Deletion of the chd7 Hinders Oligodendrocyte Progenitor Cell Development and Myelination in Zebrafish
Abstract
:1. Introduction
2. Results
2.1. Developmental Expression and Characterization of chd7 in Zebrafish
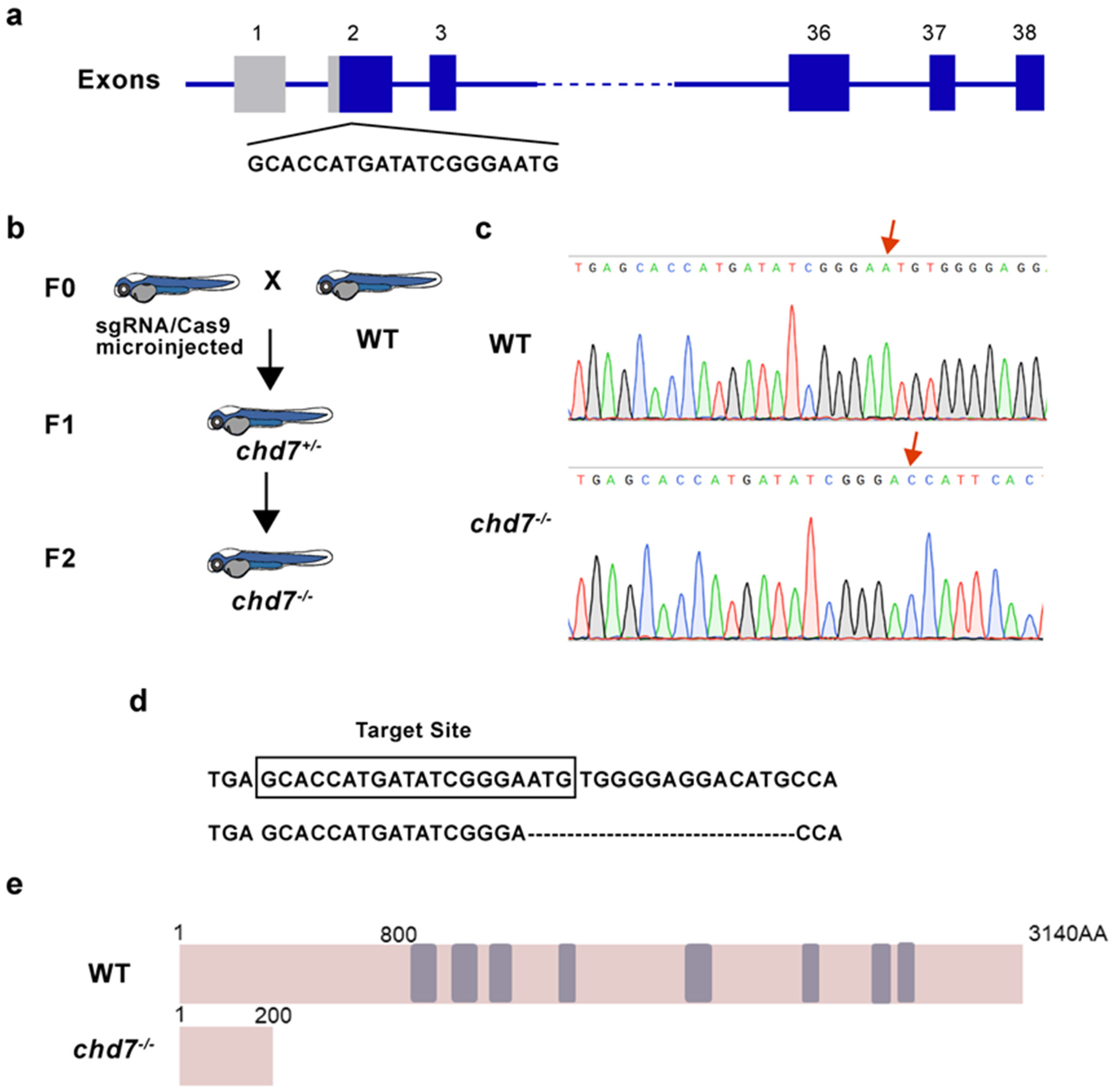
2.2. Construction of chd7 Homozygous Mutant Zebrafish
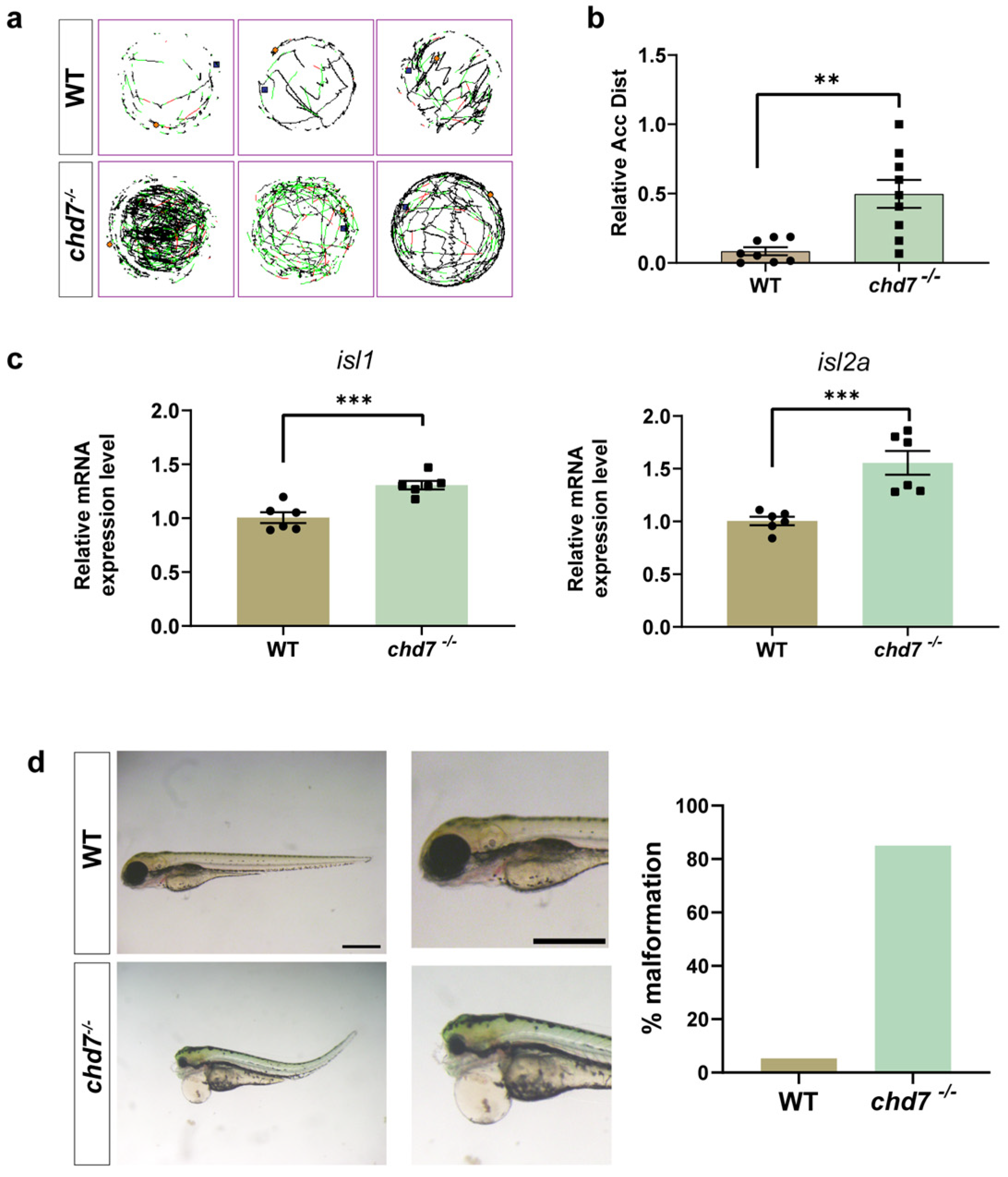
2.3. Knockout of chd7 Caused Hyperactivity and Malformation
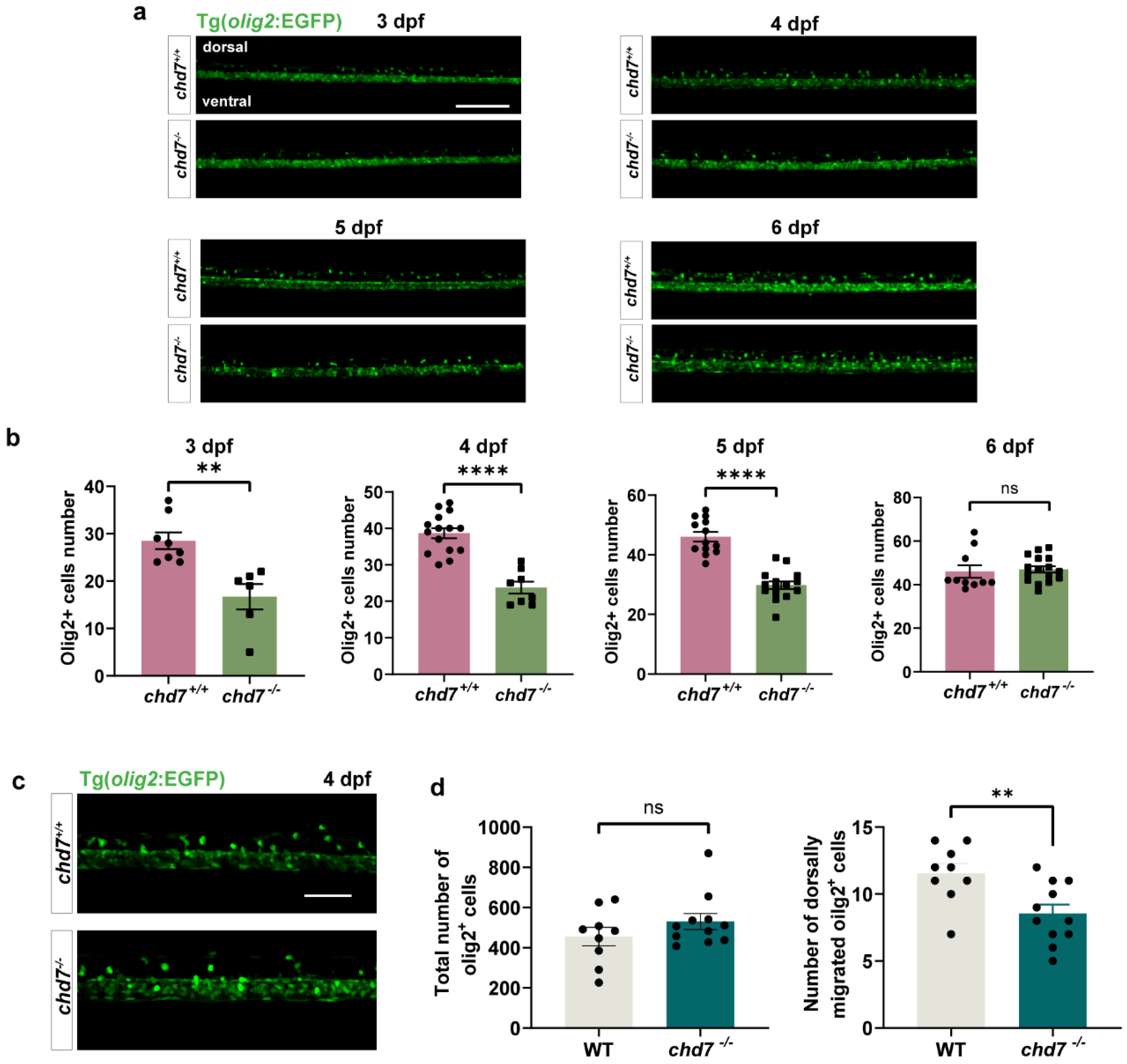
2.4. Knockout of chd7 Slowed down the Migration of OL to the Dorsal Side
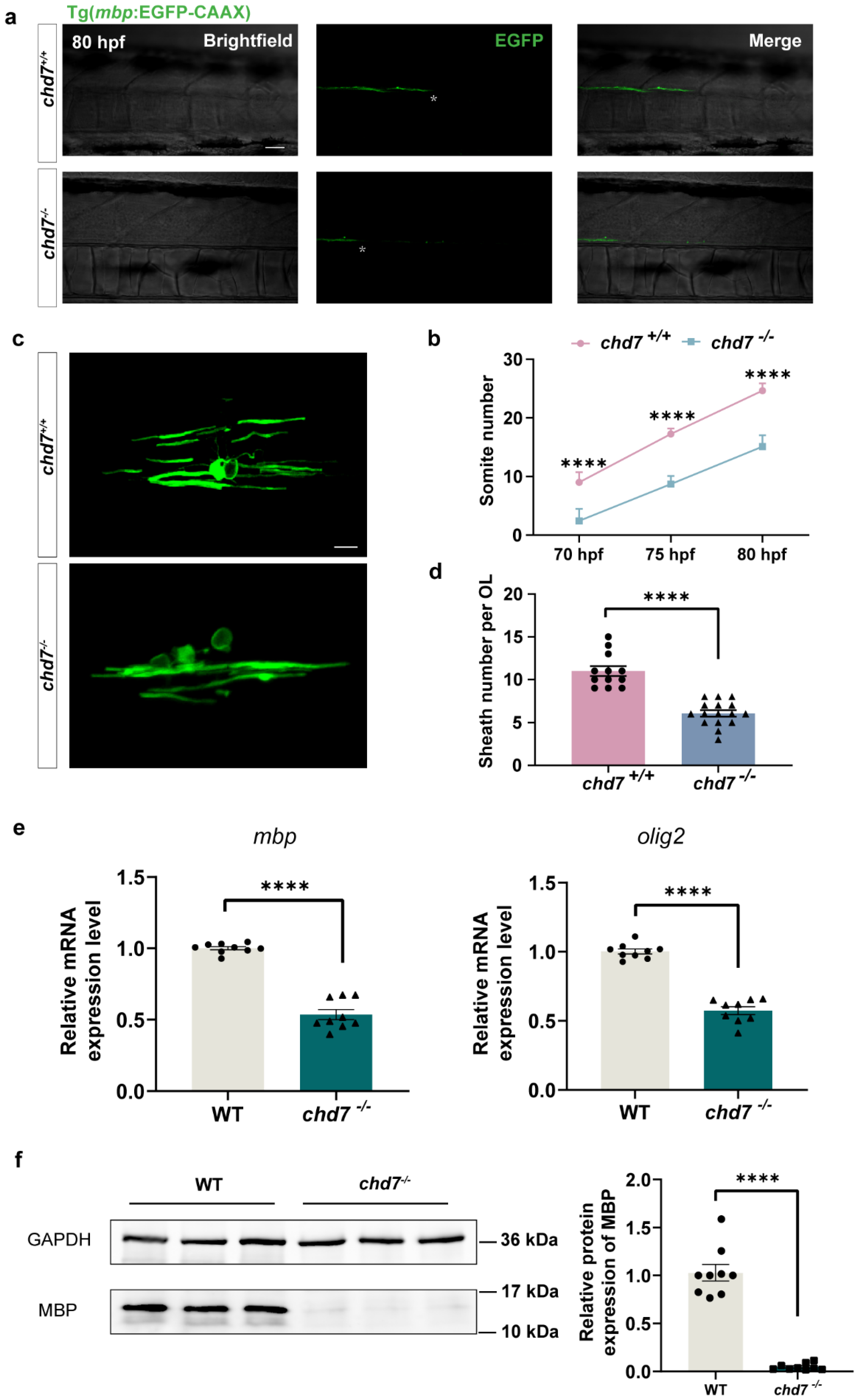
2.5. Chd7 Knockout Hindered Myelination of OL
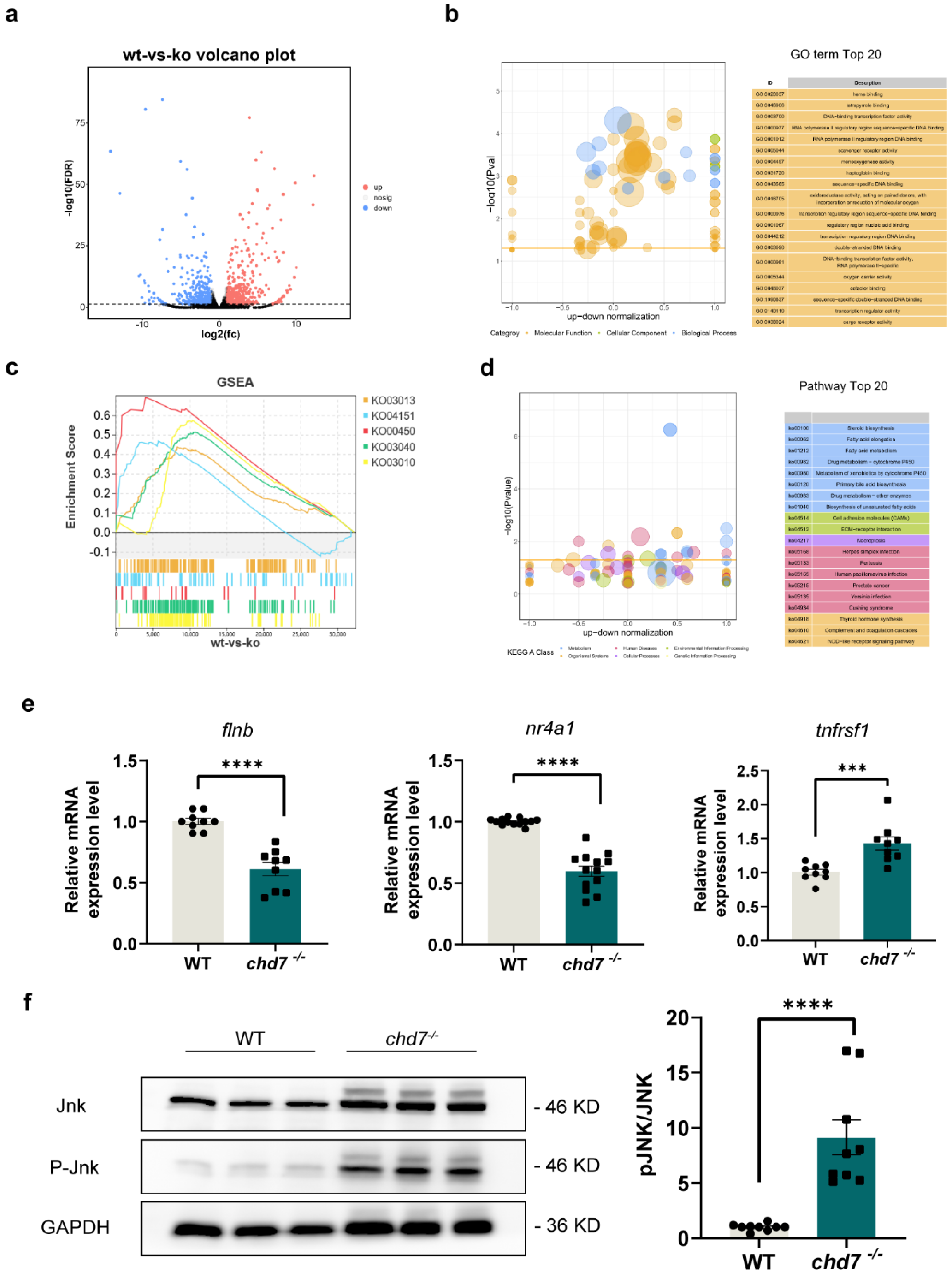
2.6. CHD7 Might Affect Myelin Development through MAPK Signaling Pathway
3. Discussion
4. Methods and Materials
4.1. Zebrafish Strains and Maintenance
4.2. Genome Editing
4.3. Quantitative Real-Time PCR
4.4. Western Blot
4.5. In Vivo Imaging
4.6. Whole-Mount In Situ Hybridization
4.7. Fluorescence-Activated Cell Sorting
4.8. Spontaneous Movement Assay
4.9. Transmission Electron Microscopy Imaging
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lalani, S.R.; Safiullah, A.M.; Fernbach, S.D.; Harutyunyan, K.G.; Thaller, C.; Peterson, L.E.; McPherson, J.D.; Gibbs, R.A.; White, L.D.; Hefner, M.; et al. Spectrum of CHD7 mutations in 110 individuals with CHARGE syndrome and genotype-phenotype correlation. Am. J. Hum. Genet. 2006, 78, 303–314. [Google Scholar] [CrossRef]
- Allen, M.D.; Religa, T.L.; Freund, S.M.; Bycroft, M. Solution structure of the BRK domains from CHD7. J. Mol. Biol. 2007, 371, 1135–1140. [Google Scholar] [CrossRef]
- Aramaki, M.; Udaka, T.; Kosaki, R.; Makita, Y.; Okamoto, N.; Yoshihashi, H.; Oki, H.; Nanao, K.; Moriyama, N.; Oku, S.; et al. Phenotypic spectrum of CHARGE syndrome with CHD7 mutations. J. Pediatr. 2006, 148, 410–414. [Google Scholar] [CrossRef]
- Graham, J.M., Jr.; Rosner, B.; Dykens, E.; Visootsak, J. Behavioral features of CHARGE syndrome (Hall-Hittner syndrome) comparison with Down syndrome, Prader-Willi syndrome, and Williams syndrome. Am. J. Med. Genet. Part A 2005, 133a, 240–247. [Google Scholar] [CrossRef]
