.D. The Dormancy Regulator DosR Controls Ribosome Stability in Hypoxic Mycobacteria. J. Biol. Chem. 2012, 287, 24053–24063. [Google Scholar] [CrossRef] [Green Version]
- Lang, M.; Krin, E.; Korlowski, C.; Sismeiro, O.; Varet, H.; Coppée, J.-Y.; Mazel, D.; Baharoglu, Z. Sleeping Ribosomes: Bacterial Signaling Triggers RaiA Mediated Persistence to Aminoglycosides. iScience 2021, 24, 103128. [Google Scholar] [CrossRef]
- Kumar, S.; Suyal, D.C.; Yadav, A.; Shouche, Y.; Goel, R. Psychrophilic Pseudomonas Helmanticensis Proteome under Simulated Cold Stress. Cell Stress Chaperones 2020, 25, 1025–1032. [Google Scholar] [CrossRef] [PubMed]
- Suyal, D.C.; Kumar, S.; Yadav, A.; Shouche, Y.; Goel, R. Cold Stress and Nitrogen Deficiency Affected Protein Expression of Psychrotrophic Dyadobacter Psychrophilus B2 and Pseudomonas Jessenii MP1. Front. Microbiol. 2017, 8, 430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Griffiths, M.W. Induced Expression of the Heat Shock Protein Genes UspA and GrpE during Starvation at Low Temperatures and Their Influence on Thermal Resistance of Escherichia Coli O157:H7. J. Food Prot. 2003, 66, 2045–2050. [Google Scholar] [CrossRef] [PubMed]
- Porankiewicz, J.; Clarke, A.K. Induction of the Heat Shock Protein ClpB Affects Cold Acclimation in the Cyanobacterium Synechococcus Sp. Strain PCC 7942. J. Bacteriol. 1997, 179, 5111–5117. [Google Scholar] [CrossRef] [Green Version]
- Chattopadhyay, M.K.; Jagannadham, M.V. A Branched Chain Fatty Acid Promotes Cold Adaptation in Bacteria. J. Biosci. 2003, 28, 363–364. [Google Scholar] [CrossRef] [PubMed]
- Shivaji, S.; Prakash, J.S.S. How Do Bacteria Sense and Respond to Low Temperature? Arch. Microbiol. 2010, 192, 85–95. [Google Scholar] [CrossRef]
- Alibaud, L.; Alahari, A.; Trivelli, X.; Ojha, A.K.; Hatfull, G.F.; Guerardel, Y.; Kremer, L. Temperature-Dependent Regulation of Mycolic Acid Cyclopropanation in Saprophytic Mycobacteria: Role of the mycobacterium smegmatis 1351 gene (msmeg_1351) in cis-cyclopropanation of α-mycolates. J. Biol. Chem. 2010, 285, 21698–21707. [Google Scholar] [CrossRef] [Green Version]
- Montanari, C.; Sado Kamdem, S.L.; Serrazanetti, D.I.; Etoa, F.-X.; Guerzoni, M.E. Synthesis of Cyclopropane Fatty Acids in Lactobacillus Helveticus and Lactobacillus Sanfranciscensis and Their Cellular Fatty Acids Changes Following Short Term Acid and Cold Stresses. Food Microbiol. 2010, 27, 493–502. [Google Scholar] [CrossRef]
- Vidovic, S.; Mangalappalli-Illathu, A.K.; Korber, D.R. Prolonged Cold Stress Response of Escherichia Coli O157 and the Role of RpoS. Int. J. Food Microbiol. 2011, 146, 163–169. [Google Scholar] [CrossRef]
- Russell, N.J. Membrane Components and Cold Sensing. In Psychrophiles: From Biodiversity to Biotechnology; Margesin, R., Schinner, F., Marx, J.-C., Gerday, C., Eds.; Springer: Berlin/Heidelberg, Germany, 2008; pp. 177–190. ISBN 978-3-540-74335-4. [Google Scholar]