- Strömland, K.; Sjögreen, L.; Johansson, M.; Ekman Joelsson, B.M.; Miller, M.; Danielsson, S.; Billstedt, E.; Gillberg, C.; Jacobsson, C.; Norinder, J.A.; et al. CHARGE association in Sweden: Malformations and functional deficits. Am. J. Med. Genet. Part A 2005, 133a, 331–339. [Google Scholar] [CrossRef]
- Hartshorne, N.; Hudson, A.; MacCuspie, J.; Kennert, B.; Nacarato, T.; Hartshorne, T.; Blake, K. Quality of life in adolescents and adults with CHARGE syndrome. Am. J. Med. Genet. Part. A 2016, 170, 2012–2021. [Google Scholar] [CrossRef]
- Hartshorne, T.S.; Hefner, M.A.; Davenport, S.L. Behavior in CHARGE syndrome: Introduction to the special topic. Am. J. Med. Genet. Part A 2005, 133a, 228–231. [Google Scholar] [CrossRef]
- Hartshorne, T.S.; Stratton, K.K.; Brown, D.; Madhavan-Brown, S.; Schmittel, M.C. Behavior in CHARGE syndrome. Am. J. Med. Genetics. Part C Semin. Med. Genet. 2017, 175, 431–438. [Google Scholar] [CrossRef]
- Jamadagni, P.; Breuer, M.; Schmeisser, K.; Cardinal, T.; Kassa, B.; Parker, J.A.; Pilon, N.; Samarut, E.; Patten, S.A. Chromatin remodeller CHD7 is required for GABAergic neuron development by promoting PAQR3 expression. EMBO Rep. 2021, 22, e50958. [Google Scholar] [CrossRef]
- Chen, X.; Yan, K.; Gao, Y.; Wang, H.; Chen, G.; Wu, B.; Qin, Q.; Yang, L.; Zhou, W. Feeding difficulty is the dominant feature in 12 Chinese newborns with CHD7 pathogenic variants. BMC Med. Genet. 2019, 20, 93. [Google Scholar] [CrossRef]
- Gregory, L.C.; Gevers, E.F.; Baker, J.; Kasia, T.; Chong, K.; Josifova, D.J.; Caimari, M.; Bilan, F.; McCabe, M.J.; Dattani, M.T. Structural pituitary abnormalities associated with CHARGE syndrome. J. Clin. Endocrinol. Metab. 2013, 98, E737–E743. [Google Scholar] [CrossRef]
- Liu, L.; Yu, T.; Wang, L.; Mo, X.; Yu, Y. A novel CHD7 mutation in a Chinese patient with CHARGE syndrome. Meta Gene 2014, 2, 469–478. [Google Scholar] [CrossRef]
- Bae, H.G.; Kim, T.K.; Suk, H.Y.; Jung, S.; Jo, D.G. White matter and neurological disorders. Arch. Pharm. Res. 2020, 43, 920–931. [Google Scholar] [CrossRef] [PubMed]
- Bugiani, M.; Boor, I.; Powers, J.M.; Scheper, G.C.; van der Knaap, M.S. Leukoencephalopathy with vanishing white matter: A review. J. Neuropathol. Exp. Neurol. 2010, 69, 987–996. [Google Scholar] [CrossRef]
- Reddy, N.C.; Majidi, S.P.; Kong, L.; Nemera, M.; Ferguson, C.J.; Moore, M.; Goncalves, T.M.; Liu, H.K.; Fitzpatrick, J.A.J.; Zhao, G.; et al. CHARGE syndrome protein CHD7 regulates epigenomic activation of enhancers in granule cell precursors and gyrification of the cerebellum. Nat. Commun. 2021, 12, 5702. [Google Scholar] [CrossRef]
- Feng, W.; Kawauchi, D.; Körkel-Qu, H.; Deng, H.; Serger, E.; Sieber, L.; Lieberman, J.A.; Jimeno-González, S.; Lambo, S.; Hanna, B.S.; et al. Chd7 is indispensable for mammalian brain development through activation of a neuronal differentiation programme. Nat. Commun. 2017, 8, 14758. [Google Scholar] [CrossRef] [PubMed]
- Hurd, E.A.; Poucher, H.K.; Cheng, K.; Raphael, Y.; Martin, D.M. The ATP-dependent chromatin remodeling enzyme CHD7 regulates pro-neural gene expression and neurogenesis in the inner ear. Development 2010, 137, 3139–3150. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.E.; Hurd, E.A.; Beyer, L.A.; Swiderski, D.L.; Raphael, Y.; Martin, D.M. Defects in vestibular sensory epithelia and innervation in mice with loss of Chd7 function: Implications for human CHARGE syndrome. J. Comp. Neurol. 2007, 504, 519–532. [Google Scholar] [CrossRef]
- Hurd, E.A.; Adams, M.E.; Layman, W.S.; Swiderski, D.L.; Beyer, L.A.; Halsey, K.E.; Benson, J.M.; Gong, T.W.; Dolan, D.F.; Raphael, Y.; et al. Mature middle and inner ears express Chd7 and exhibit distinctive pathologies in a mouse model of CHARGE syndrome. Hear. Res. 2011, 282, 184–195. [Google Scholar] [CrossRef]
- Gage, P.J.; Hurd, E.A.; Martin, D.M. Mouse Models for the Dissection of CHD7 Functions in Eye Development and the Molecular Basis for Ocular Defects in CHARGE Syndrome. Investig. Ophthalmol. Vis. Sci. 2015, 56, 7923–7930. [Google Scholar] [CrossRef]