- Kumar, A.; Kumar, S.; Kumar, D.; Mishra, A.; Dewangan, R.P.; Shrivastava, P.; Ramachandran, S.; Taneja, B. The Structure of Rv3717 Reveals a Novel Amidase from Mycobacterium Tuberculosis. Acta Crystallogr. D Biol. Crystallogr. 2013, 69, 2543–2554. [Google Scholar] [CrossRef] [Green Version]
- Kołodziej, M.; Łebkowski, T.; Płociński, P.; Hołówka, J.; Paściak, M.; Wojtaś, B.; Bury, K.; Konieczny, I.; Dziadek, J.; Zakrzewska-Czerwińska, J. Lsr2 and Its Novel Paralogue Mediate the Adjustment of Mycobacterium Smegmatis to Unfavorable Environmental Conditions. mSphere 2021, 6, e00290-21. [Google Scholar] [CrossRef] [PubMed]
- Elbourne, L.D.H.; Tetu, S.G.; Hassan, K.A.; Paulsen, I.T. TransportDB 2.0: A Database for Exploring Membrane Transporters in Sequenced Genomes from All Domains of Life. Nucleic Acids Res. 2017, 45, D320–D324. [Google Scholar] [CrossRef] [PubMed]
- Klepp, L.I.; Sabio y Garcia, J. FabianaBigi Mycobacterial MCE Proteins as Transporters That Control Lipid Homeostasis of the Cell Wall. Tuberculosis 2022, 132, 102162. [Google Scholar] [CrossRef] [PubMed]
- Gierga, G.; Voss, B.; Hess, W.R. Non-Coding RNAs in Marine Synechococcus and Their Regulation under Environmentally Relevant Stress Conditions. ISME J. 2012, 6, 1544–1557. [Google Scholar] [CrossRef] [PubMed]
- Majdalani, N.; Cunning, C.; Sledjeski, D.; Elliott, T.; Gottesman, S. DsrA RNA Regulates Translation of RpoS Message by an Anti-Antisense Mechanism, Independent of Its Action as an Antisilencer of Transcription. Proc. Natl. Acad. Sci. USA 1998, 95, 12462–12467. [Google Scholar] [CrossRef]
- Sledjeski, D.; Gottesman, S. A Small RNA Acts as an Antisilencer of the H-NS-Silenced RcsA Gene of Escherichia Coli. Proc. Natl. Acad. Sci. USA 1995, 92, 2003–2007. [Google Scholar] [CrossRef] [PubMed]
- Hnilicová, J.; Jirát Matějčková, J.; Šiková, M.; Pospíšil, J.; Halada, P.; Pánek, J.; Krásný, L. Ms1, a Novel SRNA Interacting with the RNA Polymerase Core in Mycobacteria. Nucleic Acids Res. 2014, 42, 11763–11776. [Google Scholar] [CrossRef] [Green Version]
- Babraham Bioinformatics—FastQC. A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 8 May 2023).
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A Web Server for Functional Enrichment Analysis and Functional Annotation of Gene Lists (2021 Update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef] [PubMed]
- McClure, R.; Balasubramanian, D.; Sun, Y.; Bobrovskyy, M.; Sumby, P.; Genco, C.A.; Vanderpool, C.K.; Tjaden, B. Computational Analysis of Bacterial RNA-Seq Data. Nucleic Acids Res. 2013, 41, e140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, M.; Gu, L. TCseq: Time Course Sequencing Data Analysis; R Package Version 1.23.0. 2022. Available online: https://bioconductor.statistik.tu-dortmund.de/packages/3.12/bioc/vignettes/TCseq/inst/doc/TCseq.pdf(accessed on 3 May 2023).
- Blighe, K.; Rana, S.; Lewis, M. EnhancedVolcano: Publication-Ready Volcano Plots with Enhanced Colouring and Labeling, R Package Version 1.18.0. 2023. Available online: https://github.com/kevinblighe/EnhancedVolcano (accessed on 3 May 2023).