- Schnetz, M.P.; Bartels, C.F.; Shastri, K.; Balasubramanian, D.; Zentner, G.E.; Balaji, R.; Zhang, X.; Song, L.; Wang, Z.; Laframboise, T.; et al. Genomic distribution of CHD7 on chromatin tracks H3K4 methylation patterns. Genome Res. 2009, 19, 590–601. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; Hannum, D.F.; Zhai, Y.; Hill, S.F.; Albanus, R.D.; Lou, W.; Skidmore, J.M.; Sanchez, G.; Saiakhova, A.; Bielas, S.L.; et al. CHD7 promotes neural progenitor differentiation in embryonic stem cells via altered chromatin accessibility and nascent gene expression. Sci. Rep. 2020, 10, 17445. [Google Scholar] [CrossRef] [PubMed]
- He, D.; Marie, C.; Zhao, C.; Kim, B.; Wang, J.; Deng, Y.; Clavairoly, A.; Frah, M.; Wang, H.; He, X.; et al. Chd7 cooperates with Sox10 and regulates the onset of CNS myelination and remyelination. Nat. Neurosci. 2016, 19, 678–689. [Google Scholar] [CrossRef]
- Marie, C.; Clavairoly, A.; Frah, M.; Hmidan, H.; Yan, J.; Zhao, C.; Van Steenwinckel, J.; Daveau, R.; Zalc, B.; Hassan, B.; et al. Oligodendrocyte precursor survival and differentiation requires chromatin remodeling by Chd7 and Chd8. Proc. Natl. Acad. Sci. USA 2018, 115, E8246–E8255. [Google Scholar] [CrossRef] [PubMed]
- Asad, Z.; Pandey, A.; Babu, A.; Sun, Y.; Shevade, K.; Kapoor, S.; Ullah, I.; Ranjan, S.; Scaria, V.; Bajpai, R.; et al. Rescue of neural crest-derived phenotypes in a zebrafish CHARGE model by Sox10 downregulation. Hum. Mol. Genet. 2016, 25, 3539–3554. [Google Scholar] [CrossRef]
- Buckley, C.E.; Goldsmith, P.; Franklin, R.J. Zebrafish myelination: A transparent model for remyelination? Dis. Models Mech. 2008, 1, 221–228. [Google Scholar] [CrossRef]
- Dubois-Dalcq, M.; Williams, A.; Stadelmann, C.; Stankoff, B.; Zalc, B.; Lubetzki, C. From fish to man: Understanding endogenous remyelination in central nervous system demyelinating diseases. Brain 2008, 131, 1686–1700. [Google Scholar] [CrossRef]
- Bin, J.M.; Lyons, D.A. Imaging Myelination In Vivo Using Transparent Animal Models. Brain Plast. 2016, 2, 3–29. [Google Scholar] [CrossRef]