- Gruber, A.R.; Bernhart, S.H.; Lorenz, R. The ViennaRNA Web Services. Methods Mol. Biol. 2015, 1269, 307–326. [Google Scholar] [CrossRef]
- Darty, K.; Denise, A.; Ponty, Y. VARNA: Interactive Drawing and Editing of the RNA Secondary Structure. Bioinformatics 2009, 25, 1974–1975. [Google Scholar] [CrossRef] [PubMed] [Green Version]

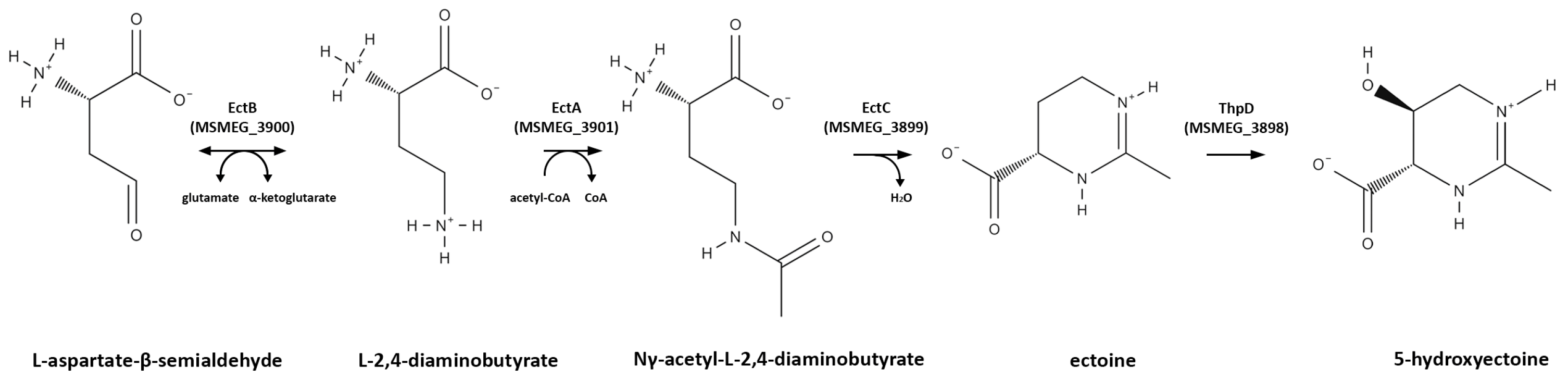
| Gene | Process | Expression Pattern (Fold Change vs. H0) | Gene Product | ||
|---|---|---|---|---|---|
| H2 | H5 | H24 | |||
| MSMEG_1540 | RNA chaperones | 9.71 | 2.04 | 3.49 | ATP-dependent RNA helicase |
| MSMEG_1930 | 5.01 | 2.52 | N/A | DEAD/DEAH box helicase | |
| MSMEG_6159 (CspA) | 2.14 | 3.41 | 2.31 | Putative cold shock protein A | |
| MSMEG_1784 | DNA conformation and repair | 4.68 | N/A | N/A | Type I topoisomerase |
| MSMEG_2389 (hup) | 1.54 | 2.39 | N/A | DNA-binding protein HU | |
| MSMEG_2943 (ruvC) | 2.47 | 1.79 | N/A | Crossover junction endodeoxyribonuclease | |
| MSMEG_2944 (ruvA) | 3.39 | 2.20 | N/A | Holliday junction DNA helicase | |
| MSMEG_2945 (ruvB) | 2.47 | 1.91 | N/A | Holliday junction DNA helicase | |
| MSMEG_6083 | N/A | 3.10 | 4.41 | Base excision DNA repair protein | |
| MSMEG_1883 (opuD) | Osmoprotection | 2.85 | 2.79 | N/A | Glycine betaine transporter |
| MSMEG_3898 (thpD) | 5.45 | 3.62 | 7.29 | Ectoine hydroxylase (ectoine biosynthesis) | |
| MSMEG_3899 (ectC) | 4.11 | 2.46 | 7.68 | Ectoine synthase (ectoine biosynthesis) | |
| MSMEG_3900 (ectB) | 4.03 | 2.39 | 6.05 | Diaminobutyrate--2-oxoglutarate aminotransferase (ectoine biosynthesis) | |
| MSMEG_3901 (ectA) | 2.72 | N/A | 2.88 | L-2,4-diaminobutyric acid acetyltransferase (ectoine biosynthesis) | |
| MSMEG_5967 | 5.36 | 2.82 | 2.26 | Glucose-methanol-choline oxidoreductase (choline metabolism) | |
| MSMEG_5305 | 4.66 | 3.98 | 14.09 | Choline dehydrogenase (choline metabolism) | |
| MSMEG_5944 | 3.36 | N/A | N/A | Glucose-methanol-choline oxidoreductase (choline metabolism) | |
| MSMEG_1878 | Translation | 3.09 | 3.05 | 5.97 | Mycobacteria-specific protein Y |
| MSMEG_0709 (dnaK) | Protein turnover | 2.96 | 2.26 | 2.67 | Chaperone |
| MSMEG_0710 (grpE) | 2.47 | 2.08 | 2.22 | Co-chaperone | |
| MSMEG_0711 (dnaJ) | 2.70 | 2.77 | 2.46 | Chaperone | |
| MSMEG_1134 (htpX) | 3.99 | 1.81 | N/A | Putative protease | |
| MSMEG_1978 (groL) | 4.57 | 2.29 | 13.16 | Chaperonin | |
| MSMEG_3582 (lon) | 5.21 | 2.60 | N/A | ATP-dependent protease La | |
| MSMEG_3761 (clpC3) | 10.94 | 4.57 | 4.31 | Clp protease subunit | |
| MSMEG_0902 | Cell wall and lipid composition | N/A | 2.67 | 2.70 | Cyclopropane-fatty-acyl-phospholipid synthase 1 (modification of mycolic acids) |
| MSMEG_1205 | 16.02 | 10.54 | 4.50 | ||
| MSMEG_1350 | N/A | 1.83 | 2.75 | ||
| MSMEG_1351 | 2.75 | 4.53 | 9.26 | ||
| MSMEG_1743 | 5.45 | N/A | N/A | Fatty acid desaturase (desaturation of fatty acids) | |
| MSMEG_3538 | 3.61 | 2.08 | 2.54 | Cyclopropane-fatty-acyl-phospholipid synthase 1 (modification of mycolic acids) | |
| MSMEG_4727 | N/A | 2.01 | 4.86 | Mycocerosic acid synthase (synthesis of lipooligosaccharides) | |
| MSMEG_5248 | 2.12 | N/A | 1.68 | Acyl-[ACP] desaturase (desaturation of fatty acids) | |
| MSMEG_5773 | 3.90 | 3.01 | 3.37 | Fatty acid desaturase (desaturation of fatty acids) | |
| MSMEG_6281 | N/A | 2.15 | 4.60 | N-acetylmuramoyl-L-alanine amidase (cell wall hydrolase) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grigorov, A.S.; Skvortsova, Y.V.; Bychenko, O.S.; Aseev, L.V.; Koledinskaya, L.S.; Boni, I.V.; Azhikina, T.L. Dynamic Transcriptional Landscape of Mycobacterium smegmatis under Cold Stress. Int. J. Mol. Sci. 2023, 24, 12706. https://doi.org/10.3390/ijms241612706
Grigorov AS, Skvortsova YV, Bychenko OS, Aseev LV, Koledinskaya LS, Boni IV, Azhikina TL. Dynamic Transcriptional Landscape of Mycobacterium smegmatis under Cold Stress. International Journal of Molecular Sciences. 2023; 24(16):12706. https://doi.org/10.3390/ijms241612706
Chicago/Turabian StyleGrigorov, Artem S., Yulia V. Skvortsova, Oksana S. Bychenko, Leonid V. Aseev, Ludmila S. Koledinskaya, Irina V. Boni, and Tatyana L. Azhikina. 2023. "Dynamic Transcriptional Landscape of Mycobacterium smegmatis under Cold Stress" International Journal of Molecular Sciences 24, no. 16: 12706. https://doi.org/10.3390/ijms241612706