- Chakraborty, C.; Hsu, C.H.; Wen, Z.H.; Lin, C.S.; Agoramoorthy, G. Zebrafish: A complete animal model for in vivo drug discovery and development. Curr. Drug Metab. 2009, 10, 116–124. [Google Scholar] [CrossRef]
- Peal, D.S.; Peterson, R.T.; Milan, D. Small molecule screening in zebrafish. J. Cardiovasc. Transl. Res. 2010, 3, 454–460. [Google Scholar] [CrossRef]
- Simons, M.; Lyons, D.A. Axonal selection and myelin sheath generation in the central nervous system. Curr. Opin. Cell Biol. 2013, 25, 512–519. [Google Scholar] [CrossRef] [PubMed]
- Lyons, D.A.; Talbot, W.S. Glial cell development and function in zebrafish. Cold Spring Harb. Perspect. Biol. 2014, 7, a020586. [Google Scholar] [CrossRef] [PubMed]
- Preston, M.A.; Macklin, W.B. Zebrafish as a model to investigate CNS myelination. Glia 2015, 63, 177–193. [Google Scholar] [CrossRef] [PubMed]
- Buckley, C.E.; Marguerie, A.; Alderton, W.K.; Franklin, R.J. Temporal dynamics of myelination in the zebrafish spinal cord. Glia 2010, 58, 802–812. [Google Scholar] [CrossRef]
- Almeida, R.G.; Czopka, T.; Ffrench-Constant, C.; Lyons, D.A. Individual axons regulate the myelinating potential of single oligodendrocytes in vivo. Development 2011, 138, 4443–4450. [Google Scholar] [CrossRef]
- Parkinson, D.B.; Bhaskaran, A.; Arthur-Farraj, P.; Noon, L.A.; Woodhoo, A.; Lloyd, A.C.; Feltri, M.L.; Wrabetz, L.; Behrens, A.; Mirsky, R.; et al. c-Jun is a negative regulator of myelination. J. Cell Biol. 2008, 181, 625–637. [Google Scholar] [CrossRef]
- Wang, L.W.; Tu, Y.F.; Huang, C.C.; Ho, C.J. JNK signaling is the shared pathway linking neuroinflammation, blood-brain barrier disruption, and oligodendroglial apoptosis in the white matter injury of the immature brain. J. Neuroinflamm. 2012, 9, 175. [Google Scholar] [CrossRef]
- Zhang, Y.; Jiang, K.; Xie, G.; Ding, J.; Peng, S.; Liu, X.; Sun, C.; Tang, X. FGF21 impedes peripheral myelin development by stimulating p38 MAPK/c-Jun axis. J. Cell. Physiol. 2021, 236, 1345–1361. [Google Scholar] [CrossRef]
- Hall, B.D. Choanal atresia and associated multiple anomalies. J. Pediatr. 1979, 95, 395–398. [Google Scholar] [CrossRef]
- Camargo, N.; Goudriaan, A.; van Deijk, A.F.; Otte, W.M.; Brouwers, J.F.; Lodder, H.; Gutmann, D.H.; Nave, K.A.; Dijkhuizen, R.M.; Mansvelder, H.D.; et al. Oligodendroglial myelination requires astrocyte-derived lipids. PLoS Biol. 2017, 15, e1002605. [Google Scholar] [CrossRef]
- Hammond, T.R.; Gadea, A.; Dupree, J.; Kerninon, C.; Nait-Oumesmar, B.; Aguirre, A.; Gallo, V. Astrocyte-derived endothelin-1 inhibits remyelination through notch activation. Neuron 2014, 81, 588–602. [Google Scholar] [CrossRef] [PubMed]
- Lampron, A.; Larochelle, A.; Laflamme, N.; Préfontaine, P.; Plante, M.M.; Sánchez, M.G.; Yong, V.W.; Stys, P.K.; Tremblay, M.; Rivest, S. Inefficient clearance of myelin debris by microglia impairs remyelinating processes. J. Exp. Med. 2015, 212, 481–495. [Google Scholar] [CrossRef] [PubMed]
- Mensch, S.; Baraban, M.; Almeida, R.; Czopka, T.; Ausborn, J.; El Manira, A.; Lyons, D.A. Synaptic vesicle release regulates myelin sheath number of individual oligodendrocytes in vivo. Nat. Neurosci. 2015, 18, 628–630. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Xiong, Q.; Li, Q.; Lin, W.; Jiang, S.; Zhang, D.; Wang, Y.; Duan, X.; Gong, P.; Kang, N. CHD7 regulates bone-fat balance by suppressing PPAR-γ signaling. Nat. Commun. 2022, 13, 1989. [Google Scholar] [CrossRef]
- Dorighi, K.M.; Tamkun, J.W. The trithorax group proteins Kismet and ASH1 promote H3K36 dimethylation to counteract Polycomb group repression in Drosophila. Development 2013, 140, 4182–4192. [Google Scholar] [CrossRef]
- Bajpai, R.; Chen, D.A.; Rada-Iglesias, A.; Zhang, J.; Xiong, Y.; Helms, J.; Chang, C.P.; Zhao, Y.; Swigut, T.; Wysocka, J. CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature 2010, 463, 958–962. [Google Scholar] [CrossRef]
- Layman, W.S.; Hurd, E.A.; Martin, D.M. Chromodomain proteins in development: Lessons from CHARGE syndrome. Clin. Genet. 2010, 78, 11–20. [Google Scholar] [CrossRef]
- Yan, S.; Thienthanasit, R.; Chen, D.; Engelen, E.; Brühl, J.; Crossman, D.K.; Kesterson, R.; Wang, Q.; Bouazoune, K.; Jiao, K. CHD7 regulates cardiovascular development through ATP-dependent and -independent activities. Proc. Natl. Acad. Sci. USA 2020, 117, 28847–28858. [Google Scholar] [CrossRef]
- Quinlivan, V.H.; Farber, S.A. Lipid Uptake, Metabolism, and Transport in the Larval Zebrafish. Front. Endocrinol. 2017, 8, 319. [Google Scholar] [CrossRef]
- Pérez-Cano, F.J.; Franch, À.; Castellote, C.; Castell, M. The suckling rat as a model for immunonutrition studies in early life. Clin. Dev. Immunol. 2012, 2012, 537310. [Google Scholar] [CrossRef]
- Corsten-Janssen, N.; van Ravenswaaij-Arts, C.M.A.; Kapusta, L. Congenital arch vessel anomalies in CHARGE syndrome: A frequent feature with risk for co-morbidity. Int. J. Cardiol. Heart Vasc. 2016, 12, 21–25. [Google Scholar] [CrossRef] [PubMed]
- Westerfield, M. The Zebrafish Book; University of Oregon Press: Eugene, OR, USA, 1993. [Google Scholar]
- Kimmel, C.B.; Ballard, W.W.; Kimmel, S.R.; Ullmann, B.; Schilling, T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 1995, 203, 253–310. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; Park, H.C.; Topczewska, J.M.; Mawdsley, D.J.; Appel, B. Neural cell fate analysis in zebrafish using olig2 BAC transgenics. Methods Cell Sci. Off. J. Soc. Vitr. Biol. 2003, 25, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.Q.; Chen, M.; Zhang, J.L.; Ren, D.L.; Hu, B. Hypoxia Delays Oligodendrocyte Progenitor Cell Migration and Myelin Formation by Suppressing Bmp2b Signaling in Larval Zebrafish. Front. Cell Neurosci. 2018, 12, 348. [Google Scholar] [CrossRef] [PubMed]
- Collins, S.C.; Vancollie, V.E.; Mikhaleva, A.; Wagner, C.; Balz, R.; Lelliott, C.J.; Yalcin, B. Characterization of Two Mouse Chd7 Heterozygous Loss-of-Function Models Shows Dysgenesis of the Corpus Callosum and Previously Unreported Features of CHARGE Syndrome. Int. J. Mol. Sci. 2022, 23, 11509. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Xu, Y.; Huang, R.; Huang, Y.; Ge, S.; Hu, B. N-Cadherin is Involved in Neuronal Activity-Dependent Regulation of Myelinating Capacity of Zebrafish Individual Oligodendrocytes In Vivo. Mol. Neurobiol. 2017, 54, 6917–6930. [Google Scholar] [CrossRef]
- Yin, W.; Hu, B. Knockdown of Lingo1b protein promotes myelination and oligodendrocyte differentiation in zebrafish. Exp. Neurol. 2014, 251, 72–83. [Google Scholar] [CrossRef]
- Baraban, M.; Koudelka, S.; Lyons, D.A. Ca2+ activity signatures of myelin sheath formation and growth in vivo. Nat. Neurosci. 2018, 21, 19–23. [Google Scholar] [CrossRef]


| chd7-sgRNA-F | TAATACGACTCACTACACCATGATATCGGGAATGGT TTTAGAGCTAGAAATAGC |
| chd7-sgRNA-R | AGCACCGACTCGGTGCCACT |
| chd7-F | ACCCAGGCATGATGAGCCTCTT |
| chd7-R | CAGGCACCTGCATTGGTTGAGCA |
| β-actin-qPCR-F | CCCTGTTCCAGCCATCCTT |
| β-actin-qPCR-R | TTGAAAGTGGTCTCGTGGATACC |
| chd7-qPCR-F | AGGGACACACAGGGACGTAT |
| chd7-qPCR-R | CGCCATGGCACATTTCGTAG |
| mbp-qPCR-R | CGAGGAGAGGACACAAAGC |
| mbp-qPCR-F | CCGTCGTGGAGACGTCAA |
| olig2-qPCR-F | ATCCGTCCAGTTGTGGCACT |
| olig2-qPCR-R | TGGTGGAAGCAGAGGATGGT |
| chd7-situ-F | TAATACGACTCACTATAGACGGTGTTGGAAGGAACTTG |
| chd7-situ-R | ACGGTGTTGGAAGGAACTTG |
| flnb-qPCR-F | GTGGACCCTGATACAGCATTAC |
| flnb-qPCR-R | ATCCCAGAAGCCTCATCTCT |
| tnfrsf1a-qPCR-F | AGACAGTGCAAATGCCAGGA |
| tnfrsf1a-qPCR-R | TCCTGGCTGACATTTATCGCA |
| nr4a1-qPCR-F | AGGCCATGTCGATCTGTGATT |
| nr4a1-qPCR-R | GTGCGTACGAAGCTTTGGTG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, L.; Wang, Z.; Li, Y.; Song, Z.; Yin, W.; Hu, B. Deletion of the chd7 Hinders Oligodendrocyte Progenitor Cell Development and Myelination in Zebrafish. Int. J. Mol. Sci. 2023, 24, 13535. https://doi.org/10.3390/ijms241713535
Shi L, Wang Z, Li Y, Song Z, Yin W, Hu B. Deletion of the chd7 Hinders Oligodendrocyte Progenitor Cell Development and Myelination in Zebrafish. International Journal of Molecular Sciences. 2023; 24(17):13535. https://doi.org/10.3390/ijms241713535
Chicago/Turabian StyleShi, Lingyu, Zongyi Wang, Yujiao Li, Zheng Song, Wu Yin, and Bing Hu. 2023. "Deletion of the chd7 Hinders Oligodendrocyte Progenitor Cell Development and Myelination in Zebrafish" International Journal of Molecular Sciences 24, no. 17: 13535. https://doi.org/10.3390/ijms241713535





